Introduction
Colorectal cancer (CRC) is the second leading cause
of cancer-related mortality and the third most prevalent cancer in
western countries (1–3). CRC is curable in its early stages,
whereas in the advanced stages, the survival time is <30 months.
Early diagnosis and treatment is therefore of high importance
(4). The current routine method of
screening is by colonoscopy. The use of colonoscopy, however, is
not an ideal method since it is an invasive technique and the use
of colonoscopy screening could be limited in high-risk populations,
in particular in low-income patients (5,6). The
fecal occult blood test for CRC is an additional screening method
that is currently performed, however this test is limited due to
its low sensitivity (7). Studies
have therefore been performed to improve the diagnostic biomarkers
available for detection of CRC (8,9).
Abnormal DNA methylation is an early event that
occurs during tumorigenesis, and numerous abnormally
hypermethylated genes have been identified in cases of CRC
(10–12). Gene methylation status could
therefore be used as a molecular diagnostic indicator (13). DNA from CRC cells can be readily
obtained from feces samples, which contain both normal and
CRC-shedded colorectal epithelial cells, from which DNA can be
extracted (14). In addition, the
alkaline environment of the intestine is conducive to the
preservation of DNA (15). NDRG4 is
a member of the NDRG gene family and is a known tumor suppressor
gene (16,17). A previous study suggested that the
NDRG4 gene is present in CRC tumor tissues and bodily fluids with
high stability and repeatability (16).
The present study analyzed the methylated NDRG4 gene
expression in 87 patients with CRC by nested methylation-specific
polymerase chain reaction (n-MSP) combined with denaturing
high-performance liquid chromatography (DHPLC). The samples were
collected from carcinoma and paracarcinoma tissues, blood, urine
and feces of patients with CRC. The sensitivity and specificity of
methylated NDRG4 gene detection in these samples were compared with
those in healthy controls, and assessed for use as a potential
candidate biomarker for the diagnosis of CRC.
Materials and methods
Patient enrollment
The present study was approved by the Institutional
Review Boards of the General Hospital of PLA (Beijing, China). The
CRC and pericarcinous tissues were obtained from surgeries
undertaken between June 2010 and August 2011. All the samples were
confirmed by pathology. All patients provided written informed
consent, and the patients did not receive chemotherapy or
radiotherapy before the surgical procedure. Stool and urine samples
were collected prior to the surgery, from which genomic DNA was
immediately extracted and then stored at −20°C. Cancer and
pericarcinous tissue specimens were stored at −80°C for further
use.
Following screening, a total of 87 CRC patients were
recruited in the study, of which 56 cases were colon cancers and 31
cases were rectal cancers. In the study group, 52 were males and 35
were females, with an age range between 38 and 73 years, and a
median age of 56. A cohort of 16 age-matched healthy subjects were
recruited as a control, of which 9 were males and 6 were females,
with an age range between 40 and 74 years, with a median age of
55.6.
DNA isolation
Homogenate-tissue DNA was extracted using the TGuide
Tissue Genomic DNA Extraction kit [Tiangen Biotech (Beijing) Co.,
Ltd, Beijing, China]. DNA from fecal samples was extracted using a
Stool DNA Extraction kit (Bioneer Corporation, Daedeok-gu, Republic
of Korea). Genomic DNA was extracted from total blood, using a
TGuide Blood Genomic DNA Extraction kit [Tiangen Biotech (Beijing)
Co., Ltd.], according to the manufacturer’s instructions.
Bisulfite modification of genomic
DNA
Following bisulfite modification, the unmethylated
cytosine in genomic DNA will be converted to uracil, whereas
methylated cytosine will not be converted. Tissue genomic DNA
methylation was performed using a Wizard DNA Clean-up system
(Promega Corporation, Madison, WI, USA) according to the
manufacturer’s instructions. Stool and blood genomic DNA
methylation were performed using an EZ DNA Methylation TM-Direct
kit (Zymo Research Corporation, Irvine, CA, USA), according to the
manufacturer’s instructions.
n-MSP
The n-MSP was performed to identify the expression
of NDRG4 genes from blood, urine, stool and tissue. The primer
sequences and polymerase chain reaction (PCR) conditions for the
n-MSP approach have been previously described (18,19).
The primer sequences, annealing temperatures, and PCR sizes are
listed in Table I. Methylated DNA
from normal human lymphocytes, which was treated by SssI
methylase (New England BioLabs, Ipswich, MA, USA) was prepared as
the positive control for methylated (M) amplifications, and DNA
from untreated lymphocytes served as a negative control for
unmethylated (U) amplifications.
 | Table IPrimer sequences, annealing
temperatures, and product sizes used for nested
methylation-specific polymerase chain reaction, to analyze the
methylation status of the NDRG4 promoter. |
Table I
Primer sequences, annealing
temperatures, and product sizes used for nested
methylation-specific polymerase chain reaction, to analyze the
methylation status of the NDRG4 promoter.
| Primer sequence | Temperature, °C | Product size, bp |
|---|
| Outer |
NDRG4-F:
5′-GGTTYGTTYGGGATTAGTTTTAGG-3′
NDRG4-R: 5′-CRAACAACCAAAAACCCCTC-3′ | 56 | 257 |
| Inner |
| Methylated |
NDRG4-UF:
5′-GATTAGTTTTAGGTTTGGTATTGTTTTGT-3′
NDRG4-UR: 5′-AAAACCAAACTAAAAACAATACACCA-3′ | 66 | 100 |
| Un-methylated |
NDRG4-MF:
5′-TTTAGGTTCGGTATCGTTTCGC-3′
NDRG4-MR: 5′-CGAACTAAAAACGATACGCCG-3′ | 66 | 88 |
The n-MSP of NDRG4 was performed in a 25-μl reaction
volume, with 0.5 μl Taq polymerase (Invitrogen Life Technologies,
Carlsbad, CA, USA) and 2 μl DNA template. The NDRG4 gene cycling
conditions for the first round were as follows: 95°C for 15 min; 25
cycles of 95°C for 30 sec, 56°C for 30 sec, and 72°C for 30 sec;
final extension at 72°C for 7 min. The cycling conditions for the
second round were as follows: Preheating at 95°C for 15 min, 35
cycles of 95°C for 30 sec, 66°C for 30 sec, and 72°C for 30 sec;
final extension at 72°C for 7 min. MSP products were analyzed by 2%
polyacrylamide gel electrophoresis, and a product size of100 bp was
expected for the NDRG4 gene.
Statistical analysis
A χ2 test was used for methylation
detection rate comparison between each sample of NDRG4. P<0.05
was considered to indicate a statistically significant difference.
SPSS 13.0 software was used for all statistical analyses (SPSS,
Inc., Chicago, IL, USA).
Results
Rate of NDRG4 methylation
nMSP detection was successfully used to assess for
methylated NDRG4 in carcinoma and paracarcinoma tissues, feces,
urine and blood of the 84 CRC cases (Fig. 1). The positive rate of methylated
NDRG4 gene expression was 81% (68/84) in carcinoma tissues, while
the positive rate was only 8.3% (7/84) in paracarcinoma tissues.
There was a significant difference in the levels of methylated
NDRG4 in carcinoma as compared with the paracarcinoma tissues
(P<0.01). The sensitivity and specificity of NDRG4 gene
detection in the diagnosis of CRC was 81 (68/84) and 91.7% (77/84)
in tissues, respectively.
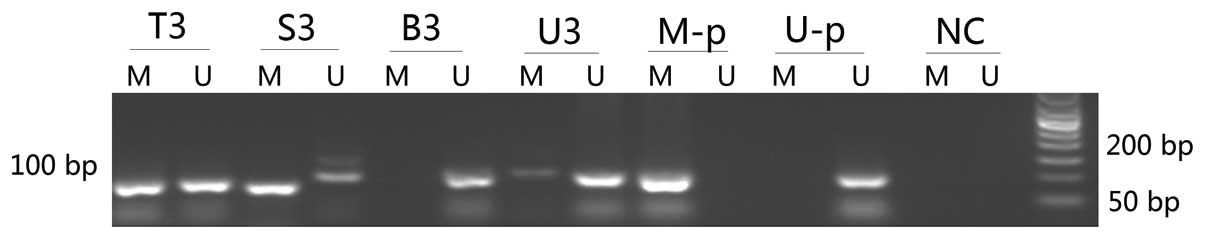 | Figure 1Nested methylation-specific polymerase
chain reaction detection of methylated NDRG4 gene in carcinoma
tissues, feces, urine and blood of a representative colorectal
cancer case (patient no. 3). T, carcinoma tissues; S, stool; B,
blood; U, urine; M, methylated products (88 bp); U, unmethylated
products (100 bp); 3, patient no. 3; M-p, positive control of
methylated DNA; U-p, positive control of unmethylated products; NC,
negative control; bp, base pairs. |
In addition to analysis in the tissues, the
sensitivity and specificity of methylated NDRG4 in feces, urine and
blood were 54.8 and 78.1% in blood; 72.6 and 85% in urine; 76.2 and
89.1% in feces. Methylated NDRG 4 genes in feces and urine showed a
higher sensitivity and specificity for diagnosis than blood. From
the present data, it could be suggested that a combination of using
urine and fecal samples to detect the methylation status of NDRG4
could be a beneficial diagnostic method for CRC.
Highly expressed methylated NDRG 4 gene
in tissue and feces DNA of CRC patients
The association between methylated NDRG4 and
clinical pathological parameters were analyzed. The data showed no
correlation between all methylated NDRG4 with age, the degree of
tumor differentiation and TNM stage. Methylated NDRG4 in
metastasis-lymph node tumors from CRC tissues and feces was
significantly highly expressed as compared with non-metastatic
samples (P<0.05). Methylated NDRG4 was not expressed highly in
urine or blood samples (Table
II).
 | Table IIAssociation between methylated NDRG4
gene expression and clinicopathological parameters of colorectal
cancer. |
Table II
Association between methylated NDRG4
gene expression and clinicopathological parameters of colorectal
cancer.
| | Tissue | Stool | Urine | Blood |
|---|
| |
|
|
|
|
|---|
| Object | Total | Methylated | Unmethylated | Methylation | Unmethylation | Methylated | Unmethylated | Methylated | Unmethylated |
|---|
| Age, years |
| <65 | 36 | 29 | 7 | 26 | 10 | 25 | 11 | 19 | 17 |
| ≥65 | 48 | 39 | 9 | 38 | 10 | 36 | 12 | 27 | 21 |
| Differentiation |
| High-middle | 57 | 49 | 8 | 45 | 12 | 44 | 13 | 29 | 28 |
| Low | 27 | 19 | 8 | 19 | 8 | 17 | 10 | 17 | 10 |
| Lymph node
metastasis |
| Exist | 55 | 50 | 5a | 48 | 7a | 42 | 13 | 30 | 25 |
| None | 29 | 18 | 11 | 16 | 13 | 19 | 10 | 16 | 13 |
| TNM stage |
| I–II | 48 | 42 | 6 | 39 | 9 | 34 | 14 | 26 | 22 |
| III–IV | 36 | 26 | 10 | 25 | 11 | 27 | 9 | 20 | 16 |
Early diagnosis of colorectal cancer by
methylated NDRG 4 gene in urine
According to the above research results, we
recruited another 76 CRC patients; they all were confirmed by
pathology diagnosis, the median age of them was 54 years (range,
39–71), and 36 non-cancer patients were selected as the normal
control group. Urine specimens were collected preoperatively in all
patients, with a collection volume of 50–100 ml, then sent to the
laboratory immediately, where genomic DNA were extracted and then
frozen at −20°C. All patients did not receive preoperative
chemotherapy or radiotherapy, and informed consent was obtained. Of
the 76 CRC patients, the proportion of positive methylated NDRG 4
gene status was 72.4% (55/76). This result showed consistency with
the former experiments; combining the convenience and sensitivity,
methylated NDRG 4 gene testing in urine could be a potential
testing method.
Discussion
The formation of a solid tumor is a multi-staged
process that results in abnormal gene expression. Epigenomic
changes are an important cause of tumorigenesis, and methylated
cytosine has been previously reported as an important factor in
this process (20,21). During the process of proliferation,
cellular nucleoside production is increased. Modified intracellular
nucleosides are difficult to degrade, and can only be excreted
through the urine. The level of urinary modified nucleosides can
therefore be used as a reflection of the in vivo metabolic
rate of nucleosides. During the process of canceration, the level
of urinary modified nucleosides markedly increase, therefore,
analysis of modified nucleosides in the urine could facilitate
preliminary tumor detection. DNA methylation is one of the
predominant mechanisms leading to loss of gene function (22). Data has shown that CRC is a tumor
type that is prone to high levels of abnormal gene methylation
(23,24). Previous studies have indicated that
methylation of multiple genes can accurately reflect CRC much more
than single gene methylation (25,26).
Previous reports have shown that DNA fragments from
tumor genes have been successfully isolated from the urine of
patients who have been diagnosed with CRC (27,28).
These studies have therefore demonstrated that DNA testing from
urine samples may be a feasible method for the diagnosis of
early-stage cancer. As compared with plasma or sera diagnoses, a
urinalysis can eliminate the possibility of blood infection
(29). In addition, the amount of
tumor DNA that can be obtained from peripheral blood serum is low
(ng), as compared with tissue samples, and the majority of DNA can
be readily degraded during the process of modification by
NaHSO3. For trace DNA samples, the sensitivity of the
conventional n-MSP method requires improvements (18).
Ahlquist et al (30) found that DNA isolated from feces has
improved sensitivity in methylated gene detection as compared with
DNA extracted from peripheral blood (30,31),
particularly in early-stage CRC. This may be associated with the
direct contact with the colorectal tissue and the alkaline
environment in which colorectal cells are exposed to. Conversely,
low DNA levels in the peripheral blood and the presence of numerous
factors in the plasma could influence the PCR reaction (32,33).
In the present study, the methylated NDRG4 gene expression rates in
the tumor and adjacent tissues, were 81 and 8.3%. This rate of
methylation at the CpG site, however, was not significantly
associated with age, differentiation level or TNM stage.
The NDRG4 transcript is 32 Kb long, consisting of 17
exons and 16 introns. The function of NDRG4 in tumor development
remains unclear, but numerous studies have indicated that NDRG4 is
associated with the growth, differentiation and metastasis of the
tumor (17,34,35).
The 5′ regulatory region of NDRG4 contains the CpG island, and is
often methylated in the occurrence and development of CRC, thus the
methylation of NDRG4 is considered to be an important biological
feature of colorectal cancer(17).
Melotte et al (16)
identified that NDRG4 is a biomarker candidate tumor suppressor
gene in CRC, and methylation of the NDRG4 promoter can be used as a
biological marker for the detection of colorectal cancer. The
sensitivity and specificity of NDRG4 methylation were 61 and 93% in
the fecal samples analyzed.
In the present study, n-MSP was used to detect
methylated NDRG4. The sensitivity and specificity was 76.2 and
89.15% in stool samples and 72.6 and 85% in urine samples. This
indicated that, as compared with blood samples, methylated NDRG4 in
DNA isolated from feces and urine, can be used as a non-invasive
biomarker for the detection of early stage CRC. This method could
be used to enrich the current diagnostic methods of CRC. Urine
contains significantly less protein than that of blood, therefore
the isolation of DNA fragments from urine is facilitated. Urine
samples are easy to obtain, and have high stability,
reproducibility and specificity. The methylation of NDRG4 may
therefore become a standard measure in the clinical diagnosis of
CRC.
In conclusion, the present study has demonstrated
that the detection of NDRG4 methylation status in feces and urine
has sufficient sensitivity and specificity as a diagnostic measure
in CRC. Considering the advantages of easy acquisition of urine
samples, detection of NDRG4 methylation in urine could be suitable
for detecting low levels of methylated genes, to facilitate the
early diagnosis of CRC.
Acknowledgements
The authors would like to thank the Research Fund of
the PLA General Hospital (Beijing, China) for financial
support.
References
|
1
|
Todosi AM, Gavrilescu MM, Aniţei GM, et
al: Colon cancer at the molecular level - usefulness of
epithelial-mesenchymal transition analysis. Rev Med Chir Soc Med
Nat Iasi. 116:1106–1111. 2012.
|
|
2
|
Wallace PM and Suzuki R: Regional, racial,
and gender differences in colorectal cancer screening in
middle-aged African-Americans and Whites. J Cancer Educ.
27:703–708. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zauber AG, Winawer SJ, O’Brien MJ, et al:
Colonoscopic polypectomy and long-term prevention of
colorectal-cancer deaths. N Engl J Med. 366:687–696. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nagasaka T, Mori Y, Umeda Y and Fujiwara
T: Biomarker for colorectal cancer. Nihon Rinsho. 70:802–808.
2012.(In Japanese). PubMed/NCBI
|
|
5
|
Imperiale TF, Ransohoff DF, Itzkowitz SH,
et al: Fecal DNA versus fecal occult blood for colorectal-cancer
screening in an average-risk population. N Engl J Med.
351:2704–2714. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sonnenberg A and Delcò F:
Cost-effectiveness of a single colonoscopy in screening for
colorectal cancer. Arch Intern Med. 162:163–168. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lind GE, Danielsen SA, Ahlquist T, et al:
Identification of an epigenetic biomarker panel with high
sensitivity and specificity for colorectal cancer and adenomas. Mol
Cancer. 10:852011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mori Y, Olaru AV, Cheng Y, et al: Novel
candidate colorectal cancer biomarkers identified by methylation
microarray-based scanning. Endocr Relat Cancer. 18:465–478. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jin X, Lin M, Zhang H, et al: Serum
biomarkers of colorectal cancer with AU and NP20 chips including a
diagnosis model. Hepatogastroenterology. 59:124–129.
2012.PubMed/NCBI
|
|
10
|
Lenhard K, Bommer GT, Asutay S, et al:
Analysis of promoter methylation in stool: a novel method for the
detection of colorectal cancer. Clin Gastroenterol Hepatol.
3:142–149. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lange CP, Campan M, Hinoue T, et al:
Genome-scale discovery of DNA-methylation biomarkers for
blood-based detection of colorectal cancer. PloS One. 7:e502662012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cassinotti E, Melson J, Liggett T, et al:
DNA methylation patterns in blood of patients with colorectal
cancer and adenomatous colorectal polyps. Int J Cancer.
131:1153–1157. 2012. View Article : Google Scholar :
|
|
13
|
Lange CP and Laird PW: Clinical
applications of DNA methylation biomarkers in colorectal cancer.
Epigenomics. 5:105–108. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ahlquist DA, Zou H, Domanico M, et al:
Next-generation stool DNA test accurately detects colorectal cancer
and large adenomas. Gastroenterology. 142:248–256. 2012. View Article : Google Scholar
|
|
15
|
Yamamoto H, Kokame K, Okuda T, et al:
NDRG4 protein-deficient mice exhibit spatial learning deficits and
vulnerabilities to cerebral ischemia. J Biol Chem. 286:26158–26165.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Melotte V, Lentjes MH, van den Bosch SM,
et al: N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor
suppressor gene and potential biomarker for colorectal cancer. J
Natl Cancer Inst. 101:916–927. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kotipatruni RP, Ferraro DJ, Ren X, et al:
NDRG4, the N-Myc downstream regulated gene, is important for cell
survival, tumor invasion and angiogenesis in meningiomas. Integr
Biol (Camb). 4:1185–1197. 2012. View Article : Google Scholar
|
|
18
|
Fan LP, Shen JZ, Ye BG, et al: Detection
of p16 gene methylation status in adult patients with acute
leukemia by using n-MSP. Zhongguo Shi Yan Xue Ye Xue Za Zhi.
15:258–261. 2007.PubMed/NCBI
|
|
19
|
Carraway HE, Wang S, Blackford A, et al:
Promoter hypermethylation in sentinel lymph nodes as a marker for
breast cancer recurrence. Breast Cancer Res Treat. 114:315–325.
2009. View Article : Google Scholar
|
|
20
|
Ryan HE, Lo J and Johnson RS: HIF-1 alpha
is required for solid tumor formation and embryonic
vascularization. EMBO J. 17:3005–3015. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yi JM, Dhir M, Guzzetta AA, et al: DNA
methylation biomarker candidates for early detection of colon
cancer. Tumour Biol. 33:363–372. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ziyab AH, Karmaus W, Holloway JW, et al:
DNA methylation of the filaggrin gene adds to the risk of eczema
associated with loss-of-function variants. J Eur Acad Dermatol
Vernereol. 27:e420–e423. 2013. View Article : Google Scholar
|
|
23
|
Morioka Y, Hibi K, Sakai M, et al:
Aberrant methylation of the CHFR gene in digestive tract cancer.
Anticancer Res. 26:1791–1795. 2006.PubMed/NCBI
|
|
24
|
de la Chapelle A and Hampel H: Clinical
relevance of microsatellite instability in colorectal cancer. J
Clin Oncol. 28:3380–3387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Uehara E, Takeuchi S, Yang Y, et al:
Aberrant methylation in promoter-associated CpG islands of multiple
genes in chronic myelogenous leukemia blast crisis. Oncol Lett.
3:190–192. 2012.PubMed/NCBI
|
|
26
|
Zhao YF, Zhang YG, Tian XX, Juan D and Jie
Z: Aberrant methylation of multiple genes in gastric carcinomas.
Int J Surg Pathol. 15:242–251. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Samejima K, Hiramatsu K, Takahashi K, et
al: Identification and determination of urinary acetylpolyamines in
cancer patients by electrospray ionization and time-of-flight mass
spectrometry. Anal Biochem. 401:22–29. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cai H, Thomasset SC, P-Berry D, et al:
Determination of anthocyanins in the urine of patients with
colorectal liver metastases after administration of bilberry
extract. Biomed Chromatogr. 25:660–663. 2011. View Article : Google Scholar
|
|
29
|
Jackson C, Best N and Elliott P: UK
Biobank Pilot Study: stability of haematological and clinical
chemistry analytes. Int J Epidemiol. 37(Suppl 1): i16–i22. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ahlquist DA, Taylor WR, Mahoney DW, et al:
The stool DNA test is more accurate than the plasma septin 9 test
in detecting colorectal neoplasia. Clin Gastroenterol Hepatol.
10:272–277. 2012. View Article : Google Scholar
|
|
31
|
deVos T, Tetzner R, Model F, et al:
Circulating methylated SEPT9 DNA in plasma is a biomarker for
colorectal cancer. Clin Chem. 55:1337–1346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lacks S and Greenberg B: Complementary
specificity of restriction endonucleases of Diplococcus pneumoniae
with respect to DNA methylation. J Mol Biol. 114:153–168. 1977.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hastings ML, Palma J and Duelli DM:
Sensitive PCR-based quantitation of cell-free circulating
microRNAs. Methods. 58:144–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ding W, Zhang J, Yoon JG, Shi D, Foltz G
and Lin B: NDRG4 is downregulated in glioblastoma and inhibits cell
proliferation. OMICS. 16:263–267. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang X, An L and Li X: NDRG3 and NDRG4,
two novel tumor-related genes. Biomed Pharmacother. 67:681–684.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hayashi T, Yoshikawa T, Bonam K, et al:
The superiority of the seventh edition of the TNM classification
depends on the overall survival of the patient cohort: comparative
analysis of the sixth and seventh TNM editions in patients with
gastric cancer from Japan and the United Kingdom. Cancer.
119:1330–1337. 2013. View Article : Google Scholar : PubMed/NCBI
|















