Introduction
In developing countries, cervical cancer is a
leading cause of mortality among females and is the second most
common malignancy in females worldwide (1). Each year, >270,000 females succumb
to cervical cancer, of which 53,000 are located in China (2). Studies have demonstrated that early
sexual intercourse, promiscuity, infection with high-risk types of
human papillomavirus (HPV) and a number of other factors may cause
the normal cervical epithelium to become pre-neoplastic cervical
intraepithelial neoplasia, which may later transform into invasive
cervical cancer (2). However there
has not been a viable explanation for the pathogenesis of cervical
cancer.
MicroRNAs (miRs/miRNAs) are non-coding small RNAs
composed of 19–25 ribonucleic acid molecules. They regulate the
stability and expression of target mRNAs and serve as
post-transcriptional regulators, which determine cell identity and
fate. These small RNAs regulate gene expression by interacting with
the 3′UTR of target mRNA (3–5). The
expression levels of the members of the miR-17-92 cluster have been
shown to be altered in numerous types of cancer. The miR-17-92
cluster encodes six miRNAs, miR-17, miR-18a, miR-19a, miR-19b,
miR-20a and miR-92, which are located in a coding region of the
open reading frame (ORF) of the C13orf25 gene (6–8). The
human miR-17-92 cluster gene is mapped to chromosome 13q31. Data
have indicated that the mir-17-92 cluster is involved in the
regulation of cell growth, cell differentiation, apoptosis, cell
motility, cell adhesion, cell invasion and angiogenesis (9,10).
Previous evidence has shown that a number of miRNAs play
significant roles in cancer by altering the expression of target
oncogenes and tumor suppressor genes, including those affecting
lung cancer, breast cancer, human glioma and lymphoproliferative
disease (11–13). The phosphatase and tensin homologue
(PTEN) protein is an important target protein. The significance of
the PTEN protein is indicated by the fact that it is frequently
disrupted in numerous cancers (14). TargetScan (http://www.targetscan.org/) has predicted hundreds of
miR-92 targets and the tumor suppressor PTEN is one of the most
prominent. PTEN has been shown to play a role in several human
cancers (15,16). However, the correlation between
miR-92 and PTEN has not previously been reported in cervical
cancer. Whether miR-92 has any association with HPV remains
undetermined. The present study aimed to examine miR-92 expression
in HPV16-positive squamous cervical carcinomas (SCCs). The
correlation between miR-92 expression and clinical status was also
evaluated. miR-92-mimics and anti-miR-92 were transfected into
cells from the SCC cell line, SiHa, and the contribution of miR-92
to tumor cell growth, apoptosis and migration was consequently
investigated. The role of miR-92 in tumor formation was also
evaluated using subcutaneously-inoculated immunocompromised mice.
HPV16 E6 siRNA was transfected into SiHa cells and the
pEGFP-N1-HPV16E6 plasmid and HPV16 E6 siRNA were transfected into
C33A cells in order to detect the correlation between HPV16 E6 and
miR-92. The roles of miR-92 and PTEN were investigated in cervical
cancer cell lines and the introduction of miR-92 was analyzed with
regard to PTEN protein and mRNA expression.
Materials and methods
Tissue collection
A total of 34 cervical cancer tissue samples, 23 of
which were HPV16-positive, were obtained from patients (mean age,
43.1±6.0 years) by biopsy during colposcopy. Another 34 normal,
HPV-negative cervical tissues were also collected from patients
(mean age, 45.7±4.71 years) during hysterectomies. The samples were
all obtained from the Shengjing Hospital of China Medical
University (Shenyang, Liaoning, China). All the cervical cancer
tissues were obtained from cervical squamous carcinomas that were
confirmed by pathological analysis. All tissues were obtained with
informed consent in accordance with the requirements of the China
Medical University Research Ethics Committee, as stipulated in the
Helsinki Declaration. This study was approved by the ethics
committee of Shengjing Hospital of China Medical University.
Cell lines and culture conditions
SiHa cells were obtained from the Shengjing Hospital
of China Medical University. C33A cells were obtained from the
American Type Culture Collection (ATCC, Manassas, VA, USA). The
SiHa cells were cultured in Dulbecco’s modified Eagle’s medium
(DMEM; Hyclone, Logan, UT, USA). The C33A cells were cultured in
ATCC-formulated Eagle’s minimum essential medium (EMEM). The media
were contained in 10% fetal bovine serum (FBS), 100 IU/ml
penicillin and 100 mg/ml streptomycin. All cells were cultured at
37°C in a humidified atmosphere of 5% CO2.
Transfection
The miR-92-mimic, anti-miR-92, negative control
(NC), E6 siRNA and E6 siRNA-NC were purchased from Shanghai
GenePharma Co., Ltd and Shanghai Sangon Co., Ltd, (Shanghai,
China), and all contained green fluorescent tags. The SiHa cells
were transfected with miR-92-mimic, anti-miR-92 and NC using
Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer’s instructions. The medium was
replaced with fresh growth medium following 6 h of transfection. A
qPCR analysis of miR-92 was used to detect the transfection
efficiency.
pEGFP-N1-16E6 and pEGFP-N1-neo plasmids were stably
transfected into the C33A cells. Briefly, the C33A cells were
seeded in 24-well plates with 500 μl EMEM medium and without
antibiotics. The cells were transfected with 0.8 μg
pEGFP-N1-neo and pEGFP-N1-16E6 plasmids and selected in medium
containing G418 (250 μg G418 per ml). At two weeks
post-transfection, the G418-resistant colonies were pooled and
expanded.
SYBR-green-based qPCR
Total RNA was extracted using the TRIzol reagent
(Invitrogen). The cDNA was reverse-transcribed from 1 ng total RNA
with a miRNA reverse transcription kit (SYBR Green-based Real Time
PCR kit, Shanghai GenePharma Co., Ltd.). This cDNA was used at
3-fold dilutions for each run of qPCR on an ABI 7500 thermal-cycler
system, according to the manufacturer’s instructions. The procedure
was normalized using U6 as the endogenous control. The reverse
transcription reaction was performed at 16°C for 30 min, 42°C for
30 min and 85°C for 10 min and then stored at 4°C. The SYBR Green
qPCR reaction conditions consisted of an initial denaturation cycle
at 95°C for 3 min, followed by 40 cycles at 95°C for 12 sec and
annealing and extension at 62°C for 50 sec. Each experiment was
repeated three times. All PCR products were resolved in agarose gel
to confirm PCR specificity.
Western blot analysis
The SiHa cells were transfected with miR-92-mimic
and NC. At 48 h post-transfection, RIPA buffer (1 lg/ml leupeptin
and 1 lg/ml PMSF; Beyotime, Nanjing, China) was used to isolate the
total protein from the transfected cells in order to detect PTEN
protein expression, in accordance with the manufacturer’s
instructions. The proteins were separated using 10% sodium
dodecylsulfate-polyacrylamide gel electrophoresis and transferred
to polyvinylidene difluoride membranes. The membranes were blocked
using 5% skimmed powdered milk for 2 h at room temperature. The
membranes were washed with TBST and incubated with the primary
antibody for PTEN (Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA), overnight at 4°C. The membranes were then washed using TBST
and incubated with horseradish peroxidase (HRP)-labeled goat
anti-mouse secondary antibodies (Beyotime). The bands were analyzed
using the SuperSignal West Pico kit (Pierce, Rockford, IL, USA) and
examined using ImageQuant enhanced chemiluminescence (ECL; GE
Healthcare, Buckinghamshire, UK). The band intensities were
analyzed using Quantity One software (Bio-Rad, Hercules, CA). Each
experiment was repeated three times.
Immunohistochemistry
The HPV16 E6 mouse polyclonal antibody was purchased
from Santa Cruz Biotechnology, Inc. The cervical cancer tissue
sections were deparaffinized and rehydrated through a series of
graded alcohols and xylene, then washed with PBS (0.02 M).
Endogenous peroxidase activity was blocked by 3%
H2O2 for 15 min at room temperature, then
exposure to the antigen. The tissue sections were incubated with
the primary antibody for HPV16 E6 (HPV16 E6 antibody diluted to
1:50 and applied to each slide) overnight at 4°C. The tissue
sections that were incubated with DAB (GSGB Biotechnology, Beijing,
China) developed coloration. The HPV16 E6 protein was mainly
expressed in the nucleus.
Cell counting kit (CCK)-8 assay
Cell survival was assessed using a CCK-8 assay
(Dojindo, Kumamoto, Japan). SiHa cells were transfected for 48 h in
96-well plates and incubated with CCK-8 for 24, 48, 72 and 96 h.
Optical density was read at 450 nm (A450) using Easy Reader 340 AT
(SLT-Lab Instruments, Bath, UK). Each experiment was repeated three
times.
Transwell cell migration assay
The SiHa cells were transfected with the
miR-92-mimic and NC in 6-well plates. At 48 h post-transfection,
the cells were collected and 5×104 were placed in the
upper chambers of the Transwell inserts with 5% FBS (8-μm
pore size; Corning Inc., Corning, NY, USA). The lower compartments
contained DMEM medium with 20% FBS. Subsequent to 24 h, the cells
that had migrated were fixed with 4% paraformaldehyde and crystal
violet stain. The cells were then counted and images were
captured.
Caspase-3 activity
Caspase-3 activity was assessed using the
colorimetric CaspACE Assay System (Promega, Mannheim, Germany),
according to the manufacturer’s instructions. Briefly, equal
amounts of cell extract were incubated with the substrate
(Ac-DEVD-pNA) in the assay buffer for 24 h at room temperature.
Absorbance was measured at 405 nm using a microplate reader
(SLT-Lab Instruments). Each experiment was performed in
triplicate.
Terminal deoxynucleotidyl
transferase-mediated dUTP nick end labeling (TUNEL) assay
The cells were washed in PBS supplemented in 0.1%
bovine serum albumin and treated with an in situ detection
kit, according to the manufacturer’s instructions (Boehringer
Mannheim Biochemicals, Indianapolis, IN, USA). Nuclei with
fragmented DNA were visualized using a fluorescence microscope.
Tumor xenografts
Four-week-old female nude mice were cared for in
accordance with the Guide for the Care and Use of Laboratory
Animals (NIH publication no. 80-23, revised 1996), and the
experiments were performed according to the Shengjing Hospital of
China Medical University ethical guidelines for animal experiments.
SiHa cells (1×106) were transfected with anti-miR-92 or
anti-miRNA-NC for 48 h. The cells were suspended in 200 μl
PBS and subcutaneously injected into the right and left posterior
flanks of the same female BALB/c athymic nude mouse. A total of 20
nude mice were used in the experiment. Tumor growth was examined
every third day for four weeks. The tumor volume (V) was calculated
by measuring the tumor length (L) and width (W) using calipers and
calculated using the following formula (L × W2) × 0.5.
The tumor xenografts were harvested and snap-frozen. Cryosections
(4 μm) were stained using hemotoxylin and eosin.
Statistics
All statistical analyses were carried out using SPSS
for Windows, version 18.0 (SPSS, Inc., Chicago, IL, USA). All
values are presented as the mean ± SD from at least three separate
experiments. All tests that were performed were two-sided Student’s
t-tests. P<0.05 was considered to indicate a statistically
significant difference. Standard curves were generated and the
relative amount of miR-92 expression was normalized to U6 snRNA.
The miR-92 expression fold change was evaluated using the
2−ΔΔCt method.
Results
miR-92 expression in cervical cancer
tissues
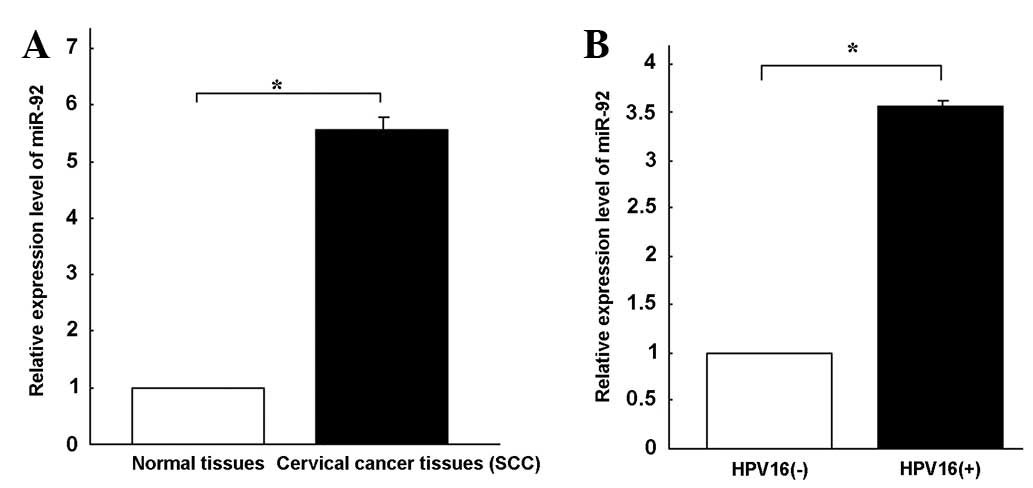
miR-92 expression was quantified in 34 tumor samples
and 34 normal cervical tissues using qPCR (Fig. 1A). miRNA expression was relatively
stable in the adjacent normal cervical tissues. miR-92 expression
was 5.56-fold higher in the SCC tissues compared with the
corresponding non-tumor tissues. These results suggest that miR-92
upregulation may play a role in the malignant progression of
cervical cancer.
Correlation between HPV16 infection and
the upregulation of miR-92 expression in cervical cancer
In order to further investigate the role of miR-92
in cervical cancer, miR-92 expression levels were assessed in
HPV16-positive and HPV16-negative cervical cancer tissues. An
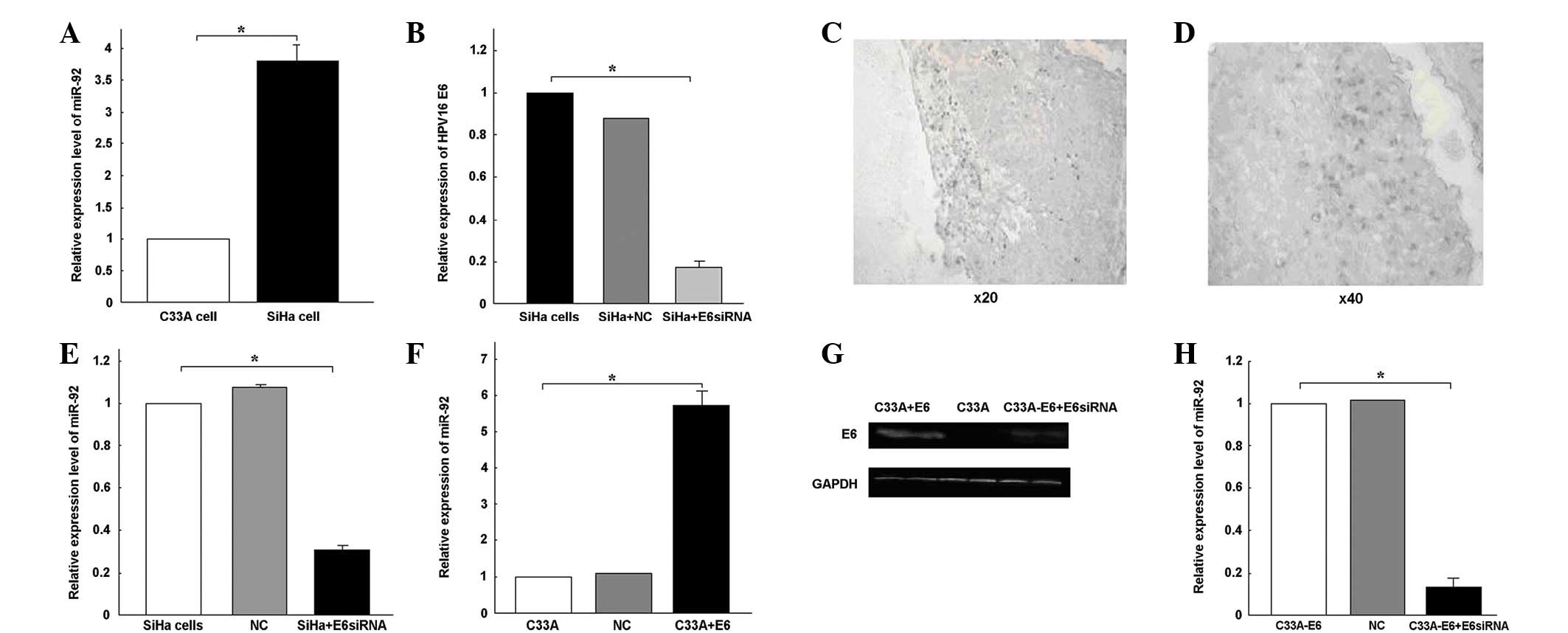
immunohistochemical assay was used to confirm the HPV16-E6-positive
nature of 34 cervical cancer tissues, 23 of which were positive for
HPV16 E6 (Fig. 2C and D). qPCR
revealed that miR-92 expression was 3.56-fold higher (Fig. 1B) in the HPV16-positive cervical
cancer tissues compared with the HPV16-negative tissues. The C33A
and SiHa cervical cancer cell lines were then analyzed. miR-92
expression was 3.79-fold higher in the SiHa cells than in the C33A
cells (Fig. 2A).
The overexpression of miR-92 in the HPV16-positive
cervical cancer tissues and SiHa cells prompted an investigation
into the possible roles that miR-92 may play in tumorigenesis. E6
siRNA was transfected into the SiHa cells. qPCR revealed that the
transfection with E6 siRNA specifically knocked down E6 expression
(P<0.05; Fig. 2B). miR-92
expression was observed to be downregulated following the
transfection with E6 siRNA (Fig.
2E). The pEGFP-N1-16E6 plasmid was transfected into the C33A
cells to increase HPV16 E6 expression, and E6 protein expression
was detected using a western blot analysis (Fig. 2G). At 6 h post-transfection, the
upregulation of E6 expression was confirmed using a fluorescence
microscope (Fig. 3B). The
expression of miR-92 increased 5.74-fold following the transfection
of pEGFP-N1-16E6 into the C33A cells (P<0.05; Fig. 2F). When E6 siRNA was transfected
into the C33A-pEGFP-N1-16E6 cells, the E6 protein was blocked
(Fig. 2G). The expression of miR-92
was downregulated following transfection with the E6 siRNA
(P<0.05; Fig. 2H). From these
results, it was concluded that the overexpression of miR-92 may be
partially caused by HPV16 infection.
Effects of miR-92 on SCC cell
proliferation in vitro
Since cell proliferation facilitates the development
of malignancy, the present study evaluated whether miR-92
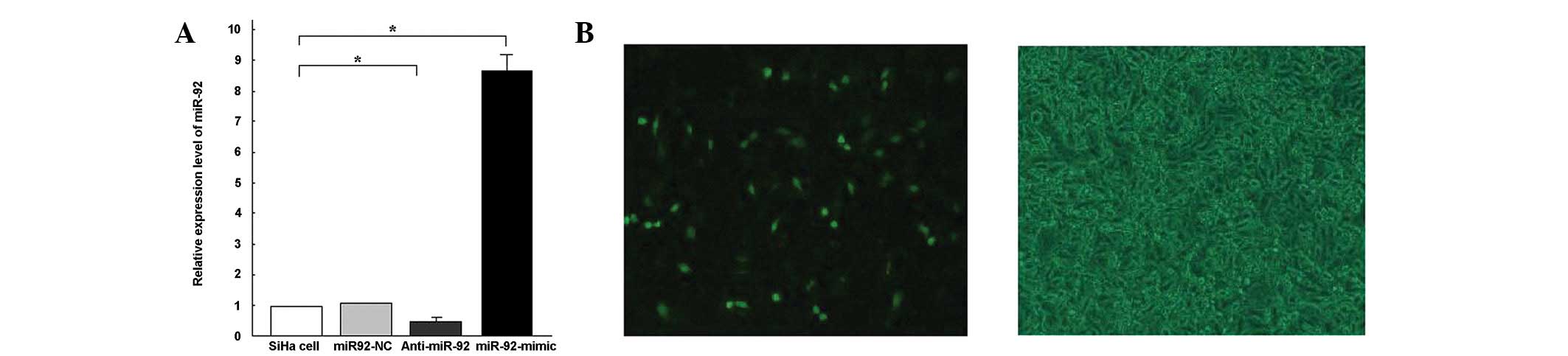
contributed to SCC cell survival. Following 6 h of transfection,
miR-92 expression was detected using qPCR. miR-92 expression levels
were shown to be 8.67-fold higher in the SiHa-miR-92 mimic cells
than in the SiHa cells (Fig. 3A).
Following 48 h of transfection, a CCK-8 assay (Fig. 4A) demonstrated that cell
proliferation was significantly increased upon transfection with
the miR-92-mimic and decreased upon transfection with anti-miR-92
(P<0.05), but not upon transfection with the unrelated NC. These
findings indicate that miR-92 is involved in the proliferation of
SCC.
Effects of miR-92 on cell migration
The Transwell method was used to determine the
effects of miR-92 on cell migration. The invasive activity of the
SiHa cells increased following transfection with the miR-92-mimic.
Use of the miR-92-mimic group increased the number of invasive
cells by ∼2-fold compared with the miR-92-NC group (Fig. 4B and C). These results indicate that
miR-92 increases proliferation and invasion in cervical cancer
cells.
Effects of miR-92-knockdown on
caspase-3-dependent apoptosis in SiHa cells
To further analyze the possible mechanisms
underlying the increased sensitivity of the SiHa cells caused by
anti-miR-92, the rate of apoptosis at 48 h post-transfection was
detected in the SiHa cells that were transfected with anti-miR-92
and NC. A TUNEL assay indicated a significant increase in the
number of apoptotic nuclei in the SiHa cells that were transfected
with anti-miR-92 compared with those that were transfected with NC
(Fig. 5C). Caspase-3 activity was
shown to have increased 4.18-fold in the SiHa cells that were
transfected with anti-miR-92 compared with the control cells
(P<0.05; Fig. 5A). The
downregulation of miR-92 may lead to an increase in the apoptosis
of SiHa cells, which correlates with the activation of
caspase-3.
Effects of miR-92 on the regulation of
PTEN protein expression in SiHa cells
PTEN protein expression is significantly
downregulated in cervical cancer. miR-92 has been shown to
post-transcriptionally inhibit PTEN expression in various types of
human cancer cells. However, the correlation between PTEN and
miR-92 in cervical cancer remains unknown. To determine the effects
of miR-92 on PTEN protein expression in cervical cancer,
anti-miR-92 and the miR-92 mimic were transfected into the SiHa
cells. The effects of miR-92 on the endogenous expression of PTEN
were then examined. qPCR demonstrated that the inhibition of
endogenous miR-92 by anti-miR-92 resulted in an upregulation of
PTEN mRNA (Fig. 5B). A western blot
analysis revealed that the expression levels of the PTEN protein in
the SiHa cells that were transfected with anti-miR-92 were higher
than in those that were transfected with the miR-92-mimic (Fig. 4D).
Effects of anti-miR-92 on tumorigenesis
in cervical cancer xenografts
To further examine the effects of miR-92 on the
in vivo growth of cervical carcinoma, anti-miR-92 and
NC-transfected SiHa cells were independently injected
subcutaneously into the two anterior flanks of the same nude mouse.
The tissue structure and cell morphology of the SiHa cells that
were transfected with anti-miR-92 did not differ from those that
were transfected with the NC miRNAs (Fig. 6A), however, fewer tumors formed
(7/10 vs. 2/10; Fig. 6B and C).
Discussion
Evidence suggests that the aberrant expression of
miRNA is involved in tumor progression, metastasis and
chemoradio-resistance (17).
Oncogenic miR-92 has been reported to be frequently overexpressed
in a variety of human cancers (18). However, our understanding of the
potential role of miR-92 in cervical cancer remains limited.
To confirm whether miR-92 expression may affect
cervical cancer cells, the present study examined the expression of
miR-92 in 34 cervical cancer tissue samples and 34 normal cervical
tissue samples The results revealed the level of miR-92 expression
in the cervical cancer tissues to be significantly higher than in
the normal cervical tissues. As in previous studies conducted on
other types of cancer, miR-92 was observed to be able to
dramatically increase cell proliferation, inhibit apoptosis and
promote cell migration in cervical cancer lines. To elucidate the
mechanisms underlying the increase in chemosensitivity induced by
miR-92, the effects of miR-92 on apoptosis were analyzed using SiHa
cells. TUNEL assays indicated that miR-92 was able to significantly
enhance apoptosis among the SiHa cells, which may be correlated
with the increase in caspase-3 activity. The tumor suppressor gene,
PTEN, was significantly downregulated in the SiHa-miR-92-mimic
cells compared with the SiHa-NC cells. PTEN is a potential target
of miR-92, which has been reported in a number of human tumors. It
has been reported that the decreased expression of miR-92 may
decrease cancer cell proliferation and increase the levels of PTEN,
BCL2L11 and CDKN1A expression (19). Therefore, the present data indicate
that miR-92 plays a number of roles in the development of cervical
cancer.
HPVs are a group of small DNA tumor viruses of ∼55
nm in diameter. The DNA molecule of these viruses contains early
ORFs, E1, E2, E4, E5, E6 and E7 and late ORFs, L1 and L2. The
proteins that are coded by the E6 ORFs of high-risk HPVs are small
nuclear proteins with transforming activities. HPV regulates the
expression of numerous cellular miRNAs through the HPV E6 protein.
This regulation has been shown in the reduced expression of
miR-34a, miR-106b/93/25 and miR-23b through the use of E6 (20–25).
High-risk E6 is able to interact with several dozen, or even
hundreds of cellular factors (26–28).
These interactions may lead to an increase or decrease in the
expression of cellular miRNAs. In the present study, plasmid
pEGFP-N1-16E6 was stably transfected into C33A cells in order to
enhance E6 expression. E6 siRNA was also transfected into C33A and
SiHa cells to block E6 expression. qPCR was used to detect miR-92
expression. The results suggested that the expression of miR-92 was
upregulated in C33A-pEGFP-N1-16E6 cells and downregulated in HPV16
E6-knockdown cells. This indicated that HPV16 infection induces
carcinogenesis, most likely by altering the expression of specific
miRNAs, including miR-92. However, miR-92 may be the superior
biomarker due to its broad impact on several targets and pathways
involved in cervical cancer. Elucidating the role of miR-92
requires further research with regard to the number of tumor
vessels that are involved in cervical disease.
In conclusion, miR-92 is overexpressed in SCC
tissues and cervical cancer cell lines. Increases or decreases in
the expression of miR-92 may alter multiple biological processes in
human cervical cancer cells, including proliferation, apoptosis and
migration, most likely through the regulation of the PTEN protein.
HPV16 E6 is able to increase miR-92 expression in SiHa- and
C33A-pEGFP-N1-16E6 cells. The identification of oncogenic miR-92
may serve as a biomarker for SCC. HPV may be necessary for the
prevention of SCC.
References
|
1.
|
Bosch FX and de Sanjosé S: Chapter 1:
Human papillomavirus and cervical cancer - burden and assessment of
causality. J Natl Cancer Inst Monogr. 31:3–13. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Parkin DM, Pisani P and Ferlay J: Global
cancer statistics. CA Cancer J Clin. 49:33–64. 1999. View Article : Google Scholar
|
|
3.
|
Rana TM: Illuminating the silence:
understanding the structure and function of small RNAs. Nat Rev Mol
Cell Biol. 8:23–36. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Rao PH, Houldsworth J, Dyomina K, et al:
Chromosomal and gene amplification in diffuse large B-cell
lymphoma. Blood. 92:234–240. 1998.PubMed/NCBI
|
|
7.
|
Ota A, Tagawa H, Karnan S, et al:
Identification and characterization of a novel gene, C13orf25, as a
target for 13q31–q32 amplification in malignant lymphoma. Cancer
Res. 64:3087–3095. 2004.
|
|
8.
|
Tagawa H and Seto M: A microRNA cluster as
a target of genomic amplification in malignant lymphoma. Leukemia.
19:2013–2016. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Olive V, Bennett MJ, Walker JC, Ma C, et
al: miR-19 is a key oncogenic component of mir-17-92. Genes Dev.
23:2839–2849. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Zhang X, Yu H, Lou JR, Zheng J, et al:
MicroRNA-19 (miR-19) regulates tissue factor expression in breast
cancer cells. J Biol Chem. 286:1429–1435. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Cho WC, Chow AS and Au JS: Restoration of
tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung
adenocarcinoma patients with epidermal growth factor receptor
mutation. Eur J Cancer. 45:2197–2206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Eades G, Yang M, Yao Y, Zhang Y and Zhou
Q: miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in
breast cancer cells. J Biol Chem. 286:40725–40733. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Xiao C, Srinivasan L, Calado DP, Patterson
HC, Zhang B, Wang J, et al: Lymphoproliferative disease and
autoimmunity in mice with increased miR-17-92 expression in
lymphocytes. Nat Immunol. 9:405–414. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Bonneau D and Longy M: Mutations of the
human PTEN gene. Hum Mutat. 16:109–122. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Petrocelli T and Slingerland JM: PTEN
deficiency: a role in mammary carcinogenesis. Breast Cancer Res.
3:356–360. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Byun D, Cho K, Ryu B, et al: Frequent
monoallelic deletion of PTEN and its reciprocal association with
PIK3CA amplification in gastric carcinoma. Int J Cancer.
104:318–327. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar
|
|
18.
|
Shigoka M, Tsuchida A, Matsudo T, et al:
Deregulation of miR-92a expression is implicated in hepatocellular
carcinoma development. Pathol Int. 60:351–357. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Al-Nakhle H, Burns PA, Cummings M, et al:
Estrogen receptor (beta)1 expression is regulated by miR-92 in
breast cancer. Cancer Res. 70:4778–4784. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Li B, Hu Y, Ye F, Li Y, Lv W and Xie X:
Reduced miR-34a expression in normal cervical tissues and cervical
lesions with high-risk human papillomavirus infection. Int J
Gynecol Cancer. 20:597–604. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Brosh R, Shalgi R, Liran A, Landan G,
Korotayev K, Nguyen GH, Enerly E, Johnsen H, Buganim Y, et al:
p53-Repressed miRNAs are involved with E2F in a feed-forward loop
promoting proliferation. Mol Syst Biol. 4:2292008. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J,
Lu W, Wan X, Ma D and Xie X: Progressive miRNA expression profiles
in cervical carcinogenesis and identification of HPV-related target
genes for miR-29. J Pathol. 224:484–495. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Rao Q, Shen Q, Zhou H, Peng Y, Li J and
Lin Z: Aberrant microRNA expression in human cervical carcinomas.
Med Oncol. 29:1242–1248. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Au Yeung CL, Tsang TY, Yau PL and Kwok TT:
Human papillomavirus type 16 E6 induces cervical cancer cell
migration through the p53/microRNA-23b/urokinase-type plasminogen
activator pathway. Oncogene. 30:2401–2410. 2011.PubMed/NCBI
|
|
26.
|
Zheng ZM: Viral oncogenes, noncoding RNAs,
and RNA splicing in human tumor viruses. Int J Biol Sci. 6:730–755.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Narisawa-Saito M and Kiyono T: Basic
mechanisms of high-risk human papillomavirus-induced
carcinogenesis: roles of E6 and E7 proteins. Cancer Sci.
98:1505–1511. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
McLaughlin-Drubin ME and Münger K: The
human papillomavirus E7 oncoprotein. Virology. 384:335–344. 2009.
View Article : Google Scholar : PubMed/NCBI
|