Introduction
The discovery that complete cellular reprogramming
may be achieved by introducing the defined transcription factors
c-Myc, Sox2, Oct3/4 and Klf4
into terminally differentiated somatic fibroblasts of mouse and
human origins was an important breakthrough (1,2). The
generation of induced pluripotent stem (iPS) cells by the
introduction of defined factors, which are generally expressed in
embryonic stem (ES) cells, results in the reconstitution of organs
in chimeric mice and contributes to the regeneration of human
tissues (3). We previously showed
that gastrointestinal cancer cells acquired multipotential
differentiation ability upon the introduction of defined factors;
the gene expression profiles of mesodermal and ectodermal cells
appeared in gastrointestinal cancer cells of endodermal origin
[termed induced multipotent cancer (iPC) cells] (4). Whether the iPC cells were generated
via a state of pluripotency remains to be investigated, although
the iPC cells expressed ES-like genes and possessed the ability to
differentiate from cells of endodermal origin into other endoderm
and mesoderm lineages (4). Notably,
in vitro differentiation resulted in sensitization to
therapeutic reagents such as vitamins A and D and the
chemotherapeutic agent 5-fluorouracil (5-FU), as well as reduced
tumorigenicity, suggesting that altering the cancer cell lineage
through reprogramming in vivo may be a promising concept for
novel and efficient cancer therapy (4). However, at present, there are a
limited number of studies concerned with reprogramming in
vivo, and thus the mechanism involved in reprogramming in
vivo remains unknown.
Epithelial tumor tissues are composed of various
types of mesenchymal cells, such as myofibroblasts, fibroblasts,
endothelial cells, lymphocytes, monocytes and macrophages, certain
of which are known to be components of a microenvironment (niche).
These components are involved in tumorigenesis at the early stages,
support cancers cells and provide resistance against exposure to
chemotherapeutic reagents. Overall, although it is assumed that
mesenchymal cells are important in the process of reprogramming in
the complex system in vivo, no investigations on how
reprogramming factors affect the mesenchymal components have been
conducted. To assess this, the effect of direct injection of a
Sendai virus (SeV) vector encoding four defined factors into the
liver was studied using transgenic and knockout mice with various
genetic backgrounds, and the effect was compared with that of
direct injection of microRNAs (miRNAs) diluted with cationic lipid.
The in vivo bioluminescence imaging data revealed
transformation hot spots for p53 (also known as TP53
in humans and Trp53 in mice) deficiency or conditional
activation of mutant Kras, and the sizes were consistent
with those in immunodeficient
NOD.CB17-Prkdcscid/J (NOD/SCID) mice and
NOD.Cg-PrkdcscidIl2rgtmSug/Jic
(NOG) mice expressing transgenic urokinase-type plasminogen
activator (uPA) in the liver (uPA-NOG), as well as larger
compared with those in the control mice. The present results
suggested that the effect of reprogramming-based, novel therapeutic
approaches was enhanced by Kras activation. The effect was
more apparent with Kras activation than with tumor
suppressor p53 deficiency, suggesting a distinct role for
the Kras pathway in direct reprogramming in the liver.
Furthermore, immunodeficiency may increase the effect of
reprogramming, presumably by blocking the immunosurveillance of
transformed cells.
Subjects and methods
Experimental animals
NOD/SCID mice were purchased from Charles River
Japan (Osaka, Japan). All animal experiments were performed with
approval from the Animal Experiments Committee of Osaka University.
The NOD/SCID mice lack B cells, T cells and the complement system,
and possess severely reduced natural killer (NK) cells. More
severely immunodeficient uPA-NOG mice were produced by
extra-uterine fertilization, resulting in zygotes that expressed
transgenic uPA in the liver; the extracellular matrix in the
liver was modified to activate the hemolytic system, which
facilitated xenogeneic engraftment or growth of transformed cells
in the present experiment in mice with an immunodeficient
background (5). Heterozygous
B6.129S4-Krastm4Tyj/J mice (Jackson
Laboratory, Bar Harbor, ME, USA), which carry an allele with the
most common point mutation whose expression is blocked by the
presence of a loxP-flanked stop codon in the ROSA loci, were
crossed with B6129-Tg(MMTV-Cre)4Mam/J mice (Jackson Laboratory),
which express P1 Cre recombinase under the control of the mouse
mammary tumor virus (MMTV) long terminal repeat (LTR) promoter. The
MMTV LTR promoter directs a widespread pattern of expression to
produce CMV-Cre/Krasmut mice; and when
expressed in B6.Cg-Tg(Alb-Cre)21Mgn/J mice (Jackson Laboratory), is
efficient in achieving liver-specific recombination to produce
Alb-Cre/Krasmut mice.
B6.129S2-Trp53tm1Tyj/J mice (Jackson
Laboratory), from which a mutant allele was produced by a targeted
neo insertion into the p53 locus, were mated with STOCK
Tg(Nanog-GFP, Puro)1 Yam mice, which express the green fluorescent
protein under the control of the Nanog gene promoter (RIKEN
BioResource Center, Tsukuba, Japan), to produce
Nanog-GFP/Trp53+/−
(KO) mice. Overall, two immunodeficient mice were used in
the experiments, NOD/SCID and uPA-NOG, as well as
CMV-Cre/Krasmut,
Alb-Cre/Krasmut and
Nonog-GFP/Trp53KO mice. miRNAs were also
used to assess the effect.
In vivo administration of viral construct
mixture
SeV vectors replicate in the form of negative-sense
single-stranded RNA in the cytoplasm of infected cells and do not
undergo a DNA phase or integrate into the host genome (6). It was shown that the efficient
induction of transgene-free human pluripotent stem cells was
achieved using a vector based on SeV, an RNA virus that does not
integrate into the host genome; iPS induction could be achieved by
the SeV-mediated gene-transfer introduction of the defined
transcription factors c-Myc, Sox2,
Oct3/4 and Klf4 from terminally differentiated
somatic cells (7). A viral
construct mixture consisting of: i) 5 μl lentiviral vector
and ii) SeV vectors (2.5 μl per each transcription factor)
or 10 μl miRNAs was prepared. Co-transfection of the
lentiviral luciferase gene was performed to trace the cell
populations in which the genes were introduced. The SeV vectors
were mixed according to the transcription factors to be introduced,
such as SeV vectors encoding c-Myc, Sox2,
Oct3/4 and Klf4 (MSOK); Sox2, Oct4 and
Klf4 (SOK); or c-Myc alone (M). With regard to
miRNAs, 60 pmol of double-stranded mature miRNAs (20 pmol of
mmu-miR-200c; 5 pmol of mmu-miR-302a, -302b, -302c and -302d; and
10 pmol of mmu-miR-369-3p and -5p) was diluted with 10 μl
siPORT (Ambion, Austin, TX, USA). Median laparotomy was performed
in each mouse under sevoflurane anesthesia and the viral construct
mixture was directly injected into the median lobe of the
liver.
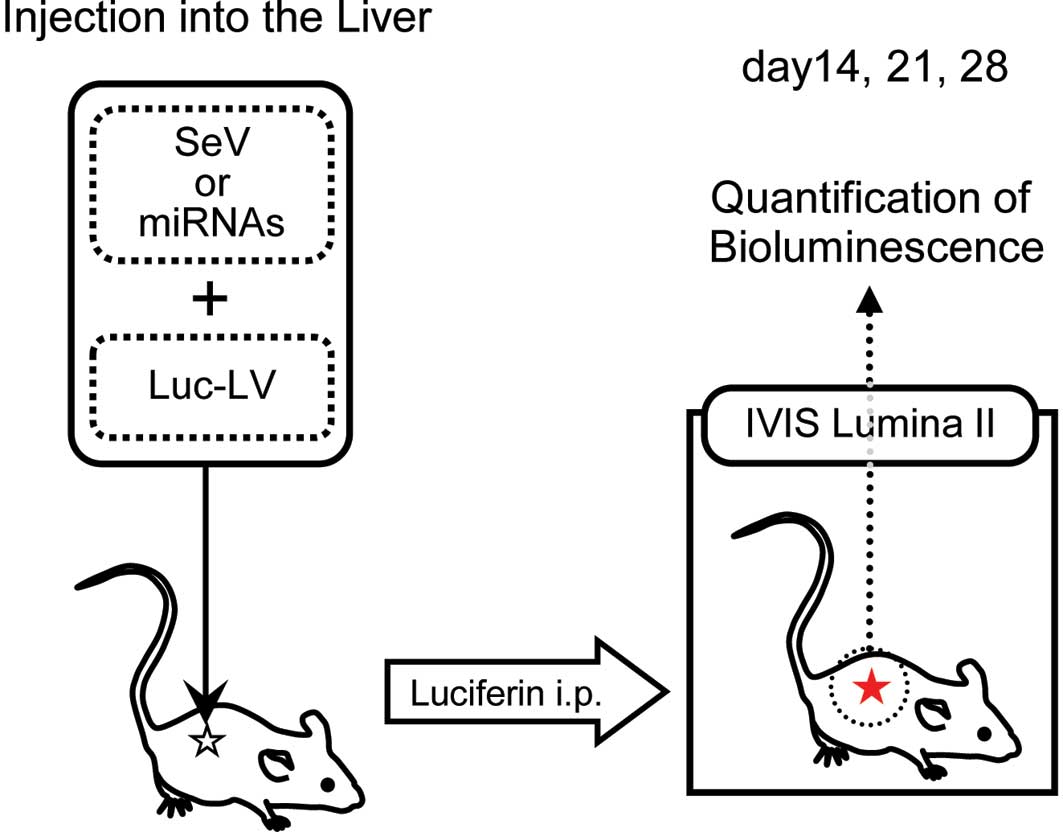
In vivo imaging
To trace the behavior of the injected viral
construct, the animals were examined at days 14, 21 and 28 using
the IVIS Lumina II imaging system (Caliper Life Sciences,
Hopkinton, MA, USA) (Fig. 1). Each
mouse received luciferin intraperitoneally at 4 mg/kg and was then
anesthetized with 2% isoflurane; the mice were left undisturbed for
10 min thereafter. Subsequently, the mice were imaged under the
following conditions: Exposure, 2 min; f-stop, 1; binning, medium;
field of view, 12.5 cm. Bioluminescence values were calculated as
photons/s/cm2/sr in the region of interest.
Results
Immunodeficient mice
In the NOD/SCID mice, the luciferase-positive area
was detected 14, 21 and 28 days following the injection of viral
construct mixture (Fig. 2A). The
mice showed no apparent health problems. The uPA-NOG mice showed a
more apparent luciferase-positive area, which was negative at day
14, but positive at day 21 and more apparent compared with day 28.
The data suggested that liver-specific modification of the
extracellular matrix under immunodeficient conditions may induce a
more apparent effect. By contrast, direct injection of miRNAs
indicated that the luciferase-positive area was relatively small in
NOD/SCID mice, but was increased in uPA-NOG mice at day 28
(Fig. 2B), suggesting that the
effects of the SeV vector infection were more apparent than the
in vivo transfection of miRNAs, and that the extracellular
structure of the liver and immunosurveillance may alter the
effect.
Oncogenic Kras activation in mice
To investigate the effect of oncogenic Kras
activation in mice, CMV-Cre/Krasmut mice
were produced, which expressed the oncogenic Kras allele
with a point mutation (G12D; Fig.
3A). The luciferase-positive area was detected at days 14, 21
and 28. Another luciferase-positive area, in the right thoracic
region, was also noted. The data suggested that oncogenic
Kras may be involved in accelerating the cellular
reprogramming process. The effect was marginal in miRNA-injected
mice (Fig. 3B), presumably due to
the relatively low gene transfection efficiency compared with SeV
vector injection.
To clarify whether hepatocytes or non-hepatocytes
(such as mesenchymal cells) in the liver were involved in the
effect, Alb-Cre/Krasmut mice were produced
and SeV vector encoding c-Myc, Sox2, Oct3/4
and Klf4 (MSOK) was directly injected (Fig. 3C). The luciferase-positive area was
limited compared with that of
CMV-Cre/Krasmut mice. The data were
similar following the injection of SeV vector encoding Sox2,
Oct3/4 and Klf4 but not c-Myc (SOK;
Fig. 3D), suggesting that
Alb-positive hepatocytes were unlikely to be targets of cellular
reprogramming.
To study the effect of c-Myc in oncogenic
Kras mutation, SeV vector encoding c-Myc (M) was
injected into CMV-Cre/Krasmut mice
(Fig. 3E). The luciferase-positive
area was detected at days 14, 21 and 28, while the injection of SeV
vector encoding c-Myc, Sox2, Oct3/4 and
Klf4 (MSOK) into the control CMV-Cre mice showed a similar
luciferase-positive area (Fig. 3F).
The data suggested that the oncogenic Kras mutation was
compatible with the administration of Sox2,
Oct3/4 and Klf4.
Tumor suppressor p53-deficient mice
Previous studies have shown that the inhibition or
absence of p53 significantly increased the reprogramming
efficiency of somatic cells to reach a pluripotent state (8–10).
Further studies have demonstrated that decreasing the level of the
tumor suppressor p53 protein enables the development of iPS
cells from murine fibroblasts; these iPS cells are capable of
generating germ-line-transmitting chimeric mice, suggesting that
p53 may not be necessary for reprogramming. The inhibition
or absence of p53 significantly increases the reprogramming
efficiency of human somatic cells (8–10).
To assess the effect of this observation in
vivo, Nanog-GFP/Trp53KO mice were
produced and infected with SeV vector encoding c-Myc,
Sox2, Oct3/4 and Klf4 (Fig. 3G). Although the efficiency was low,
it was possible to detect the luciferase-positive area at days 14,
21 and 28. The administration of miRNAs did not produce a
luciferase-positive area, suggesting that the efficiency of this
approach was low or undetectable (Fig.
3H). The data showed that although the effect of p53 was
significant in cellular reprogramming, its effect in direct
reprogramming in the liver was limited.
Discussion
Although there is little knowledge concerning the
mechanism of reprogramming in vivo, it is known that certain
types of gene alterations have significant effects on cellular
reprogramming in vitro. For example, the absence of
p53, which is critical in epithelial tumors, increases the
efficiency of iPS cell generation (8–10). We
previously demonstrated that the reprogramming efficiency was
enhanced by co-transfection of key tumor suppressor gene mutants
(11, data not shown). The results support the theory that mutations
involved in DNA contact may be critical in the efficiency of iPS
generation, and suggest two roles for p53 mutations in
reprogramming. Structural mutations may contribute to the
maintenance of genomic stability, while DNA contact mutations
define the downstream target genes, which may be distinct from
wild-type p53 function. Moreover, in a further reprogramming
study using other cancer cells with gain-of-function mutations,
such as p53R175H and KrasG12D, we
demonstrated the multipotency of differentiation and temporal
suppression of tumorigenicity. However, the cells subsequently
resumed growth in long-term culture (>2 months) and also showed
increased tumorigenicity. After iPS factor-mediated reprogramming,
the expression of ES-like genes, with the exception of activated
endogenous c-Myc, was downregulated in long-term cultures of
iPC cells derived from cholangiocellular carcinoma HuCC-T1 cells
with gain-of-function mutations. This suggests a role for such
oncogenic mutations in the reactivation of a malignant phenotype in
long-term culture, presumably via the accumulation of further
mutations or increased genomic instability during in vitro
culture (11).
The present study showed that the following factors
were involved in the efficiency of the causal effects due to
directly administered reprogramming factors in the liver in
vivo: i) immunodeficiency; ii) extracellular components such as
uPA; and iii) activation of oncogenic Kras in
mesenchymal cells.
Severely immunodeficient NOG mice are utilized as
recipients for human tissue transplantation, which produces
chimeric mice with various types of human tissue. In the present
study, uPA-NOG mice were used. Human hepatocytes injected into
uPA-NOG mice repopulated the recipient livers with human cells, and
the uPA-NOG model has a number of advantages over previously
produced chimeric mouse models of the human liver (5). The immunodeficient condition
facilitates this process by the elimination of transformed cells.
In the present study, uPA-NOG mice showed larger
luciferase-positive areas in comparison with NOD/SCID mice,
suggesting that the extracellular matrix has a critical effect on
reprogramming. Furthermore, the tissues were examined and an
irregular arrangement of hepatocytes was observed, although no
cancerous cells or teratoma were detected, suggesting that the
cells directly affected by reprogramming factors in vivo may
be altered or adapted in tissues with a supportive surrounding
microenvironment.
Oncogenic Kras has a pivotal role in the
carcinogenesis and progression of gastrointestinal tumors, such as
those of the pancreas and colon, and in novel treatment options in
Kras-mutant metastatic colorectal cancer. However,
Kras mutations associated with vinyl chloride exposure and
the observed mutations in liver cancers are relatively rare in
direct DNA-sequencing analyses following microdissection,
suggesting that activation of the oncogenic Kras is unlikely
to have a significant role in liver cancer (12–15).
This is in agreement with the present observation that
Alb-Cre/Krasmut mice, in which the
oncogenic Kras is activated in Alb-positive hepatocytes,
developed a weak luciferase signal. The present data showed a low
frequency of luciferase-positive cells in
Alb-Cre/Krasmut mice compared with
CMV-Cre/Krasmut mice, suggesting that
Alb-negative cells may be targets of in vivo reprogramming.
Activating mutations in the Kras gene are commonly detected
in certain, but not all, types of epithelial cancer. Ray et
al studied a Cre-mediated KrasG12D
mutation, which has the same position of amino acid substitution as
in the present study, during recombination in tissues expressing
cytokeratin 19 to understand the susceptibility of various
epithelial tissues to Kras-induced tumorigenesis (16). The study showed that exposure to
extracellular components promoted
KrasG12D-initiated tumorigenesis, although
environmental exposure did not consistently correlate with tumor
formation, such as that in the small intestine, suggesting the
presence of intrinsic differences in susceptibility to Kras
activation and that tumor susceptibility is not limited to the
epithelial cells but is different depending on the cellular context
(16). To the best of our
knowledge, the present study is the first to demonstrate that the
effect of reprogramming factors in vivo is not dominant in
epithelial cells; instead, the effect is more likely to be
transformed in non-epithelial, mesenchymal cells, demonstrating
that the efficiency at the same dose is dependent on the cell of
origin. However, tumor suppressor p53 deficiency had limited
significance in the present study. Given that the data indicated
Alb-negative cell involvement in direct reprogramming in the liver
in the present system, genomic surveillance of p53 may be
limited in mesenchymal cells. It is reasonable to consider that the
genotype of the p53-deficient mice was heterogeneous for
p53
(p53+/−);
thus, the remaining intact allele may be involved in the
suppression of the transformation in mice with this genetic
background.
The present data indicated that the activation of
oncogenic signals, such as KrasG12D, in
mesenchymal tissues may be critical in the generation of the effect
of directly administered reprogramming factors in the liver in
vivo. This may provide answers to queries regarding
reprogramming, including efficiency and tumorigenicity, to
establish experimental models of organ/tissue/cell-specific
oncogenic gain-of-function with various types of immunodeficient
mice. Therefore, in the future, a reprogramming-based, novel
therapeutic approach may be applied clinically.
Abbreviations:
|
iPS cells
|
induced pluripotent stem cells;
|
|
ES cells
|
embryonic stem cells;
|
|
iPC cells
|
induced multipotent cancer cells;
|
|
5-FU
|
5-fluorouracil;
|
|
SeV
|
Sendai virus;
|
|
miRNA
|
microRNA;
|
|
NOD/SCID mice
|
NOD.CB17-Prkdcscid/J mice;
|
|
NOG mice
|
NOD.Cg-PrkdcscidIl2rgtmSug/Jic
mice;
|
|
uPA
|
urokinase-type plasminogen
activator;
|
|
MMTV
|
mouse mammary tumor virus;
|
|
LTR
|
long terminal repeat
|
Acknowledgements
The present study was partly
supported by a grant from the Core Research for Evolutional Science
and Technology (CREST); a Grant-in-Aid for Scientific Research on
Priority Areas; Grants-in-Aid for Scientific Research from the
Ministry of Education, Culture, Sports, Science and Technology;
Grants-in-Aid for the 3rd Comprehensive 10-Year Strategy for Cancer
Control from the Ministry of Health, Labor and Welfare; and a grant
from the Tokyo Biochemical Research Foundation, Tokyo, Japan.
References
|
1.
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Takahashi K, Tanabe K, Ohnuki M, Narita M,
Ichisaka T, Tomoda K and Yamanaka S: Induction of pluripotent stem
cells from adult human fibroblasts by defined factors. Cell.
131:861–872. 2007. View Article : Google Scholar
|
|
3.
|
Yamanaka S: Elite and stochastic models
for induced pluripotent stem cell generation. Nature. 460:49–52.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Miyoshi N, Ishii H, Nagai K, et al:
Defined factors induce reprogramming of gastrointestinal cancer
cells. Proc Natl Acad Sci USA. 107:40–45. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Suemizu H, Hasegawa M, Kawai K, et al:
Establishment of a humanized model of liver using NOD/Shi-scid
IL2Rgnull mice. Biochem Biophys Res Commun. 377:248–252. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Lamb RA and Kolakofsky D: Paramyxoviridae;
the viruses and their replication. Fields Virology. Knipe DM and
Howley PM: 1. 4th edition. Lippincott Williams & Wilkins;
Philadelphia: pp. 1305–1340. 2001
|
|
7.
|
Fusaki N, Ban H, Nishiyama A, Saeki K and
Hasegawa M: Efficient induction of transgene-free human pluripotent
stem cells using a vector based on Sendai virus, an RNA virus that
does not integrate into the host genome. Proc Jpn Acad Ser B Phys
Biol Sci. 85:348–362. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Zhao Y, Yin X, Qin H, et al: Two
supporting factors greatly improve the efficiency of human iPSC
generation. Cell Stem Cell. 3:475–479. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Kawamura T, Suzuki J, Wang YV, et al:
Linking the p53 tumour suppressor pathway to somatic cell
reprogramming. Nature. 460:1140–1144. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Hong H, Takahashi K, Ichisaka T, et al:
Suppression of induced pluripotent stem cell generation by the
p53–p21 pathway. Nature. 460:1132–1135. 2009.
|
|
11.
|
Nagai K, Ishii H, Miyoshi N, et al:
Long-term culture following ES-like gene-induced reprogramming
elicits an aggressive phenotype in mutated cholangiocellular
carcinoma cells. Biochem Biophys Res Commun. 395:258–263. 2010.
View Article : Google Scholar
|
|
12.
|
Feldmann G, Beaty R, Hruban RH and Maitra
A: Molecular genetics of pancreatic intraepithelial neoplasia. J
Hepatobiliary Pancreat Surg. 14:224–232. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Prenen H, Tejpar S and Van Cutsem E: New
strategies for treatment of KRAS mutant metastatic colorectal
cancer. Clin Cancer Res. 16:2921–2926. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Laurent-Puig P and Zucman Rossi J:
Genetics of hepatocellular tumors. Oncogene. 25:3778–3786. 2006.
View Article : Google Scholar
|
|
15.
|
Tannapfel A, Sommerer F, Benicke M, et al:
Mutations of the BRAF gene in cholangiocarcinoma but not in
hepatocellular carcinoma. Gut. 52:706–712. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Ray KC, Bell KM, Yan J, Gu G, Chung CH,
Washington MK and Means AL: Epithelial tissues have varying degrees
of susceptibility to Kras(G12D)-initiated tumorigenesis in a mouse
model. PLoS One. 6:e167862011. View Article : Google Scholar : PubMed/NCBI
|