Introduction
Solute carrier family 22 (organic cation
transporter) member 18 (SLC22A18) is located within the 11p15.5
cluster, and has been assigned a number of different nomenclatures
(1,2). Blast homology analysis suggests that
SLC22A18 is a member of a family of polyspecific transporters and
multidrug resistance genes (2).
SLC22A18 has been shown to be a tumor suppressor candidate, as well
as a substrate for RING105 (3).
Structural mutations in SLC22A18 are rare, with isolated reports of
point mutations in a breast cancer cell line, a rhabdomyosarcoma
cell line (2), and Wilms’ and lung
tumors (1). Exonic deletions in
Wilms’ tumors and loss of heterozygosity in hepatoblastomas were
also reported (1,4), indicating that SLC22A18 may play a
role in tumorigenesis.
In this study, a eukaryotic expression vector of
human SLC22A18 (pcDNA3.1-SLC22A18) was constructed and the vector
was stably transfected into the U251 glioma cell line, which did
not express SLC22A18 under normal growth conditions, in order to
investigate the role of SLC22A18 in glioma cells. The elevated
expression of SLC22A18 was found to increase the sensitivity of
U251 cells to the anticancer drug BCNU in vitro.
Materials and methods
Construction of the expression
vector
The 890-bp SLC22A18 cDNA was amplified using reverse
transcription-polymerase chain reaction (RT-PCR) (5). The PCR reaction (10 μl)
contained 1 μl cDNA, l μl 10X buffer
(MgCl2), 0.4 mM dNTPs, 1 μmol primer and 1 unit
TaqDNA polymerase. After denaturation at 95°C for 5 min, PCR was
performed for 35 cycles (30 sec at 95°C, 30 sec at 50°C and 30 sec
at 72°C) and extended at 72°C for 5 min. The linear
NHeI-EcoRI fragment containing the SLC22A18 cDNA was
subcloned into pcDNA3.1 (Invitrogen Corp., Carlsbad, CA, USA),
which yielded pcDNA3.1-SLC22A18 by T4 ligase (Takara Bio Inc.,
Otsu, Shiga, Japan ). The insertion of SLC22A18 in pcDNA3.1 was
confirmed by PCR, restriction enzyme digestion analysis
(NHeI and EcoRI) and DNA sequencing.
Cell culture and determination of the
optimal concentration of G418
The U251 human glioma cells, obtained from the Wuhan
University of China, Wuhan, China, were incubated at 37°C in
RPMI-1640 supplemented with 10% calf serum, 100 μg/ml
penicillin and 100 μg/ml streptomycin, in an atmosphere of
5% CO2 at a saturation humidity. The culture medium was
changed every 48 h. G418 is an aminoglycoside and is commonly used
as a selective agent for the bacterial neo r/kan r genes. To
determine the optimal concentration of G418 for the selection of
resistance, U251 cells were plated at the same concentration of
5×104/well, in 24-well plates containing 2 ml culture
medium per well. G418 (Sigma, St. Louis, MO, USA) was added to the
wells at 10 different concentrations (50, 100, 150, 200, 300, 400,
500, 600, 700 and 800 μg/ml). The culture media were changed
once every 48 h. The lowest G418 concentration, in which all cells
died after 12–14 days of culture, was selected as the optimal
concentration for resistance selection.
Transfection of U251 cells with
pcDNA3.1-SLC22A18 and PCR confirmation of SLC22A18 in U251
cells
For the stable transfection of the SLC22A18 gene,
U251 cells (1×106) were plated in 6-well plates 24 h
prior to transfection. Lipofectamine 2000 (Invitrogen) was used to
mediate transfection using 5.0 μg of pcDNA3.1-SLC22A18
vector or 5.0 μg of the empty pcDNA3.1 vector as a control
according to the manufacturer’s instructions. After 48 h of
transfection, the cells were suspended in media supplemented with
G418 (150 μg/ml). The medium was changed every 48 h.
Non-transfected U251 cells died within 2 weeks. G418-resistant
cells were selected and denoted as U251-SLC22A18. Cells with empty
vector pcDNA3.1 were named as U251-EV. DNA from cells of normal
U251, U251-EV and U251-SLC22A18 was isolated using a DNA extraction
kit (Puregene® DNA isolation kit, Gentra Systems, Inc.,
Minneapolis, MN, USA). A section of the SLC22A18 gene was used to
design the primers (6). The
upstream primer sequence was 5′-AGCTGAGCAGCCACTTCTC-3′ and the
downstream was 5′-AAAGCTGCGGTACAGGAGG-3′. The PCR reaction system
(50 μl) was: 3 μl cDNA, 5 μl 10X buffer, 4
μl MgC12, 1 μl dNTP, 1 μl primer
and 0.3 μl TaqDNA polymerase. The conditions for the PCR
reaction comprised an initial denaturation step of 94°C for 7 min,
followed by 30 cycles of a three-step program of 94°C for 30 sec,
56°C for 30 sec, 72°C for 45 sec and a final extension step of 72°C
for 7 min. All products were electrophoresed on agarose gel.
RT-PCR of SLC22A18 mRNA
The analysis of SLC22A18 mRNA expression in the
three cell groups (normal U251, U251-EV and U251-SLC22A18) was
performed by RT-PCR. Total RNA was extracted from glioma tissues,
their adjacent brain tissues and the glioma cell line U251 with
TRIzol-reagent (Invitrogen) following the manufacturer’s
instructions. The RT reaction was performed on 2 μg of total
RNA with the SuperScript II first-strand synthesis using an
oligo(dT) primer system (Life Technologies, Corp., Carlsbad, CA,
USA). Primer sequences and conditions for the RT-PCR product were
previously described (forward, 5′-GCTTCGGCGTCGGAGT CAT-3′ and
reverse, 5′-AGCCTGGGCGTCAGTTTT-3′) (7). The housekeeping gene GAPDH was used as
an internal control of the RT reaction (forward primer, 5′-GGGAGC
CAAAAGGGTCATCATCTC-3′ and reverse primer, 5′-CCA
TGCCAGTGAGCTTCCCGTTC-3′) (7). PCR
consisted of 35 cycles at 94°C for 1 min, at 62°C for 1 min and at
72°C for 1 min followed by a final extension at 72°C for 5 min. PCR
products were analyzed on 2% agarose gels.
Western blot analysis
The three cell groups (normal U251, U251-EV and
U251-SLC22A18) were washed in ice-cold phosphate-buffered saline
(PBS) and lysed in a buffer using standard methods (8). After centrifugation at 10,000 × g for
10 min, the supernatants were stored at −70°C. Lysate equalized for
protein content was separated in 150 g/l sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto a polyvinylidine fluoride membrane. The membranes
were blocked for 1 h at room temperature in 10 g/l bovine serum
albumin and incubated overnight at 4°C with SLC22A18 antibody,
followed by incubation with sheep antirabbit second antibody
conjugated to HRP (Dako, Glostrup, Denmark) for 120 min. Finally,
the membranes were developed with DAB and incubated until color
developed sufficiently.
Apoptosis analysis
Normal U251, U251-EV and U251-SLC22A18 cells were
incubated in 6-well plates at 1×106 cells/well in medium
with or without 50 μM BCNU (Medical Isotopes, Inc., Pelham,
NH, USA ) for 24 h. Apoptosis was detected using the Annexin V-FITC
apoptosis detection kit (Jingmei Biotech Co., Ltd., Shenzhen,
Guangdong, China) (9). Briefly,
cells were harvested and then resuspended in 1 ml buffer prior to
the addition of 5 μl Annexin V and 10 μl propidium
iodite (PI). Cells were incubated in the dark at room temperature
for 15 min. Cell death was determined using a flow cytometer
(Backman Co.). Data were obtained and analyzed using CellQuest
software (Largo Co.).
Immunohistochemical analysis for
caspase-3 and bcl-2
Normal U251, U251-EV and U251-SLC22A18 cells were
incubated in 6-well plates at 1×106 cells/well in medium
with or without 50 μM BCNU (Medical Isotopes, Inc.). AC for
24 h. Cells were washed in 0.05 M (PBS) (pH 7.4) for 15 min, then
fixed in 4% paraformaldehyde for 20 min. The
streptavidin/biotin-peroxidase method was used for
immunohistochemical staining. The primary antibodies, i.e.,
antibodies against caspase-3 and bcl-2 (Wuhan Boster Biological
Technology, China), were diluted at a concentration of 1:100. PBS
was used as a control. Labeled cells were photographed and the
integrated optical density (IOD) was measured using Image-Pro Plus
5.02 (Media Cybernetics, Inc., Bethesda, MD, USA).
Statistical analysis
The differences between the controls and treated
groups were analyzed using the χ2-test. SPSS 10.0
statistical software was used to analyze the results. P<0.05 was
considered to be statistically significant.
Results
SLC22A18 cDNA amplification and
identification of the expression vector
First, the primers were designed and then the 890-bp
SLC22A18 gene was amplified. The PCR product was collected and
purified. The purified SLC22A18 fragment and the pcDNA3.1 vector
were digested by NHeI and EcoRI, followed by ligation
with T4 ligase at 16°C overnight. The ligated plasmid was
transformed into the DH5α strain of E. coli. Single colonies
were selected and PCR amplification confirmed a single band of 890
bp. The plasmids were isolated from DH5α and digested by
NHeI and EcoRI. DNA gel showed two bands, one
corresponding to the 890-bp fragment and the second corresponding
to the vector pcDNA3.1. DNA sequencing showed that the 890-bp
fragment corresponded with the DNA sequence of the SLC22A18
gene.
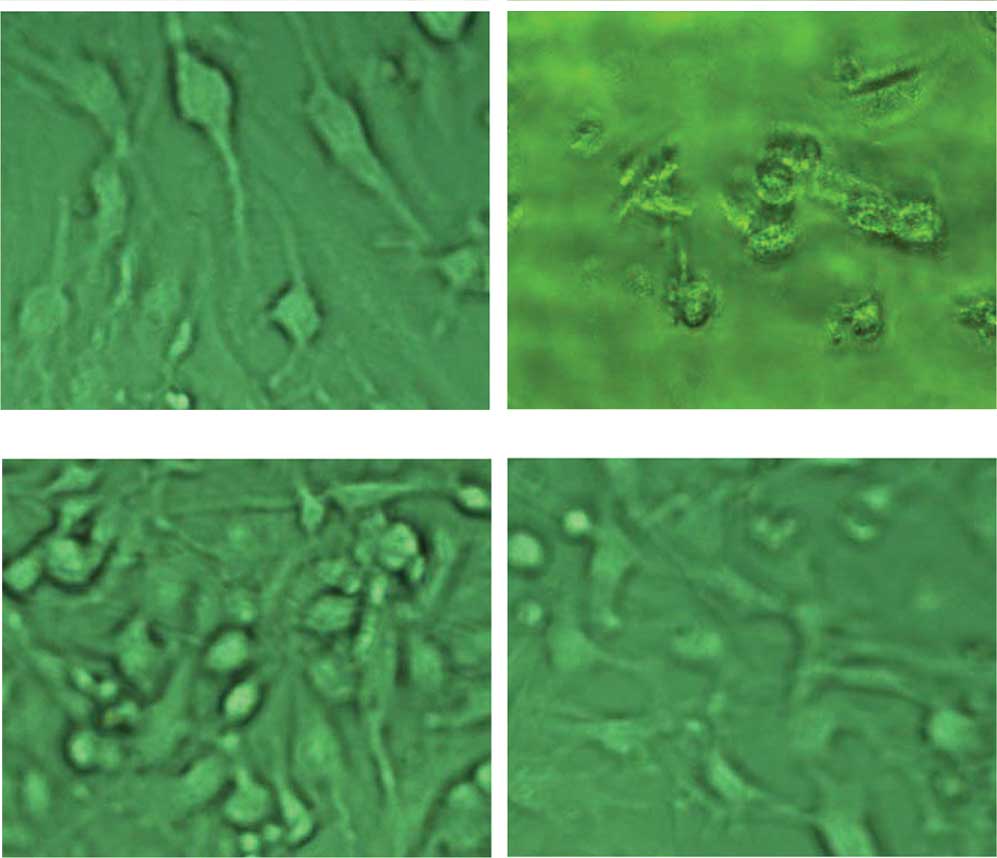
Cell morphology and U251-SLC22A18
screening
Normal U251 cells were usually elongated and
elliptical (Fig. 1A). The cells
died within 2 weeks when cultured in the presence of 150
μg/ml G418 (Fig. 1B). Cells
transfected with pcDNA3.1-SLC22A18 were resistant to G418 and were
screened under G418 for ~3 weeks and denoted as U251-SLC22A18. The
U251-SLC22A18 cells had a similar appearance to the non-transfected
cells and occasionally formed clusters (Fig. 1C and D).
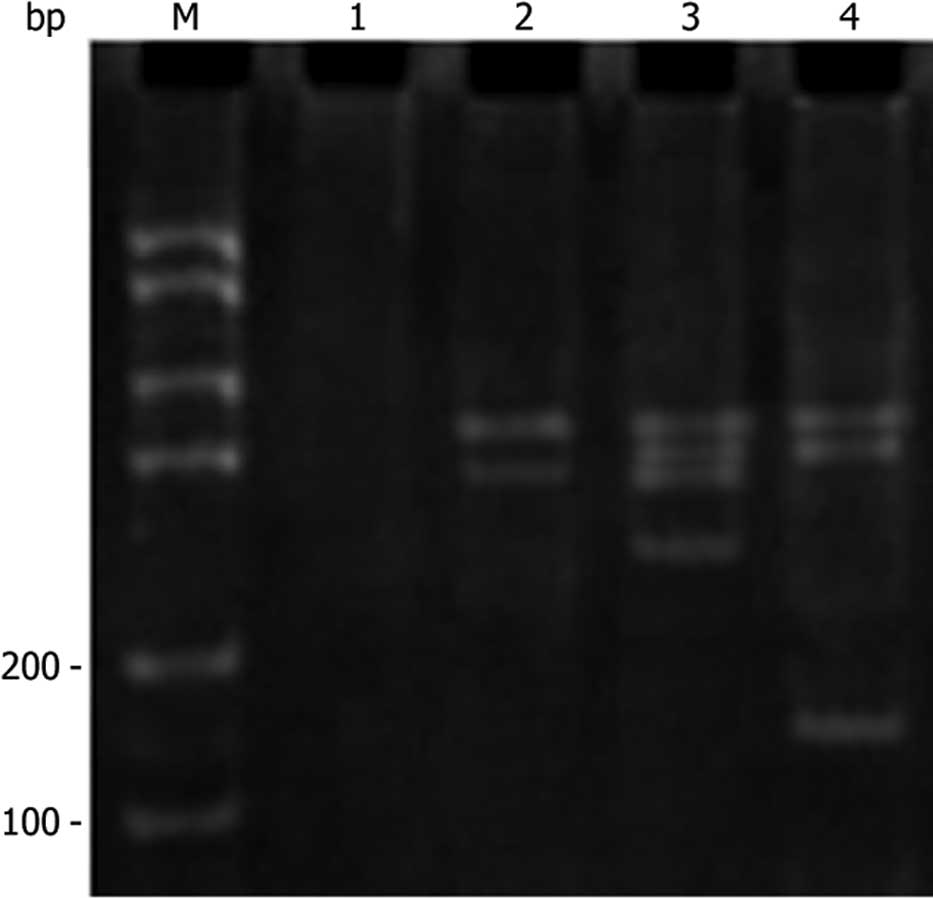
PCR analysis of SLC22A18 in U251
cells
DNA was extracted from the cells of normal U251,
U251-EV and U251-SLC22A18. The extracted DNA was amplified by PCR
using the primer pair described above. As expected, a 170-bp
fragment was observed in U251-SLC22A18 cells, but not in normal
U251 or U251-EV cells (Fig. 2).
This result further confirmed the specific transfection of the
SLC22A18 gene into U251 cells.
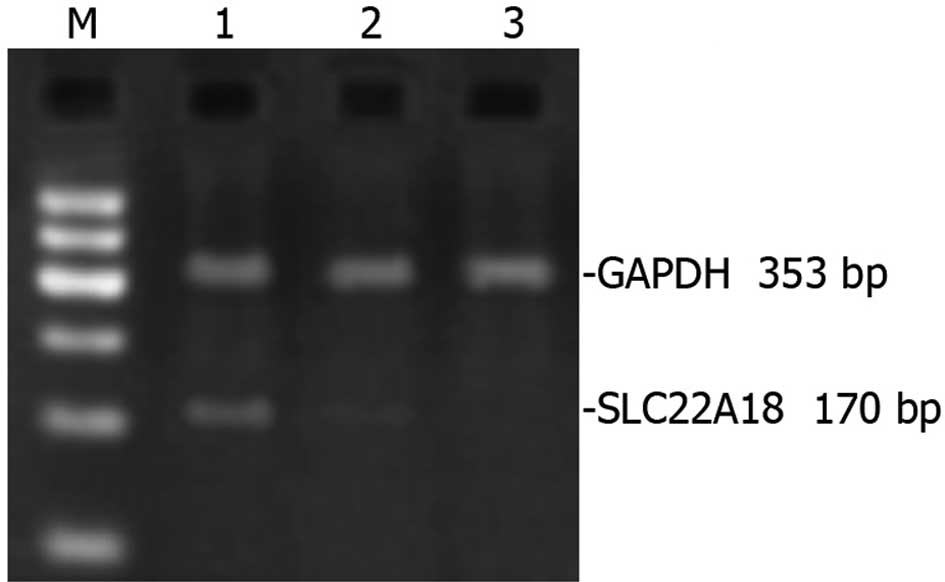
SLC22A18 mRNA expression in U251
cells
RNA extracted from normal U251, U251-EV and
U251-SLC22A18 cells was amplified by RT-PCR and subsequently
analyzed using DNA gel. A prominent 170-bp band was detected in
U251-SLC22A18 cells, but was non-detectable in U251-EV cells or
normal U251 cells (Fig. 3).
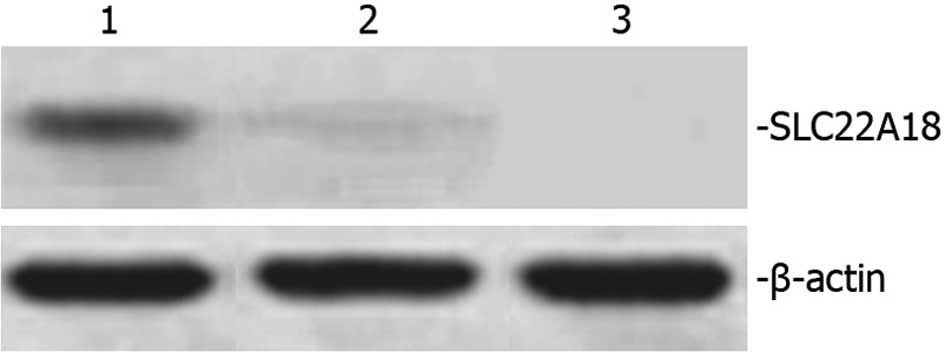
SLC22A18 protein expression in U251
cells
Total protein extracted from normal U251, U251-EV
and U251-SLC22A18 cells was separated using 12% SDS-PAGE and was
subsequently analyzed by Western blot analysis. A 50-kDa band
corresponding to the size of the SLC22A18 protein was observed in
U251-SLC22A18 cells, but not in U251-EV or normal U251 cells
(Fig. 4).
BCNU-induced apoptosis
BCNU is an anticancer drug and an inducer of
apoptotic cell death. BCNU was used to further assess the role of
SLC22A18 in U251 cells. Apoptosis was observed in the three cell
groups (normal U251, U251-EV and U251-SLC22A18). The average
apoptosis ratio of normal U251, U251-EV and U251-SLC22A18 cells was
2.4±0.2, 2.2±0.3 and 9.2±0.3%, respectively. The apoptosis ratio of
U251-SLC22A18 cells was significantly (P<0.05) higher than that
of normal U251 or U251-EV cells. Minimal apoptosis was observed in
all three cell groups in the absence of BCNU.
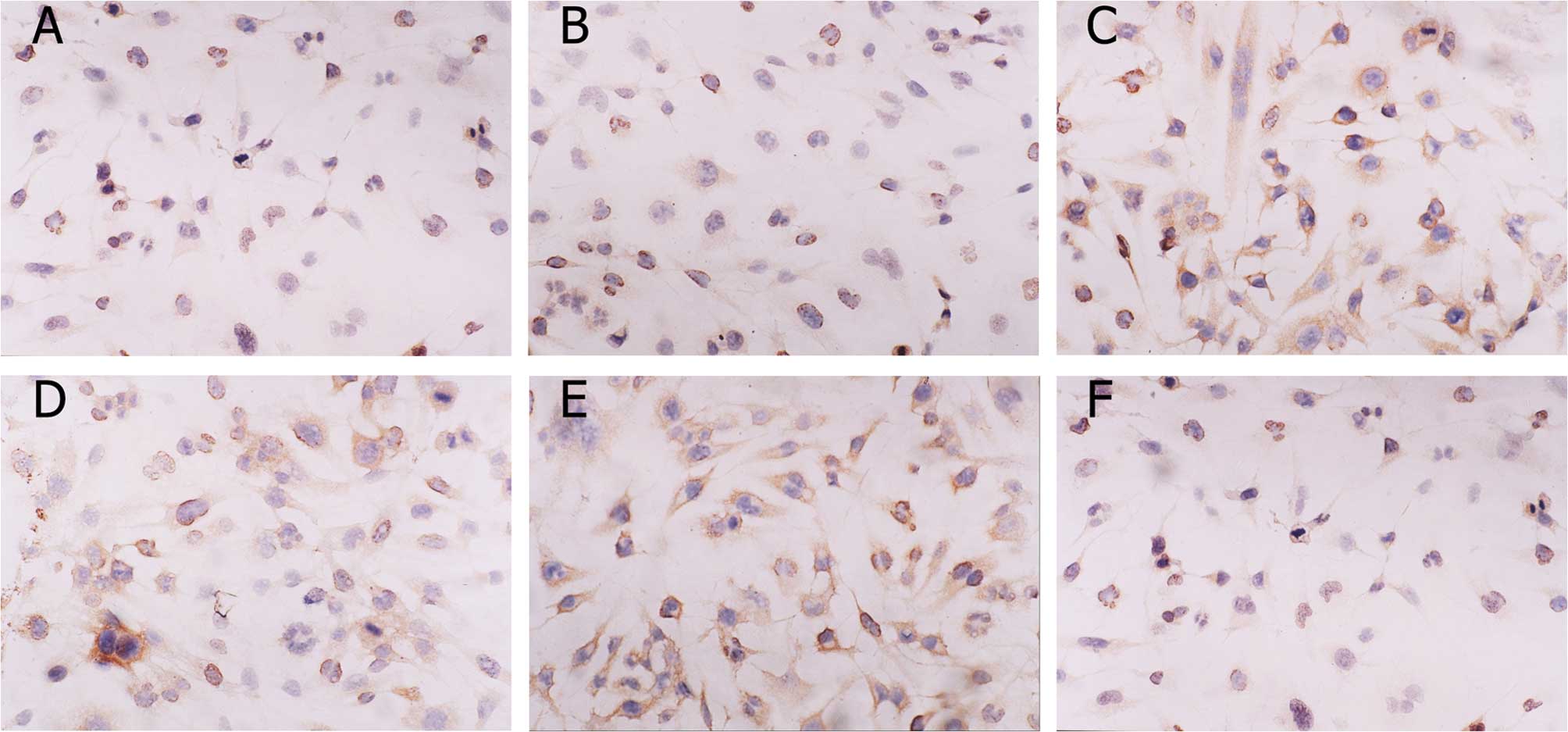
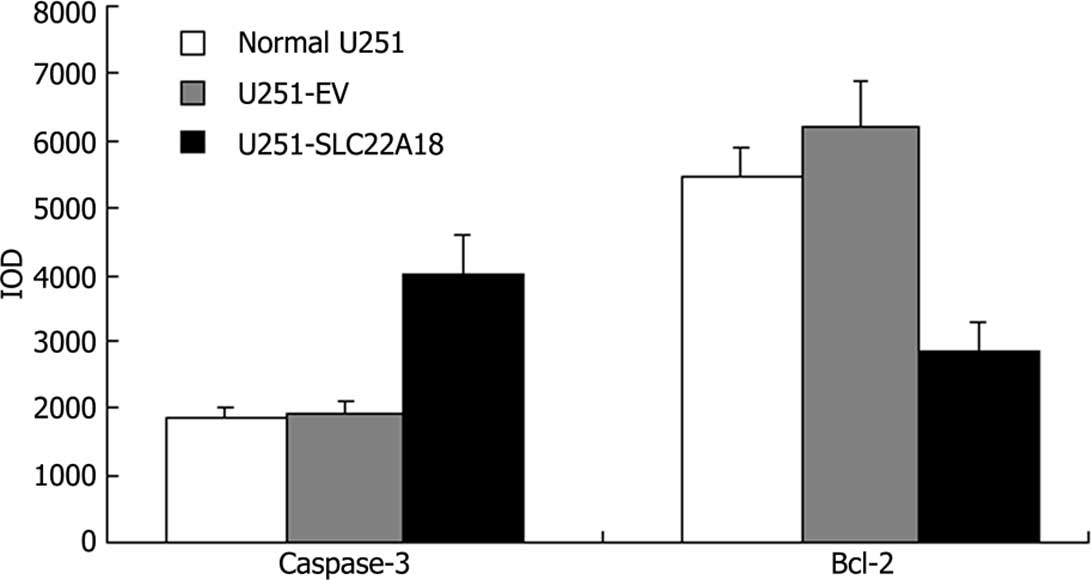
Immunohistochemistry
Images of caspase-3 and bcl-2 staining are shown in
Fig. 5. The positive reaction
located in cytosol was stained brown. The color of the stain is
positively correlated to the protein expression. The IOD of each
group revealed that in U251-SLC22A18 cells the expression of
caspase-3 increased, whereas the expression of bcl-2 decreased
(Fig. 6).
Discussion
SLC22A18 is located within the 11p15.5 cluster and
has been assigned a number of different nomenclatures (1,2). Blast
homology analysis suggests that SLC22A18 is a member of a family of
polyspecific transporters and multidrug resistance genes (2). SLC22A18 has also been shown to be a
tumor suppressor candidate and a substrate for RING105 (3). Structural mutations in SLC22A18 are
infrequent, with isolated reports of point mutations in a breast
cancer cell line, a rhabdomyosarcoma cell line (2), and Wilms’ and lung tumors (1). Exonic deletions in Wilms’ tumors and
loss of heterozygosity in hepatoblastomas have also been reported
(1,4). Thus, SLC22A18 may be involved in
tumorigenesis.
The role SLC22A18 plays in tumorigenesis is
functionally unclear. In this study, a eukaryotic expression vector
of human SLC22A18 (pcDNA3.1-SLC22A18) was constructed and the
vector was stably transfected into the U251 glioma cell line, which
did not express SLC22A18 under normal growth conditions. Data from
the present study revealed that apoptotic cell death observed in
the SLC22A18-transfected cells was more abundant than in the
non-transfected cells or in cells with the empty vector. Moreover,
the expression of caspase-3 increased, whereas the expression of
bcl-2 decreased in the U251-SLC22A18 cells, indicating that the
expression of SLC22A18 increased the sensitivity of U251 cells to
the anticancer drug BCNU in vitro, and that SLC22A18 has a
pro-apoptotic function in glioma. We speculate that the function of
SLC22A18 may be tissue- or cell type-specific. Further studies, in
particular a direct comparison of SLC22A18 in different cell types
under the same conditions, are required in order to better
understand the complex functions of SLC22A18 in relation to tumor
development.
Acknowledgements
This study was supported by grants from the Natural
Science Foundation of China (30901535), the Research Fund of Xinhua
Hospital Affiliated to Shanghai Jiao Tong University School of
Medicine (10XHJT01), and the New One Hundred Person Project of
Shanghai Jiao Tong University School of Medicine (10XBR01).
References
|
1
|
Lee MP, Reeves C, Schmitt A, Su K, Connors
TD, Hu RJ, Brandenburg S, Lee MJ, Miller G and Feinberg AP: Somatic
mutation of TSSC5, a novel imprinted gene from human chromosome
11p15.5. Cancer Res. 58:4155–4159. 1998.PubMed/NCBI
|
|
2
|
Schwienbacher C, Sabbioni S, Campi M,
Veronese A, Bernardi G, Menegatti A, Hatada I, Mukai T, Ohashi H,
Barbanti-Brodano G, Croce CM and Negrini M: Transcriptional map of
170-kb region at chromosome 11p15.5: identification and mutational
analysis of the BWR1A gene reveals the presence of mutations in
tumor samples. Proc Natl Acad Sci USA. 95:3873–3878. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamada HY and Gorbsky GJ: Tumor suppressor
candidate TSSC5 is regulated by UbcH6 and a novel ubiquitin ligase
RING105. Oncogene. 25:1330–1339. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Albrecht S, Hartmann W, Houshdaran F, Koch
A, Gärtner B, Prawitt D, Zabel BU, Russo P, von Schweinitz D and
Pietsch T: Allelic loss but absence of mutations in the
polyspecific transporter gene BWR1A on 11p15.5 in hepatoblastoma.
Int J Cancer. 111:627–632. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mohammad F, Mondal T, Guseva N, Pandey GK
and Kanduri C: Kcnq1ot1 noncoding RNA mediates transcriptional gene
silencing by interacting with Dnmt1. Development. 137:2493–2499.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gallagher E, McGoldrick A, Chung WY,
McCormack O, Harrison M, Kerin M, Dervan PA and McCann A: Gain of
imprinting of SLC22A18 sense and antisense transcripts in human
breast cancer. Genomics. 88:12–17. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu HM, Zhang HW, He HY, Hou YY and Zhao
ZQ: Methylation and mRNA expression of imprinted gene
SLC22A18/TSSC5/BWR1A in breast cancer. Chin J Pathophysiol.
24:1007–1012. 2008.
|
|
8
|
Chu SH, Yuan XH, Li ZQ, Jiang PC and Zhang
J: C-Met antisense oligodeoxynucleotide inhibits growth of glioma
cells. Surg Neurol. 65:533–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou Y, Li W, Xu Q and Huang Y: Elevated
expression of Dickkopf-1 increases the sensitivity of human glioma
cell line SHG44 to BCNU. J Exp Clin Cancer Res. 29:1312010.
View Article : Google Scholar : PubMed/NCBI
|