Introduction
Lung cancer is the most common form of cancer
worldwide. Non-small cell lung cancer (NSCLC) accounts for ∼80% of
all lung cancer cases and is typically associated with frequent
development of resistance towards chemotherapy. Deficiency in
apoptosis is considered to be a major cause of the therapeutic
resistance of NSCLC, as the chemotherapeutic drug cisplatin is one
of the most frequently used agents in NSCLC treatment, and its
cytotoxic effects are speculated to be due to the DNA damage and
apoptosis it induces in cells (1,2).
Apoptosis is activated and inactivated by a variety
of genes. Deregulation of genes involved in the activation or
execution of the apoptotic process may lead to chemoresistance in
cells. p53-dependent apoptosis is a significant mechanism whereby
DNA-damaging cisplatin exerts its biological effects (3). However, apoptosis is capable of
occurring in p53-deficient cells. It has been demonstrated that p73
functionally replaces p53 in adriamycin-treated, p53-deficient
breast cancer cells (4,5). The data suggest that p53-independent
pathways exist and these are involved in the cellular response to
anti-cancer drugs.
p73 shares a marked homology in DNA sequence and
protein structure with p53. As with p53, p73 is a tumor suppressor
gene. The p73 gene maps to chromosome 1p36 and encodes two
different proteins that are expressed under the control of two
independent promoters, and that have opposite functions: The
transcriptionally active full-length TAp73 and the
NH2-terminally truncated dominant-negative ΔNp73. TAp73
has been demonstrated to be involved in cellular responses to DNA
damage induced by chemotherapeutic agents. The promoter was capable
of inhibiting cell growth in a p53-like manner by inducing
apoptosis. However, in contrast with p53, mutation of p73 has
rarely been found in the majority of human cancers (6,7). The
transcription of p73 is regulated by the promoter and exon 1, which
is rich in CpG dinucleotides. Methylation of the cytosine residues
at the CpG dinucleotides within this region plays a critical role
in inactivating gene expression (8,9).
As p53 function is often impaired in NSCLC, it would
be valuable to understand whether p73 in NSCLC is capable of
compensating for the impaired p53 function and thus triggering
p53-independent apoptosis of cancer cells in response to
chemotherapy. Therefore, the aim of this study was to investigate
the correlation between p73 gene expression and chemosensitivity in
NSCLC cell lines.
Materials and methods
Cell lines and cultures, and
5-aza-2-deoxycytidine (5-aza-dC) treatment
Two cell lines derived from squamous cell carcinomas
of the lung (SK-MES-1 and C57), three cell lines derived from
adenocarcinomas of the lung (A549, GLC and P15) and one cell line
derived from a large cell lung carcinoma (NCI-H460) were obtained
from the American Type Culture Collection (Manassas, VA, USA) or
the Animal Experiment Center (Sun Yat-sen University, Guangzhou,
China). The breast cancer cell line MCF7 was also included to
validate the reverse transcription-polymerase chain reaction
(RT-PCR) approach to p73 expression. The five cell lines (C57, GLC,
P15, NCI-H460 and A549) were maintained in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS) in 5% CO2 at 37°C.
SK-MES-1 cells were grown in α-Minimum Essential medium
supplemented with 10% FBS and antibiotics. Cells were harvested
with 0.25% trypsin. The human lung squamous cell carcinoma cell
line C57 was exposed to different concentrations (2, 5 and 10
μmol/l) of the demethylation agent 5-aza-dC (Sigma, St.
Louis, MO, USA) for 72 h, to assess restoration of p73 gene
expression.
The study was approved by the Ethics Committee of
the School of Medicine, Jinan University, Guangzhou, Guangdong,
China.
Total RNA extraction and RT-PCR
Total RNA was extracted from cells using TRIzol
reagent (Omega Bio-Tek, Norcross, GA, USA). After purification, RNA
was dissolved in diethylpyrocarbonate (DEPC)-treated distilled
water. The cDNA was synthesized with random hexamer primers and
stored at −20°C until use. Total RNA (1.0 μg) was
reverse-transcribed by AMV Reverse Transcriptase (Takara
Biotechnology; Dalian, China), from which the cDNA was obtained for
PCR amplification. The PCR reaction was performed using primers
spanning exons 5 and 6 of the p73 gene without amplification of
genomic sequences (p73–715 forward, 5′-ACTTCAACGAAGGACAGTCTGCT and
p73–856 reverse, 5′-AATTCCGTCCCCACCTGTG) (10). RT-PCR was performed for 1 cycle at
94°C for 2 min, followed by 30 cycles at 94°C for 30 sec, 63°C for
30 sec and 72°C for 1 min. The length of the PCR product for the
p73 transcript was 142 bp. β-actin gene was selected as an
endogenous reference.
DNA extraction, sodium bisulfite
modification of DNA and methylation-specific PCR (MSP)
Genomic DNA was prepared from the cultured cells.
DNA was extracted by OB Protease and Tissue Isolation Reagent
(Omega Bio-Tek) according to the manufacturer’s instructions. The
methylation status within the CpG island of the p73 gene in exon 1
(sequence 110-42 bp relative to translation start, GenBank
Accession number Y11416) was determined by MSP using
bisulfite-modified DNA (11).
Bisulfite modification was conducted using the EZ DNA
Methylation-Gold kit (Zymo Research Co.; Beijing, China). Following
this reaction, all unmethylated cytosines were deaminated and
converted to uracil, while methylated cytosines remained unchanged.
Following bisulfite conversion, methylated and unmethylated genomic
regions were distinguished by PCR using each sequence-specific pair
of primers. Primer sequences for methylated and unmethylated
alleles of p73 are presented in Table
I. Unmethylated and methylated fragments were amplified under
the following PCR reactions: 35 cycles of 30 sec at 94°C, 30 sec at
59°C and 30 sec at 72°C for the unmethylated products; whereas
methylated products were amplified in 40 cycles of 30 sec at 94°C,
30 sec at 60°C and 30 sec at 72°C. In both cases an initial
denaturation of 7 min at 94°C and a final extension of 5 min at
72°C were conducted. PCR products were electrophoresed on 3%
agarose gel and visualized by ethidium bromide staining. Human
placental DNA treated in vitro with Sss I methylase (New
England Biolabs, Inc.; Ipswich, MA, USA) served as a positive
control for the methylated reaction. Control reactions without DNA
were performed alongside each PCR.
 | Table I.Primer sequences for methylated and
unmethylated alleles of p73. |
Table I.
Primer sequences for methylated and
unmethylated alleles of p73.
| p73 allele | Primer pairs |
|---|
| Methylated | 5′-Primer
5′-GGACGTAGCGAAATCGGGGTTC-3′ |
| 3′-Primer
5′-ACCCCGAACATCGACGTCCG-3′ |
| Unmethylated | 5′-Primer
5′-AGGGGATGTAGTGAAATTGGGGTTT-3′ |
| 3′-Primer
5′-CCATCACAACCCCAAACATCA-3′ |
Immunohistochemistry
After washing with phosphate-buffered saline (PBS),
slides were incubated in diluted primary antibody at room
temperature for 2 h. The p73 (E-4) antibody used was a mouse
monoclonal antibody (Santa Cruz Biotechnology, Inc.; Santa Cruz,
CA, USA) raised against amino acids 1–80 and mapping at the
N-terminus of p73 of human origin, which recognizes all human TAp73
isoforms. The mouse anti-human p73 monoclonal antibody was used at
a 1/50 dilution for a final concentration of 4.0 mg/l. Slides were
subsequently incubated in 10% normal horse serum for 20 min. Then,
the sections were added to poly-horse, rabbit-peroxidase-anti
mouse/rabbit IgG (Maixin, Biotechnology Development Co., Ltd;
Fuzhou, China). After 30 min, the sections were rinsed with PBS.
Development of the slides was performed using 3,
3′-diaminobenzidine (DAB) solution. Hematoxylin was used as the
nuclear counterstain. Immunoreactivity was confirmed as positive
for the p73 nuclear identification. In all runs, negative controls
were included and PBS was substituted for the primary antibody;
staining was not observed.
Clonogenic survival assay
Following treatment with 5 μmol/l 5-aza-dC
for 72 h, the exponentially growing cells were trypsinized into a
single cell suspension. Cell viability was assessed by trypan blue
dye exclusion. Viable cells (5x102) were plated in 60 mm
Petri dishes and exposed to the chemotherapeutic drug cisplatin
(1.25 μmol/l) for 48 h. Following exposure, cells were
incubated at 37°C in 5% CO2 to facilitate colony
formation. Following growth for 10–14 days, the colonies were fixed
with methanol and stained with Giemsa dye (Huamei Biotechnology
Co., Ltd., Beijing, China). Colonies exhibiting a minimum of 50
viable cells were counted. Colony plating efficiency was calculated
to be the number of viable plated cells, and was expressed as a
percentage of inoculated cells.
Statistical analysis
The results were expressed as the mean ± standard
deviation. MS Excel 7.0 computer software was used to perform
statistical analyses. A two-tailed Student’s t-test was used for
statistical comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of p73 in human lung carcinoma
cell line
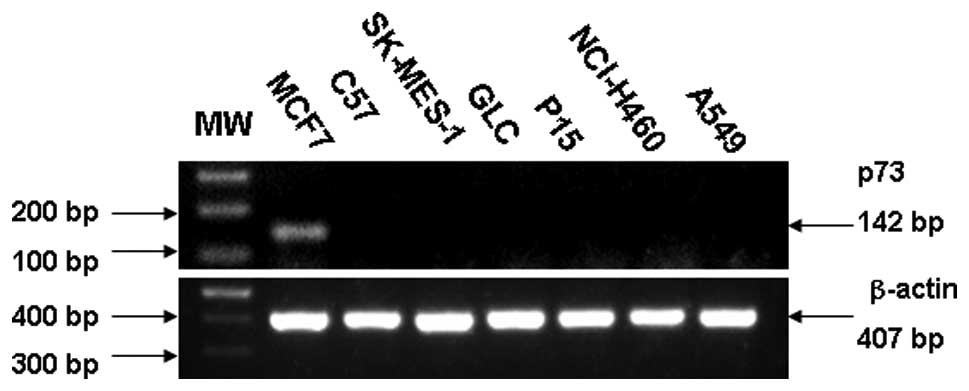
Expression of p73 at the RNA level is illustrated in
Fig. 1. A sample was considered
negative when it exhibited positive staining for β-actin and
negative staining for p73. SK-MES-1 and C57 derived from human lung
squamous cell carcinoma did not express any p73 mRNA. GLC, P15,
A549 and NCI-H460, derived from adenocarcinoma of lung and large
cell lung carcinoma respectively, failed to express p73 at the RNA
level.
p73 methylation status
To determine whether aberrant methylation of p73
occurred in human NSCLC, we first investigated six NSCLC cell
lines. By MSP, all six cell lines including C57, SK-MES-1, GLC,
P15, A549 and NCI-H460 demonstrated evidence of methylation in exon
1 (Fig. 2). The observed
methylation patterns correlated with transcriptional silencing of
the p73 gene. The p73 transcript was undetectable in the six
methylated cell lines by RT-PCR (Fig.
1).
Restoration of p73 gene expression
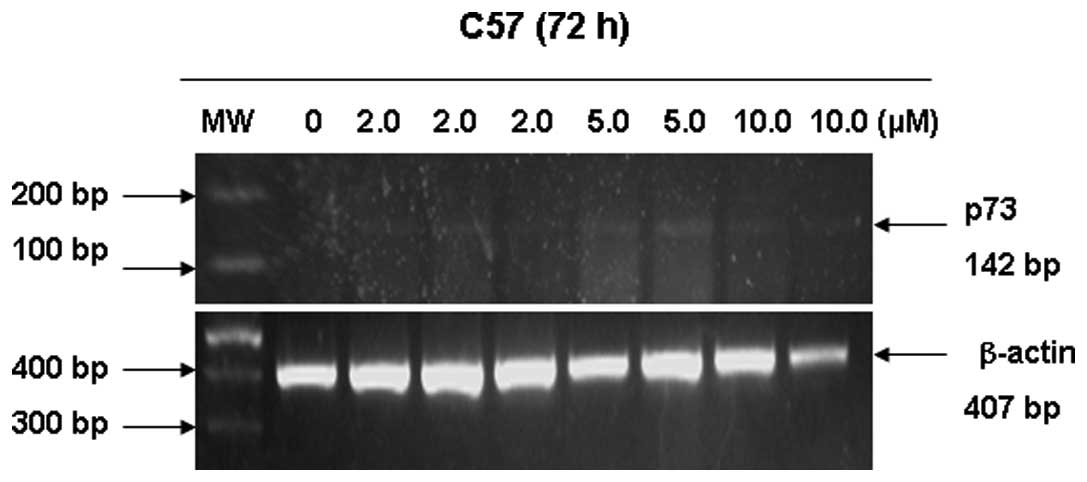
Human lung squamous cell carcinoma cell line C57 was
exposed to different concentrations of 5-aza-dC to restore
expression of p73 transcripts. As demonstrated in Fig. 3, 5-aza-dC induced re-expression of the p73
gene at the mRNA level by RT-PCR. p73 mRNA level was abundant at 5
μmol/l 5-aza-dC, and was weakly detected at 2 and 10
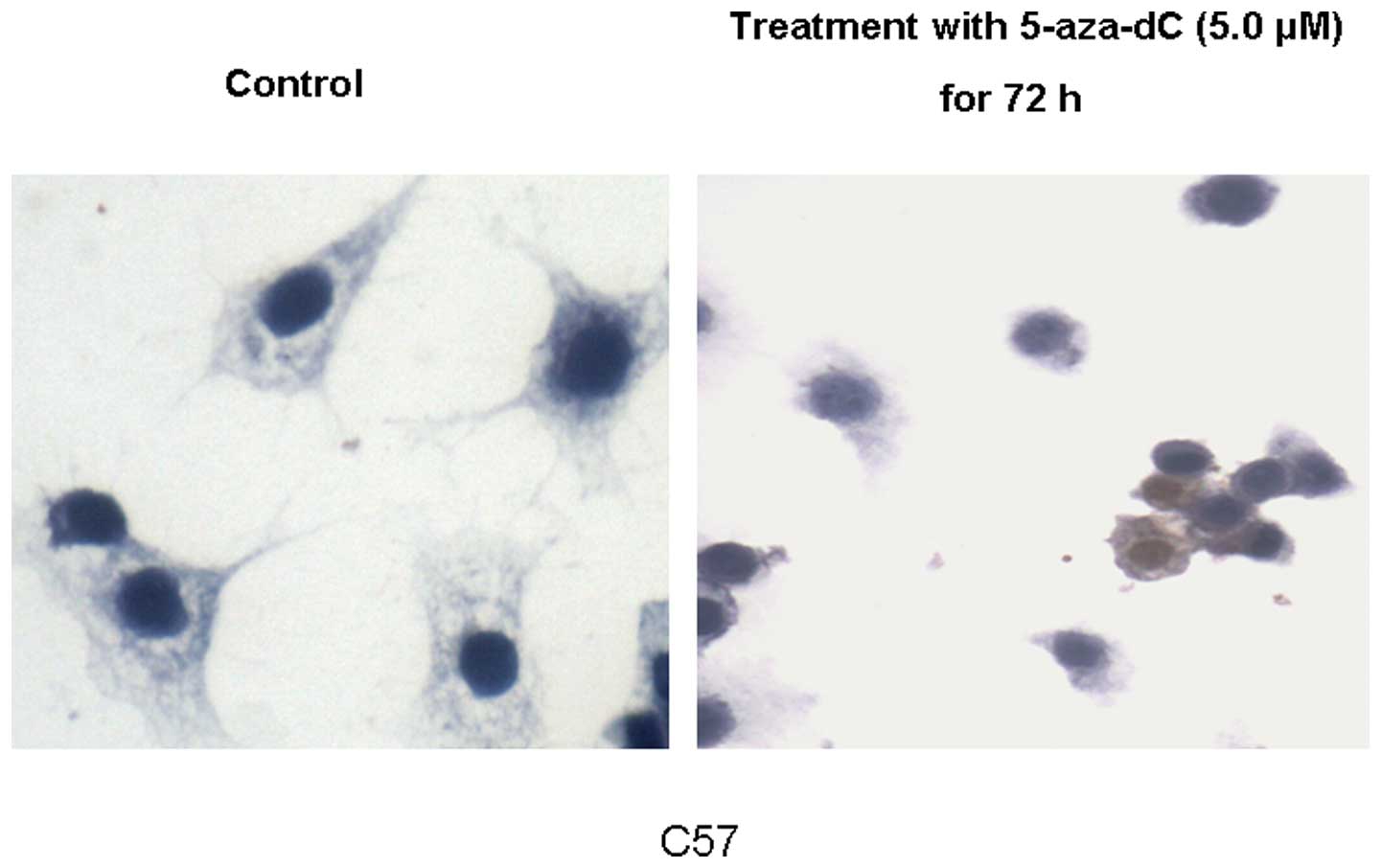
μmol/l 5-aza-dC. Immunohistochemistry revealed that low and
high expression of the p73 protein was restored in the C57 cell
line following exposure to 5 μmol/l 5-aza-dC (Fig. 4).
Demethylation of p73 in the C57 cell line
increases chemotherapeutic drug-(cisplatin-) induced cell
death
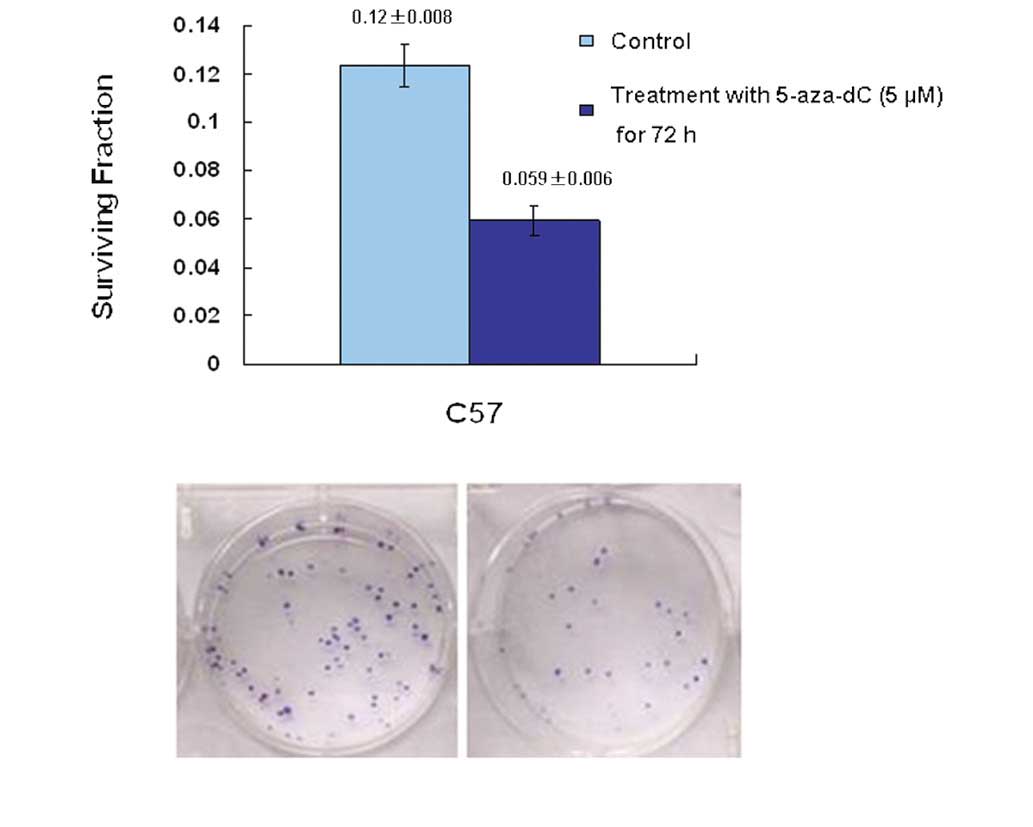
In order to explore how modulation of p73 status in
the C57 cell line influenced chemosensitivity, C57 cells were
treated with 5-aza-dC and subsequently assayed for clonogenic
survival following treatment with 1.25 μmol/l cisplatin. The
results indicated that upregulation of the level of p73 in cells,
induced by 5-aza-dC, significantly increased the chemosensitivity
of C57 cells; the p73 promoter was demethylated and p73 gene
expression was restored. The survival fraction for colony formation
in cells pre-treated with 5-aza-dC was 0.059±0.006. This was
significantly different from that of the control group (0.12±0.008;
P<0.05) (Fig. 5).
Discussion
Although chemotherapeutic agents including cisplatin
are widely used in the treatment of lung cancer, chemoresistance
remains a significant problem. The cytotoxic effect of cisplatin is
attributed to the formation of bulky DNA adducts, causing DNA
damage and finally inducing cancer cell apoptosis. Therefore, the
cellular apoptosis pathway may play a key role in chemosensitivity
(12).
p73 is a member of the p53 tumor suppressor protein
family. As with p53, p73 elicits cancer cell apoptosis in response
to DNA damage caused by cisplatin-based chemotherapy. Also, p73 is
capable of inducing apoptosis in tumor cells that lack functional
p53. Combination of p73 gene therapy and chemotherapy has been
demonstrated to be more effective compared with either of these
agents alone in a clinical trial for patients with NSCLC (13). However, epigenetic silencing of the
p73 gene via promoter and exon 1 hypermethylation has been
demonstrated to be a common event in types of lymphoma, brain tumor
and cervical cancer (14–16). In the present study, the epigenetic
modification of p73 via CpG island methylation represented a
critical mechanism for inactivation of this gene in six NSCLC cell
lines (Fig. 2). Hypermethylation
was significantly correlated with loss of p73 expression in these
cell lines (Fig. 1). In
vitro demethylation assay by the DNA methyltransferase
inhibitor, 5-aza-dC was successful in restoring p73 mRNA (Fig. 3) and protein (Fig. 4) expression levels in the human lung
squamous cell carcinoma cell line C57. Furthermore, following
restoration of p73 expression by 5-aza-dC treatment,
chemotherapeutic agent cisplatin-induced cell death was increased
in the C57 cell line (Fig. 5).
Although 5-aza-dC was not a p73-specific demethylation agent, the
change in p73 expression induced by 5-aza-dC influenced the
cisplatin-based chemosensitivity in the C57 cell line.
At present, a large number of genes have been
investigated for their methylation status in lung cancer patients.
Genes that have been intensively studied in primary NSCLC include
p16 (cyclin-dependent kinase inhibitor 2A), RASSF1A (Ras
association domain family protein 1A), APC (adenomatous polyposis
coli), RARβ-2 (retinoic acid receptor β), DAPK (death-associated
protein kinase) and MGMT (O6-methylguanine DNA methyltransferase).
While p16 and RASSF1A are involved in cell cycle regulation, APC
inhibits β-catenin, RARβ-2 is involved in growth regulation, DAPK
plays a role in the regulation of apoptosis and MGMT functions in
DNA repair. Overall, detecting the methylation status in these
genes has the potential to aid prediction of disease recurrence
after surgery, monitoring of the responses to therapy and early
detection of NSCLC. In addition, it may also be a therapeutic
target (17). Restoration of p73
expression is highly desirable, as p73 has been suggested to have a
role in determining cellular chemosensitivity.
In conclusion, the present study demonstrated the
significant correlation between expression of p73 and cellular
chemosensitivity in NSCLC that has been treated with cisplatin.
Therefore, p73 may play an important role in regulating the
cellular response of NSCLC to chemotherapy. The underlying
mechanism of p73 signaling in the apoptosis pathway during
chemotherapy requires further investigation. Whether the
methylation status of the p73 gene is a potential predictive marker
in evaluating the treatment regimens of NSCLC patients is a subject
that requires validation in a prospective study with a large group
of NSCLC patients.
Abbreviations:
|
NSCLC
|
non-small cell lung cancer;
|
|
5-aza-dC
|
5-aza-2-deoxycytidine;
|
|
MSP
|
methylation-specific PCR
|
Acknowledgements
This study was supported by the
Student Innovation Fund of Jinan University, China.
References
|
1.
|
Lee MW, Kim DS, Min NY and Kim HT: Akt1
inhibition by RNA interference sensitizes human non-small cell lung
cancer cells to cisplatin. Int J Cancer. 122:2380–2384. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Takenaka T, Yoshino I, Kouso H, Ohba T,
Yohena T, Osoegawa A, Shoji F and Maehara Y: Combined evaluation of
Rad51 and ERCC1 expressions for sensitivity to platinum agents in
non-small cell lung cancer. Int J Cancer. 121:895–900. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Machado-Silva A, Perrier S and Bourdon JC:
p53 family members in cancer diagnosis and treatment. Semin Cancer
Biol. 20:57–62. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Vayssade M, Haddada H, Faridoni-Laurens L,
Tourpin S, Valent A, Bénard J and Ahomadegbe JC: p73 functionally
replaces p53 in Adriamycin-treated, p53-deficient breast cancer
cells. Int J Cancer. 116:860–869. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Leong CO, Vidnovic N, De Young MP, Sgroi D
and Ellison L: The p63/p73 network mediates chemosensitivity to
cisplatin in a biologically defined subset of primary breast
cancers. J Clin Invest. 117:1370–1380. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Maas AM, Bretz AC, Mack E and Stiewe T:
Targeting p73 in cancer. Cancer Lett. Aug 22–2011.(Epub ahead of
print).
|
|
7.
|
Han S, Semba S, Abe T, Makino N, Furukawa
T, Fukushige S, Takahashi H, Sakurada A, Sato M, Shiiba K, Matsuno
S, et al: Infrequent somatic mutations of the p73 gene in various
human cancers. Eur J Surg Oncol. 25:194–198. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Corn PG, Kuerbitz SJ, van Noesel MM,
Esteller M, Compitello N, Baylin SB and Herman JG: Transcriptional
silencing of the p73 gene in acute lymphoblastic leukemia and
Burkitt’s lymphoma is associated with 5′CpG island methylation.
Cancer Res. 59:3352–3356. 1999.PubMed/NCBI
|
|
9.
|
Liu K, Zhan M and Zheng P: Loss of p73
expression in six non-small cell lung cancer cell lines is
associated with 5′ CpG island methylation. Exp and Mol Pathology.
84:59–63. 2008.PubMed/NCBI
|
|
10.
|
Peters UR, Tschan MP, Kreuzer KA,
Baskaynak G, Lass U, Tobler A, Fey MF and Schmidt CA: Distinct
expression patterns of the p53-homologue p73 in malignant and
normal hematopoiesis assessed by a novel real-time reverse
transcription-polymerase chain reaction assay and protein analysis.
Cancer Res. 59:4233–4236. 1999.
|
|
11.
|
Herman JG, Graff JR, Myohanen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Biswas AK and Johnson DG: Transcriptional
and nontranscriptional functions of E2F1 in response to DNA damage.
Cancer Res. 72:13–17. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
He Y, Fan S and Jiang Y: Effects of p73
gene overexpression on apoptosis and chemosensitivity of human lung
adenocarcinoma cell line A549. Chinese J Cancer. 25:925–932.
2006.PubMed/NCBI
|
|
14.
|
Pei JH, Luo SQ, Zhong Y, Chen JH, Xiao HW
and Hu WX: The association between non-Hodgkin lymphoma and
methylation of p73. Tumour Biol. 32:1133–1138. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Meyer G: p73: a complex gene for building
a complex brain. Cell Cycle. 10:1188–1189. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Liu SS, Leung RC, Chan KY, Chiu PM, Cheung
AN, Tam KF, Ng TY, Wong LC and Ngan HY: p73 expression is
associated with the cellular radiosensitivity in cervical cancer
after radiotherapy. Clin Cancer Res. 10:3309–3316. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Heller G, Zielinski CC and
Zöchbauer-Müller S: Lung cancer: from single-gene methylation to
methylome profiling. Cancer Metastasis Rev. 29:95–107. 2010.
View Article : Google Scholar : PubMed/NCBI
|