Introduction
According to the American Cancer Society, prostate
cancer is currently the second most common cause of cancer-related
mortality among males. An estimated >235,000 new cases of
prostate cancer are expected in the US during 2013 (1). Furthermore, in a recent study by
Arcangeli et al, it is predicted that the increase in birth
rate may correlate with an increased prevalence of prostate cancer
in the United States by 2020 (2).
However, with the establishment of diagnostic markers, including
prostate-specific antigen screening, and recent advances in
molecular imaging, clinicians are able to detect early cancer
proliferation prior to the development of apparent clinical
manifestations and, more significantly, prior to the occurrence of
metastasis. This affords clinicians more time to design the
appropriate and effective treatment procedures. The current
treatment methods for prostate cancer include the administration of
steroidal and non-steroidal anti-androgens, radiation therapy,
chemotherapy, surgery or a combination of these modalities.
Although these options may be successful in controlling the
progression of prostate cancer, they are often associated with
comorbidities that affect urinary and sexual function. Therefore
the aim of prostate cancer research is to develop innovative
treatment options to avoid such complications. Several
characteristics of prostate cancer make it useful to serve as a
model for developing new chemopreventive techniques, including its
high prevalence, heterogeneous presentation, long latency, slow
progression, preneoplastic lesions and tumor marker availability
(2,3).
Males have an equal rate of histological prostate
cancer worldwide, as assessed by volume, grade and number of
malignant foci (4). However,
disease incidence varies widely according to the geographic
location. Western nations have higher rates of mortality compared
with Asian countries, including India, China and Japan. More
notably, migrating populations from low-risk areas (Asian
countries) to high-risk areas (Western countries) also have an
increased risk of developing prostate cancer. Since genetic
predisposition accounts for only 5–10% of cases, as cited by the
American Cancer Society (1), the
uniting theme in the literature has become identifying the
environmental factors that promote or inhibit the development of
prostate cancer.
Foods or part of foods with medicinal value, termed
nutraceuticals, which are prepared and consumed variably across
cultures, may be active in the prevention and treatment of
diseases, including prostate cancer. Curcumin, a non-nutritive
yellow pigment derived from the rhizome of Curcumin longa
(turmeric), has received attention as an established nutraceutical
that is capable of anticancer activity (5). Turmeric contains three principal
components, curcumin, demethoxycurcumin and bisdemethoxycurcumin,
of which curcumin is the most abundant and potent (6–9). The
concurrence of a high consumption of turmeric in Asian countries
and a low incidence of prostate cancer suggest its role in
chemoprevention (10). Curcumin and
a number of its derivatives have been identified to exhibit
anti-inflammatory, antioxidative and anticarcinogenic properties
(11). As the compound does not
exhibit toxic, genotoxic or teratogenic properties, curcumin has
been selected for several clinical trials to be used as a possible
treatment for human cancers (3,5,11).
Curcumin has been shown to diminish the proliferation of
androgen-dependent and androgen-independent prostate cancer cell
lines (12). Furthermore, studies
have revealed a wide array of therapeutic activities against
multiple myeloma, pancreatic cancer, myelodysplastic syndromes,
colon cancer, psoriasis, Alzheimer’s disease and others (13). The pro-apototic, antioxidant and
anti-inflammatory properties of curcumin are implicated in its
anticancer activity, yet the mechanism of action of curcumin
remains unknown (8). Curcumin is a
highly pleiotropic molecule with multiple mechanisms by which it
may mediate chemotherapy and chemopreventive effects on cancer,
while remaining safe with little or no side effects. This dietary
compound has been shown to inhibit several cell signaling pathways,
including nuclear factor (NF)-κB, activating protein-1, tumor
necrosis factor and metastatic and angiogenic pathways. The
compound also inhibits certain enzymes, including cyclooxygenase
(COX)-2 and matrix metalloproteinases (MMPs) (9,13,14).
The present study randomly identified several enzymes that are
essential in carcinogenesis and are also targeted by curcumin,
aldo-keto reductase family 1 member B10 (AKR1B10),
serine/threonine-protein kinase (mTOR), protein kinase C (PKC),
MMP-9, COX-1 and epidermal growth factor receptor (EGFR), to gain
further insight into the mechanism of action (5,7,13,15–17).
Curcumin has a poor systemic bioavailability as it
is not able to reach and sustain the appropriate levels at the site
of action due to its high metabolic instability and poor aqueous
solubility (18,19). The present study aimed to identify
the anticancer activity of curcumin-like compounds with potentially
greater bioavailability, and speculate the protein targets of these
compounds that are implicated in the mechanism of action. Novel
curcumin-like compounds, E21cH and Q012095H, with greater water
solubility were tested. Molecular modeling methods were used to
identify the targets of curcumin and curcumin-like compounds by
comparing them with other anticancer drugs (Q012138 and Q012169AT),
which were used as a controls.
Materials and methods
Compounds
The small molecular chemicals with anticancer
activities were obtained from PharmaIP, LLC (Greenwich CT, USA).
Curcumin
[(1E,4Z,6E)-5-hydroxy-1,7-bis(4-hydroxy-3-methoxy-phenyl)hepta-1,4,6-trien-3-one];
Q0121138
[4-[[(1S)-1-(benzothiophen-2-ylmethyl)-2-ethoxy-2-oxo-ethyl]carbamoyl]phenyl]
methylammonium; Q012095H (1E,4Z,6E)-1,7-bis[5-(2-dimethylaminoethyl
sulfanyl)-2-thienyl]-5-hydroxy-hepta-1,4,6-trien-3-one; Q012138
[4-[[(1S)-1-(benzothiophen-2-ylmethyl)-2-ethoxy-2-oxo-ethyl]carbamoyl]
phenyl] methylammonium; and Q012169AT
(N-ethyl-5-hydroxy-2-phenoxy-benzamide; Fig. 1). All the compounds were dissolved
in dimethyl sulfoxide (DMSO) 2.5 mg/ml and stored at −20°C until
they were used.
Cell culture and clonal assay
The DU-145 human prostate cancer cell line was grown
in RPMI-1640 medium supplemented with 10% fetal bovine serum
(Atlanta Biologicals, Lawrenceville, GA, USA) and 100 U/ml
penicillin, 100 μg/ml streptomycin (Sigma-Aldrich, St. Louis, MO,
USA). A total of ~50 or ~100 viable DU-145 cells (Trypan blue
viability assay, two separate trials) were plated in 0.5 or 1 ml of
complete medium onto 12 or 24-well tissue culture dishes. The cells
were allowed to attach for 48 h. The cells were then treated for 4
h with 1.2-, 2.5-, 5.0- or 10-μl allotments of DMSO, curcumin,
E21cH, Q0121138, Q012095H or Q012169AT dissolved in 1 mg/ml DMSO.
The surviving cells were incubated for nine days to allow colony
formation and then rinsed with 10% saline, fixed with 100% methanol
and stained using Giemsa stain. The colony counts were performed
under ×10 magnification (Stereomaster, Thermo Fisher Scientific
Inc., Waltham, MA, USA). The experiments were repeated in
triplicate to determine the anticancer activity.
Molecular modeling
Two-dimensional structures (2D) of small molecular
chemicals were created by AccelrysDraw v. 4.0 (Accelrys, Inc., San
Diego, CA, USA) in an SKC format. The 2D structures were converted
into three dimensional and PDB format files using a web-based
program (http://www.molecular-networks.com/products). Docking
of the potential inhibitors to the proteins was performed using
VINA Autodock (Molecular Graphics Lab, The Scripps Research
Institute, La Jolla, CA, USA) (20). The protein structures were
downloaded from http://www.rcsb.org/pdb/home/home.do as: 1zua, AKR1B10
(21); 3oaw, mTOR (22); 1yrk, PKC (23); 2ovx, MMP-9 (24); 3ln1, COX-2 (25) and 2itx, EGFR (26). A search box was set up with
following parameters: AKR1B10 human NADPH-dependent aldo-keto
reductase (center: x, −29; y, 22; z, 0.1; size: x, 50; y, 50; z,
50), mTOR (center: x, −17.5; y, −11; z, −12; size: x, 50; y, 40; z,
46). PKC (center: x, 25; y, 40; z, 31; size: x,40; y, 44; z, 74),
MMP-9 (center: x, 27; y, 6; z, 51; size: x, 40; y, 56; z, 40),
COX-2 (center: x, 32; y, −22; z, −16; size: x, 40; y, 40; z, 40)
and EGFR (center: x, −47; y, −2; z, −22; size: x, 50; y, 40; z,
50). The inhibitors that were present in the PDB structures were
used to determine the center of the search and later removed from
structure. The small molecules were kept flexible by allowing
rotation around the single bonds. By default, VINA Autodock
analyzes eight various protein/inhibitor complexes (conformers) and
the one with the lowest free energy is considered the most
probable. Free energy is converted to Ki using the
following formula (20,27–30):
Ki = exp [ΔG / (R × T)], where ΔG is Gibbs free energy
change, R is the gas constant and T is the absolute temperature.
The final analyses of structures that were generated by Autodock
and the generation of the figures was performed using PyMOL v. 1.4
(Schrödinger, München, Germany) (31,32).
Results
In vitro anticancer activity
The present study tested the anticancer activities
of >30 curcuminoids, thiotryptophanes and 4-phenoxyphenol
derivatives. In the clonal assay, Q012095H demonstrated the
strongest anticancer activity, followed by Q012138 and Q012165H.
E21cH and curcumin activity were comparable with each other but
lower than the three others (Fig.
2). The highest concentrations of Q012095H and Q012138 showed a
complete inhibition of cancer cell growth.
Molecular modeling
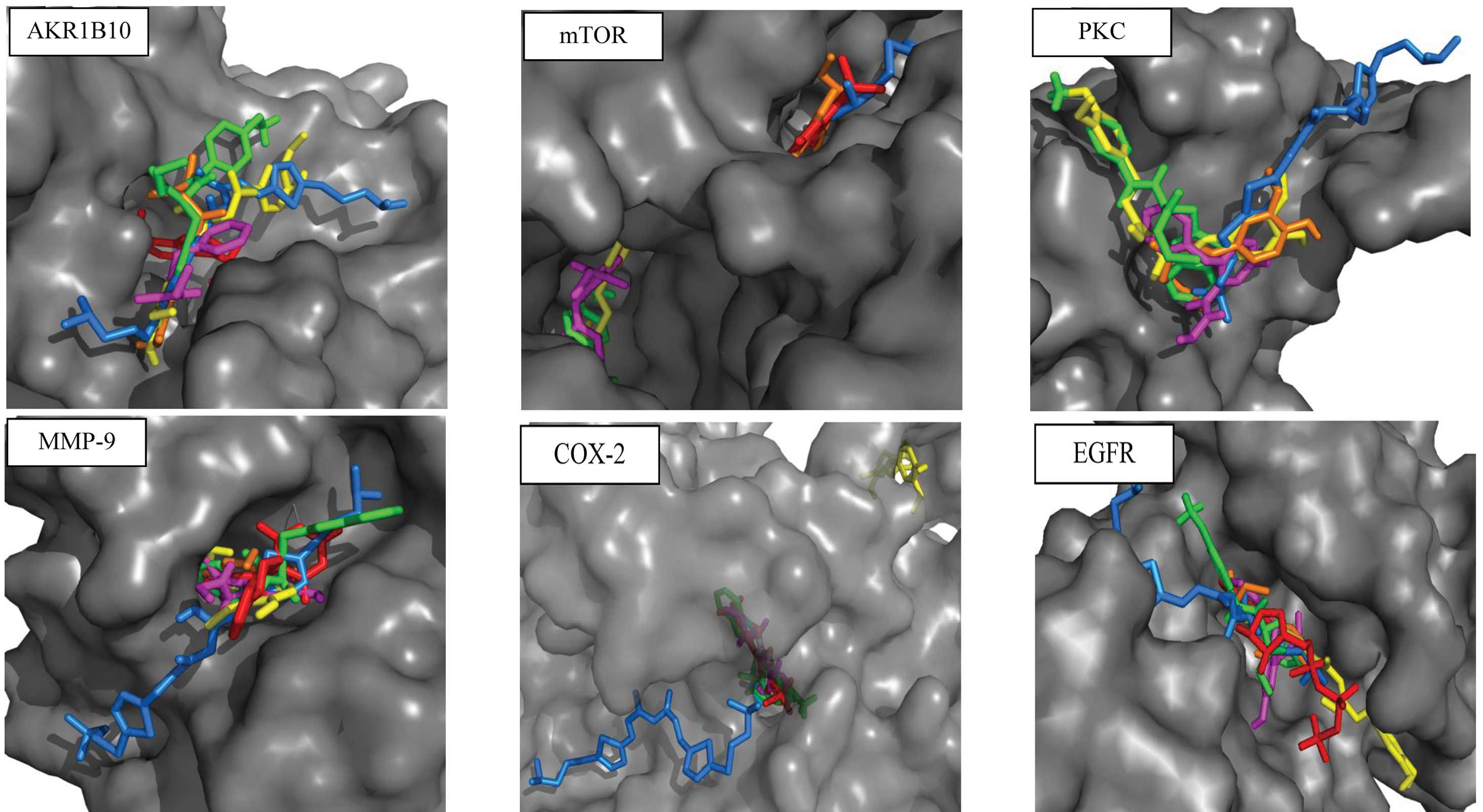
The results of the docking are illustrated in
Fig. 3 and the calculated
Ki values are provided in Table I. All the compounds that were tested
contained an aldo-keto moiety. One of the human enzymes that was
tested in the in silico experiment was AKR1B10, an
NADPH-dependent aldo-keto reductase that reduces a variety of
aldehydes and ketones. AKR1B10 has been reported to be upregulated
in number of cancers. Additionally, AKR1B10-gene silencing results
in the inhibition of colorectal cancer cell growth, suggesting that
AKR1B10 regulates cell proliferation (33). It has been proposed that AKR1B10
controls retinoic acid signaling and impacts the carcinogenesis
process. Also, tolrestat, which efficiently inhibits AKR1B10, is
suggested to have a potential application in cancer control
(34,35). Thus, the investigated chemicals were
tested for the capacity to bind to the active site of this enzyme.
All the investigated chemicals were found to bind near the active
site and functionally block its access. The calculated
Ki was in μM levels for all the tested chemicals,
indicating their relative strength of affinity (Table I).
 | Table ICalculated Ki for the
various protein and inhibitor complexes. |
Table I
Calculated Ki for the
various protein and inhibitor complexes.
| Compound | AKR1B10
(kcal/M)/Ki(M) | mTOR
(kcal/M)/Ki(M) | PKC
(kcal/M)/Ki(M) | MMP-9
(kcal/M)/Ki(M) | COX-2
(kcal/M)/Ki(M) | EGFR
(kcal/M)/Ki(M) |
|---|
| Curcumin |
−7.6/2.8×10−6 |
−6.9/9.0×10−6 NA |
−6.2/2.9×10−5 |
−9.1/2.2×10−7 |
−7.3/4.6×10−6 NA |
−6.7/1.3×10−5 |
| E21cH |
−7.4/3.9×10−6 |
−5.8/5.7×10−5 |
−4.9/2.6×10−4 |
−7.9/1.7×10−6 |
−7.6/2.8×10−6 |
−6.3/2.5×10−5 |
| Q012095H |
−5.8/5.7×10−5 |
−4.6/4.3×10−4 |
−4.5/5.1×10−4 |
−6.2/2.9×10−5 |
−6.1/3.5×10−5 NA |
−5.2/1.6×10−4 |
| Q012138 |
−7.1/6.5×10−6 |
−6.2/2.9×10−5 NA |
−5.8/5.7×10−5 |
−8.9/3.1×10−7 |
−9.2/1.9×10−7 |
−6.2/2.9×10−5 |
| Q012169AT |
−6.2/2.9×10−5 |
−6.9/9.0×10−6 NA |
−4.9/2.6×10−4 |
−7.8/2.0×10−6 |
−8.8/3.7×10−7 |
−6.6/1.5×10−5 |
Discussion
mTOR, a serine/threonine protein kinase, regulates
cell growth, cell proliferation, cell motility, cell survival,
protein synthesis and transcription (36). The inhibition of mTOR mediates the
antiproliferative effects of curcumin in numerous human and
non-human cell lines (15,37,38).
In addition, curcumin has been reported to be able to dissociate
the raptor subunit from mTOR as well as inhibit mTORC1 activity
(15). Liu et al designed
several idopyrimidinone (1)
4-methylpteridinones that bind to a small pocket within the mTOR
binding site. This inhibitor in the protein structure was used as a
center of search (22). In the
present study, the molecular modeling revealed that all the
compounds that were tested had a relatively low affinity and bound
in various locations outside the active site of mTOR. Only E21cH
and Q012095H were able to bind in proximity to where the inhibitor
was localized. mTOR may be an unlikely target of the chemicals that
were tested. By contrast, Lin et al stated that PKC and mTOR
were the major upstream molecular targets for curcumin (39). A possible explanation is that the
products of curcumin degradation act on mTOR instead of curcumin
itself (7,8,13,14,18,19,40).
Curcumin is an inhibitor of PKC. Consequently,
curcumin inhibits the activation of NF-κB and the expression of
oncogenes, including c-jun, c-fos, c-myc, NF-κB-inducing kinase
(NIK), mitogen-activated protein kinases, ERK, ELK,
phosphoinositide 3-kinase, Akt, cyclin-dependent kinases and
inducible nitric oxide synthase (39). Conboy et al performed
molecular modeling and identified that curcumin was able to dock
effectively on PKC. However, curcumin did not directly inhibit PKC
activity, but rather increased its degradation (41). The calculations in the present study
revealed all tested compounds bind to PKC in essentially the same
place, but with low affinity.
Traditionally, MMP-9 was associated with tumor
angiogenesis and metastasis by lysing proteins of connective tissue
(42,43). However, curcumin has been reported
to protect MMP-9 from proteolytic degradation (44). MMP-9 plays a critical function in
normal and pathological angiogenesis and/or controlling the
biological activity of growth factors, cytokines and chemokines
(45). Proteolytic enzymes that
stimulate angiogenesis and metastasis frequently show other
functions in carcinogenesis in addition to their traditional roles
(46–48). Ravindranath et al reported
that blocking the activity of MMP-9 may arrest cell growth and
proliferation in addition to the inhibition of invasion and
angiogenesis (49). All the
chemicals that were tested were observed to bind to the active site
of MMP-9 in the proximity of the MMP-9 inhibitor (5-(4-phenoxy
phenyl)-5-(4-pyrimidin-2-ylpiperazin-1-yl)pyrimidine-2,4,6(2H,3H)-trione)
with high affinity.
Curcumin possesses anti-inflammatory activity and is
a potent inhibitor of reactive oxygen generating enzymes, including
COX (15,37). Curcumin itself is a potent scavenger
of free radicals and the inhibition of COX potentiates its
anticancer activity (50–52). Although the specific regulation of
COX-2 by curcumin is not fully understood, the evidence suggests
that curcumin regulates COX-2 at the transcriptional and the
post-translational levels (17,53).
In the present study, molecular modeling revealed that E21cH,
Q012138 and Q012169AT bind to COX-2 with a high affinity, deep in
the tunnel of the active site where celecoxib
(4-[5-(4-methylphenyl)-3-(trifluoromethyl)-1h-pyrazol-1-yl]benzenesulfonamide)
is bound (25). However, Q012095H
and curcumin were observed to bind outside of this site. This is in
contrast with studies that state that curcumin inhibits COX
(16,54–56). A
possible explanation is that the non-enzymatic degradation of
curcumin occurs, resulting in degradation products that are formed
through cleavage of the heptadienone chain that connects the
phenolic rings (57). Dong et
al have shown that COX acts as a dimer, where one monomer with
a heme moiety is active and the other is apo, which acts as the
allosteric site, controlling activity of the active monomer
(58). Thus, the docking scenario
in its simplification in silico may not reflect the true
situation in vivo, which may be more complex.
Xu et al reported that cyclohexanone analogs
that are designed based on the curcumin structure are potential
EGFR inhibitors and exhibit antiproliferative activity in human
tumor cell lines. Cyclohexanone analogs fit in the active site of
EGFR, as shown by molecular docking (59). This was confirmed by the
experimental modeling in the present study. All the investigated
compounds also bound the EGFR active site, but with low
affinities.
Based on the results of the present study, AKR1B10
and MMP-9 have been shown to be the most likely targets of curcumin
and curcumin-like derivatives. Curcumin and the investigated
curcumin-like compounds bound the other proteins that were tested
outside of the active site or with low affinities.
Acknowledgements
This study was supported in part by a grant from
Frank Stranahan Endowed Chair.
References
|
1
|
American Cancer Society. Cancer Facts
& Figures. 2013, http://www.cancer.org/research/cancerfactsstatistics/index.
Accessed March 24, 2013
|
|
2
|
Arcangeli S, Pinzi V and Arcangeli G:
Epidemiology of prostate cancer and treatment remarks. World J
Radiol. 4:241–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cimino S, Sortino G, Favilla V, et al:
Polyphenols: key issues involved in chemoprevention of prostate
cancer. Oxid Med Cell Longev. 2012:6329592012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fleshner N and Zlotta AR: Prostate cancer
prevention: past, present, and future. Cancer. 110:1889–1899. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Agrawal DK and Mishra PK: Curcumin and its
analogues: potential anticancer agents. Med Res Rev. 30:818–860.
2010.PubMed/NCBI
|
|
6
|
Da-Lozzo EJ, Moledo RC, Faraco CD,
Ortolani-Machado CF, Bresolin TM and Silveira JL:
Curcumin/xanthan-galactomannan hydrogels: rheological analysis and
biocompatibility. Carbohydr Polym. 93:279–284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hsu CH and Cheng AL: Clinical studies with
curcumin. Adv Exp Med Biol. 595:471–480. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park W, Amin AR, Chen ZG and Shin DM: New
perspectives of curcumin in cancer prevention. Cancer Prev Res
(Phila). 6:387–400. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Strimpakos AS and Sharma RA: Curcumin:
preventive and therapeutic properties in laboratory studies and
clinical trials. Antioxid Redox Signal. 10:511–545. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin L, Shi Q, Nyarko AK, et al: Antitumor
agents. 250. Design and synthesis of new curcumin analogues as
potential anti-prostate cancer agents. J Med Chem. 49:3963–3972.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mimeault M and Batra SK: Potential
applications of curcumin and its novel synthetic analogs and
nanotechnology-based formulations in cancer prevention and therapy.
Chin Med. 6:312011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dorai T, Gehani N and Katz A: Therapeutic
potential of curcumin in human prostate cancer-I. curcumin induces
apoptosis in both androgen-dependent and androgen-independent
prostate cancer cells. Prostate Cancer Prostatic Dis. 3:84–93.
2000. View Article : Google Scholar
|
|
13
|
Hatcher H, Planalp R, Cho J, Torti FM and
Torti SV: Curcumin: from ancient medicine to current clinical
trials. Cell Mol Life Sci. 65:1631–1652. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shehzad A, Wahid F and Lee YS: Curcumin in
cancer chemoprevention: molecular targets, pharmacokinetics,
bioavailability, and clinical trials. Arch Pharm (Weinheim).
343:489–499. 2010. View Article : Google Scholar
|
|
15
|
Beevers CS, Chen L, Liu L, Luo Y, Webster
NJ and Huang S: Curcumin disrupts the Mammalian target of
rapamycin-raptor complex. Cancer Res. 69:1000–1008. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cuendet M and Pezzuto JM: The role of
cyclooxygenase and lipoxygenase in cancer chemoprevention. Drug
Metabol Drug Interact. 17:109–157. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rao CV: Regulation of COX and LOX by
curcumin. Adv Exp Med Biol. 595:213–226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Verderio P, Bonetti P, Colombo M, Pandolfi
L and Prosperi D: Intracellular drug release from curcumin-loaded
PLGA nanoparticlesinduces G2/M block in breast cancer cells.
Biomacromolecules. 14:672–682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zlotogorski A, Dayan A, Dayan D, Chaushu
G, Salo T and Vered M: Nutraceuticals as new treatment approaches
for oral cancer - I: Curcumin. Oral Oncol. 49:187–191. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trott O and Olson AJ: AutoDock Vina:
improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading. J Comput
Chem. 31:455–461. 2010.PubMed/NCBI
|
|
21
|
Gallego O, Ruiz FX, Ardèvol A, et al:
Structural basis for the high all-trans-retinaldehyde reductase
activity of the tumor marker AKR1B10. Proc Natl Acad Sci USA.
104:20764–20769. 2007. View Article : Google Scholar
|
|
22
|
Liu KK, Bagrodia S, Bailey S, et al:
4-methylpteridinones as orally active and selective PI3K/mTOR dual
inhibitors. Bioorg Med Chem Lett. 20:6096–6099. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Benes CH, Wu N, Elia AE, Dharia T, Cantley
LC and Soltoff SP: The C2 domain of PKCdelta is a phosphotyrosine
binding domain. Cell. 121:271–280. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tochowicz A, Maskos K, Huber R, et al:
Crystal structures of MMP-9 complexes with five inhibitors:
contribution of the flexible Arg424 side-chain to selectivity. J
Mol Biol. 371:989–1006. 2007. View Article : Google Scholar
|
|
25
|
Wang JL, Limburg D, Graneto MJ, et al: The
novel benzopyran class of selective cyclooxygenase-2 inhibitors.
Part 2: the second clinical candidate having a shorter and
favorable human half-life. Bioorg Med Chem Lett. 20:7159–7163.
2010. View Article : Google Scholar
|
|
26
|
Yun CH, Boggon TJ, Li Y, et al: Structures
of lung cancer-derived EGFR mutants and inhibitor complexes:
mechanism of activation and insights into differential inhibitor
sensitivity. Cancer Cell. 11:217–227. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bikádi Z, Hazai E, Zsila F and Lockwood
SF: Molecular modeling of non-covalent binding of homochiral
(3S,3′S)-astaxanthin to matrix metalloproteinase-13 (MMP-13).
Bioorg Med Chem. 14:5451–5458. 2006.PubMed/NCBI
|
|
28
|
D’hoedt D and Bertrand D: Nicotinic
acetylcholine receptors: an overview on drug discovery. Expert Opin
Ther Targets. 13:395–411. 2009.
|
|
29
|
Hetényi C and van der Spoel D: Blind
docking of drug-sized compounds to proteins with up to a thousand
residues. FEBS Lett. 580:1447–1450. 2006.PubMed/NCBI
|
|
30
|
Iorga B, Herlem D, Barré E and Guillou C:
Acetylcholine nicotinic receptors: finding the putative binding
site of allosteric modulators using the “blind docking” approach. J
Mol Model. 12:366–372. 2006.PubMed/NCBI
|
|
31
|
Seeliger D and de Groot BL: Ligand docking
and binding site analysis with PyMOL and Autodock/Vina. J Computb
Aided Mol Des. 24:417–422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
DeLano WL: The case for open-source
software in drug discovery. Drug Discov Today. 10:213–217.
2005.PubMed/NCBI
|
|
33
|
Matsunaga T, Endo S, Soda M, et al: Potent
and selective inhibition of the tumor marker AKR1B10 by
bisdemethoxycurcumin: probing the active site of the enzyme with
molecular modeling and site-directed mutagenesis. Biochem Biophys
Res Commun. 389:128–132. 2009. View Article : Google Scholar
|
|
34
|
Balendiran GK: Fibrates in the chemical
action of daunorubicin. Curr Cancer Drug Targets. 9:366–369. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xuyu Z, Ruilan Y, Jun M, Duan-Fang L and
Deliang C: AKR1B10: A potential target for cancer therapy.
Bioscience Hypotheses. 2:31–33. 2009. View Article : Google Scholar
|
|
36
|
Tokunaga C, Yoshino K and Yonezawa K: mTOR
integrates amino acid- and energy-sensing pathways. Biochem Biophys
Res Commun. 313:443–446. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Beevers CS, Zhou H and Huang S: Hitting
the golden TORget: curcumin’s effects on mTOR signaling. Anticancer
Agents Med Chem. Dec;2012.(epub ahead of print).
|
|
38
|
Deters M, Hütten H and Kaever V:
Synergistic immunosuppressive effects of the mTOR inhibitor
sirolimus and the phytochemical curcumin. Phytomedicine.
20:120–123. 2013. View Article : Google Scholar
|
|
39
|
Lin JK: Molecular targets of curcumin. Adv
Exp Med Biol. 595:227–243. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lou JR, Zhang XX, Zheng J and Ding WQ:
Transient metals enhance cytotoxicity of curcumin: potential
involvement of the NF-kappaB and mTOR signaling pathways.
Anticancer Res. 30:3249–3255. 2010.
|
|
41
|
Conboy L, Foley AG, O’Boyle NM, et al:
Curcumin-induced degradation of PKC delta is associated with
enhanced dentate NCAM PSA expression and spatial learning in adult
and aged Wistar rats. Biochem Pharmacol. 77:1254–1265. 2009.
View Article : Google Scholar
|
|
42
|
Tartour E, Pere H, Maillere B, et al:
Angiogenesis and immunity: a bidirectional link potentially
relevant for the monitoring of antiangiogenic therapy and the
development of novel therapeutic combination with immunotherapy.
Cancer Metastasis Rev. 30:83–95. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
van Kempen LC and Coussens LM: MMP9
potentiates pulmonary metastasis formation. Cancer Cell. 2:251–252.
2002.PubMed/NCBI
|
|
44
|
Bolignano D, Donato V, Lacquaniti A, et
al: Neutrophil gelatinase-associated lipocalin (NGAL) in human
neoplasias: a new protein enters the scene. Cancer Lett. 288:10–16.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Masson V, de la Ballina LR, Munaut C, et
al: Contribution of host MMP-2 and MMP-9 to promote tumor
vascularization and invasion of malignant keratinocytes. FASEB J.
19:234–236. 2005.PubMed/NCBI
|
|
46
|
Jankun J, Aleem AM, Selman SH, Basrur V
and Skrzypczak-Jankun E: VLHL plasminogen activator inhibitor
spontaneously reactivates from the latent to active form. Int J Mol
Med. 23:57–63. 2009.PubMed/NCBI
|
|
47
|
Jankun J, Aleem AM, Specht Z, et al: PAI-1
induces cell detachment, downregulates nucleophosmin (B23) and
fortilin (TCTP) in LnCAP prostate cancer cells. Int J Mol Med.
20:11–20. 2007.
|
|
48
|
Jankun J and Skrzypczak-Jankun E: Yin and
yang of the plasminogen activator inhibitor. Pol Arch Med Wewn.
119:410–417. 2009.PubMed/NCBI
|
|
49
|
Ravindranath MH, Muthugounder S, Presser N
and Viswanathan S: Anticancer therapeutic potential of soy
isoflavone, genistein. Adv Exp Med Biol. 546:121–165. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fujisawa S, Atsumi T, Ishihara M and
Kadoma Y: Cytotoxicity, ROS-generation activity and
radical-scavenging activity of curcumin and related compounds.
Anticancer Res. 24:563–569. 2004.PubMed/NCBI
|
|
51
|
Youssef KM, El-Sherbeny MA, El-Shafie FS,
Farag HA, Al-Deeb OA and Awadalla SA: Synthesis of curcumin
analogues as potential antioxidant, cancer chemopreventive agents.
Arch Pharm (Weinheim). 337:42–54. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zingg JM, Hasan ST and Meydani M:
Molecular mechanisms of hypolipidemic effects of curcumin.
Biofactors. 39:101–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Menon VP and Sudheer AR: Antioxidant and
anti-inflammatory properties of curcumin. Adv Exp Med Biol.
595:105–125. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gafner S, Lee SK, Cuendet M, et al:
Biologic evaluation of curcumin and structural derivatives in
cancer chemoprevention model systems. Phytochemistry. 65:2849–2859.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Guo LY, Cai XF, Lee JJ, et al: Comparison
of suppressive effects of demethoxycurcumin and
bisdemethoxycurcumin on expressions of inflammatory mediators in
vitro and in vivo. Arch Pharm Res. 31:490–496. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Handler N, Jaeger W, Puschacher H, Leisser
K and Erker T: Synthesis of novel curcumin analogues and their
evaluation as selective cyclooxygenase-1 (COX-1) inhibitors. Chem
Pharm Bull (Tokyo). 55:64–71. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Griesser M, Pistis V, Suzuki T, Tejera N,
Pratt DA and Schneider C: Autoxidative and cyclooxygenase-2
catalyzed transformation of the dietary chemopreventive agent
curcumin. J Biol Chem. 286:1114–1124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dong L, Vecchio AJ, Sharma NP, Jurban BJ,
Malkowski MG and Smith WL: Human cyclooxygenase-2 is a sequence
homodimer that functions as a conformational heterodimer. J Biol
Chem. 286:19035–19046. 2011. View Article : Google Scholar
|
|
59
|
Xu YY, Cao Y, Ma H, Li HQ and Ao GZ:
Design, synthesis and molecular docking of α,β-unsaturated
cyclohexanone analogous of curcumin as potent EGFR inhibitors with
antiproliferative activity. Bioorg Med Chem. 21:388–394. 2013.
|
|
60
|
Kolev TM, Velcheva EA, Stamboliyska BA and
Spiteller M: DFT and experimental studies of the structure and
vibrational spectra of curcumin. Int J Quantum Chem. 102:1069–1079.
2005. View Article : Google Scholar
|