Introduction
Non-small cell lung cancer (NSCLC), which includes
adenocarcinoma and squamous cell carcinoma, is the predominant form
of lung cancer and remains the leading cause of cancer-associated
mortality in the world, particularly in China (1,2),
accounting for >80% of all cases of lung cancer. It is
hypothesized that NSCLC will continue to be a major health hurdle
for mankind over the next century (3). NSCLC tumorigenesis is not only
strongly associated with certain protein-coding genes, including
p53, Rb and Ras (4), but is also
regulated by ~100 microRNAs (miRNAs) (5). Therefore, an improved understanding of
the molecular mechanisms regulated by microRNAs involved in NSCLC
development is required as a basis to identify novel strategies for
the treatment of these diseases.
miRNAs are a class of endogenous non-coding RNAs,
~20–25 nucleotides long, that are widely expressed in eukaryotes
and predominantly inhibit gene expression at the
post-transcriptional level by base pairing with target mRNAs in the
3′-untranslated region, leading to mRNA cleavage or translation
repression (6–8). The levels of individual miRNAs vary
significantly between tissues, developmental stages and
physiological processes, indicating that miRNAs play a role in
cellular proliferation and differentiation, tumorigenesis and
apoptosis (9,10). Deregulation or mutation of miRNAs
has been frequently reported in numerous human malignancies,
including breast carcinoma, primary glioblastoma, lung cancer and
colon carcinoma (11–14), and has been found to contribute to
the initiation and progression of cancer (15). miRNA is able to function as either a
tumor suppressor or an oncogene (16). The tissue- and disease-specific
expression patterns of miRNAs indicate their potential as
diagnostic and prognostic cancer biomarkers and therapeutic tools
(17).
miR-99a, which is transcribed from the 21q21 region,
has been reported to be deregulated in renal cell carcinoma (RCC)
(18). miR-99a induces
G1-phase cell cycle arrest and suppresses
tumorigenicity, functioning as a tumor suppressor in RCC. It has
also been reported that, in NSCLC, the miR-99a gene locates to a
homozygous deletion region, indicating that miR-99a may play a
crucial role in tumorigenesis and cancer progression (5). miR-99a has been reported to be
downregulated in squamous cell lung carcinoma (19). However, to the best of our
knowledge, there have been no studies investigating the role of
miR-99a in adenocarcinoma.
In the present study, the expression of miR-99a in
adenocarcinoma tissues was examined, and the impact of miR-99a on
A549 and H1299 NSCLC cells was assessed.
Materials and methods
Tissue samples and cell lines
Paired NSCLC and normal adjacent lung tissues were
obtained, with informed consent, from 15 patients who underwent
primary surgical resection of NSCLC between 2011 and 2012 at the
Teaching Hospital of Chengdu University of Traditional Chinese
Medicine (Chengdu, Sichuan, China). The NSCLC A549 and H1299 cell
lines, and the normal lung HBE cell line were frozen in the
Functional Genomic Laboratory, Sichuan University (Chengdu,
Sichuan). The HBE and A549 cells were maintained in Dulbecco’s
modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA), and
the H1299 cells were maintained in RPMI-1640 medium, each
supplemented with 2 mM L-alanyl-L-glutamine, 1%
penicillin/streptomycin and 10% fetal bovine serum (Invitrogen) at
37°C and 5% CO2.
RNA extraction and stem-loop conventional
reverse transcription-polymerase chain reaction (RT-PCR)
analysis
Total RNA was isolated using TRIzol reagent
(Invitrogen) according to the manufacturer’s instructions.
Reverse-transcribed complementary DNA was synthesized using the
Prime-Script RT reagent kit (Takara Biotechnology Co., Ltd.,
Dalian, China). Conventional PCR was used to assay miRNA expression
with the specific forward primers, and the universal reverse primer
complementary to the anchor primer and U6 small nuclear RNA was
used as the internal control. The PCR primers for mature miR-99a or
U6 were designed as follows: miR-99a forward, 5′-ACAGTCGAGATGGGATAC
CCTTACCATTACT-3′ and reverse, 5′-CTGCTGACGTCGA GTGGGCAA-3′; and U6
forward, 5′-CTCGCTTCGGCAGCA CA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. The PCR cycles were performed by
initial denaturation at 95°C for 5 min, then by completing 40
cycles at 95°C for 10 sec followed by 60°C for 1 min.
Plasmid construction and miRNA
transfection
The plasmids pMSCV-miR-99a and pMSCV-miR-NC were
kindly provided by Dr R Agami (Faculty of Science, Ain Shams
University, Cairo, Egypt) (20).
Stable transfection of pMSCV-miR-99a resulted in mock A549
(A549-miR-99a) and mock H1299 (H1299-miR-99a The 2′-O-methyl
oligonucleotides were chemically synthesized by LifeTechnologies
(Guangzhou, Guangdong, China). The oligonucleotide sequences were
as follows: miR-99a mimic forward, 5′-AACCCGUAGAUCCGA UCUUGUG-3′
and reverse, 5′-CAAGAUCGGAUCUACGGG UUUU-3′; miR-negative control
(miR-NC) forward, 5′-UUC UCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGAC ACGUUCGGAGAATT-3′. The A549 (5×105) and H1299
(3×105) cells were seeded 24 h prior to 48-h
transfection with the miR-99a mimic or miR-NC, respectively. The
transfections were performed using Lipofectamine 2000 (Invitrogen)
according to the manufacturer’s instructions. The cells were
harvested for further testing 48 h after transfection.
Cell proliferation assay
Cell proliferation was detected using a 3-(4,
5-dimethylthazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT)
assay. The cells were seeded into 24-well plates
(1.2×104 cells/well) and allowed to attach overnight.
After 24, 48, 72 and 96 h, cell viability was assessed using an MTT
assay. The absorbance at 490 nm of each well was read on a
spectrophotometer. Three independent experiments were performed in
quadruplicate.
Colony formation assay
In total, ~5×103 cells from each group,
mock A549 (A549-miR-99a), stably transfected A549 (A549-miR-NC),
mock H1299 (H1299-miR-99a) and stably transfected H1299
(H1299-miR-NC) cells, were placed in a six-well plate containing
RPMI-1640 medium supplemented with 10% FBS for three weeks. The
colonies were fixed with methanol and stained with 0.1% crystal
violet (Sheng Gong, Shanghai, China) in 20% methanol for 30 min.
Each assay was performed in triplicate.
Cell cycle assay
Transfected A549 and H1299 cells in the log phase of
growth were collected and fixed in 75% ethanol at −20°C for 16 h.
For the cell cycle analysis, the transfected cells were stained
with propidium iodide (PI) and examined with a
fluorescence-activated cell sorting (FACS) flow cytometer (BD
Biosciences, San Jose, CA, USA). Each test was performed in
triplicate.
Cell migration and invasion assay
The migratory and invasive potential of the
transient transferred cells and bulk-selected A549 and H1299 cells
were examined. A scratch assay was performed to assess the
migratory potential. The cells were scratched using a pipette tip
when the cell confluence reached ~95%, and were further incubated
with fresh medium. The medium was changed every two days. Images
were captured at a ×40 magnification immediately subsequent to
scratching (0 h) and at 12 and 24 h subsequent to scratching. The
cell invasion assay was tested using Transwell plates (8-μm pore
size; 6.5-mm diameter; Corning Life Sciences, Tewksbury, MA, USA)
pre-coated with Matrigel Basement Membrane Matrix and at a
concentration of 1 mg/ml (BD Biosciences, Franklin Lakes, NJ, USA),
according to the manufacturer’s instructions. In total,
2×104 cells in 0.2 ml media, supplemented with 2% FBS,
were seeded into the upper chamber, with 0.6 ml of medium
containing 10% FBS under the upper chamber. The plates were
incubated at 37°C in a 5% CO2 atmosphere. After 48 h,
the chambers were removed and a cotton swab was used to remove the
non-invading cells from the upper side of the chamber. The cells
under the chamber were then fixed in methanol for 10 min and
stained with 0.1% crystal violet in 20% methanol for 30 min.
Western blot assay
Protein extract (50 μg) was separated by 10%
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and was
electrophoretically transferred onto a polyvinylidene fluoride
membrane (Millipore, Darmstadt, Germany). The membranes were
blocked for 30 min at room temperature with 5% non-fat dried milk
and incubated for 1 h with monoclonal antibodies against E-cadherin
(sc-8426, mouse, IgG1), N-cadherin (sc-7939, rabbit,
IgG), γ-catenin (sc-30997, goat, IgG), insulin-like growth factor 1
receptor (IGF-1R; sc-sc462, mouse, IgG1) or β-actin
(sc-1616, goat, IgG) (Santa Cruz Biotechnology, Dallas, TX, USA).
The antibodies against E-cadherin, N-cadherin and γ-catenin were at
a dilution of 1:1,000 and the antibody against β-actin was at a
dilution of 1:5,000. Subsequent to washing with PBS-T (10 mm Tris,
pH 8.0; 150 mm NaCl; 0.5% Tween 20; Sheng Gong), the membranes were
incubated for 1 h with secondary HRP-linked polyclonal goat
anti-rabbit antibody (sc-2004, IgG, Santa Cruz Biotechnology), at a
1:20,000 dilution. The membranes were washed again with PBS-T and
the proteins were visualized using ECL chemiluminescence and
exposed to X-ray film.
Statistics
Statistical analyses were performed using the
GraphPad prism software (GraphPad Software, Inc., La Jolla, CA,
USA). Statistical differences were calculated using an unpaired
two-tailed student’s t-test. P≤0.05 was considered to indicate a
statistically significant difference. The statistical significance
of a correlation was assessed using the Pearson test.
Results
miR-99a is significantly downregulated in
human NSCLC tissues
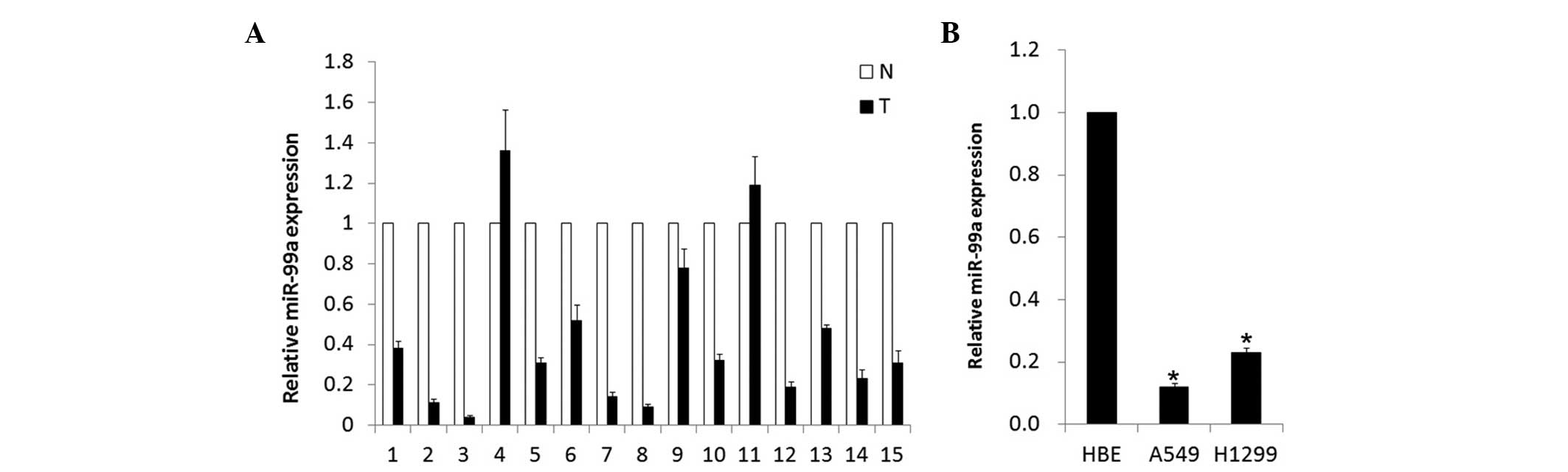
In the present study, a stem-loop RT-quantitative
PCR assay was performed to determine the expression of miR-99a in
15 pairs of matched NSCLC and normal adjacent lung tissues. As
shown in Fig. 1A, the miR-99a
expression levels were all significantly reduced by 2.1–25 times in
86.7% (13/15) of NSCLC tumor tissues compared with the
corresponding adjacent normal lung tissues. Of the 15 tested lung
cancer tissues, the expression level of miR-99a demonstrated no
difference between ages, genders or metastasis status. To further
assess the biological role of miR-99a in adenocarcinoma cell lines,
its expression level was detected in A549 and H1299 cells. Compared
with the normal human bronchial epithelial HBE cell line, the level
of miR-99a was markedly decreased (Fig.
1B). These results revealed that the expression of miR-99a was
downregulated in NSCLC adenocarcinoma tissues and cell lines,
indicating an involvement in NSCLC carcinogenesis.
miR-99a suppresses tumorigenicity by
inducing G1-phase cell cycle arrest in vitro
The downregulation of miR-99a in NSCLC
adenocarcinoma cell lines prompted the identification of its
activity as a tumor suppressor. First, miR-99a was restored in A549
and H1299 cell lines that contained a relatively low miR-99a level,
and MTT, colony formation and flow cytometric assays were then
performed. As shown in Fig. 2A, the
A549-miR-99a and H1299-miR-99a cell lines exhibited a significant
increase in cell viability compared with the mock A549 and H1299,
A549-miR-NC or H1299-miR-NC cell lines (P<0.05). As shown in
Fig. 2B, A549-miR-99a and
H1299-miR-99a cells exhibited notably fewer and smaller colonies
compared with negative controls (P<0.05). The flow cytometry
analysis revealed that A549-miR-99a and H1299-miR-99a cells
underwent a significant increase in the proportion of cells in the
G1-phase population compared with control cell lines
(Fig. 2C). Additionally, the effect
of miR-99a on apoptosis was also detected and it was found that
exogenous miR-99a did not influence apoptosis (data not shown).
Thus, the upregulation of miR-99a was able to induce growth
inhibition by blocking the cell cycle at the G1
phase.
MiR-99a suppresses the migration and
invasion of NSCLC cell lines in vitro
To test the role of miR-99a in the migration and
invasion of NSCLC adenocarcinoma cell lines, the A549-miR-99a and
H1299-miR-99a cells were analyzed by performing scratch and
Transwell assays. As expected, the A549-miR-99a and H1299-miR-99a
cells underwent a morphological change of cells to an elongated
shape and exhibited a reduction in migration (Fig. 3A and B). Western blot analysis was
then performed to detect the changes in N-cadherin, E-cadherin and
γ-catenin. A corresponding marked increase in E-cadherin and
γ-catenin and decrease in N-cadherin were also observed (Fig. 3C).
MiR-99a inhibited EMT by decreasing the
IGF-1R level
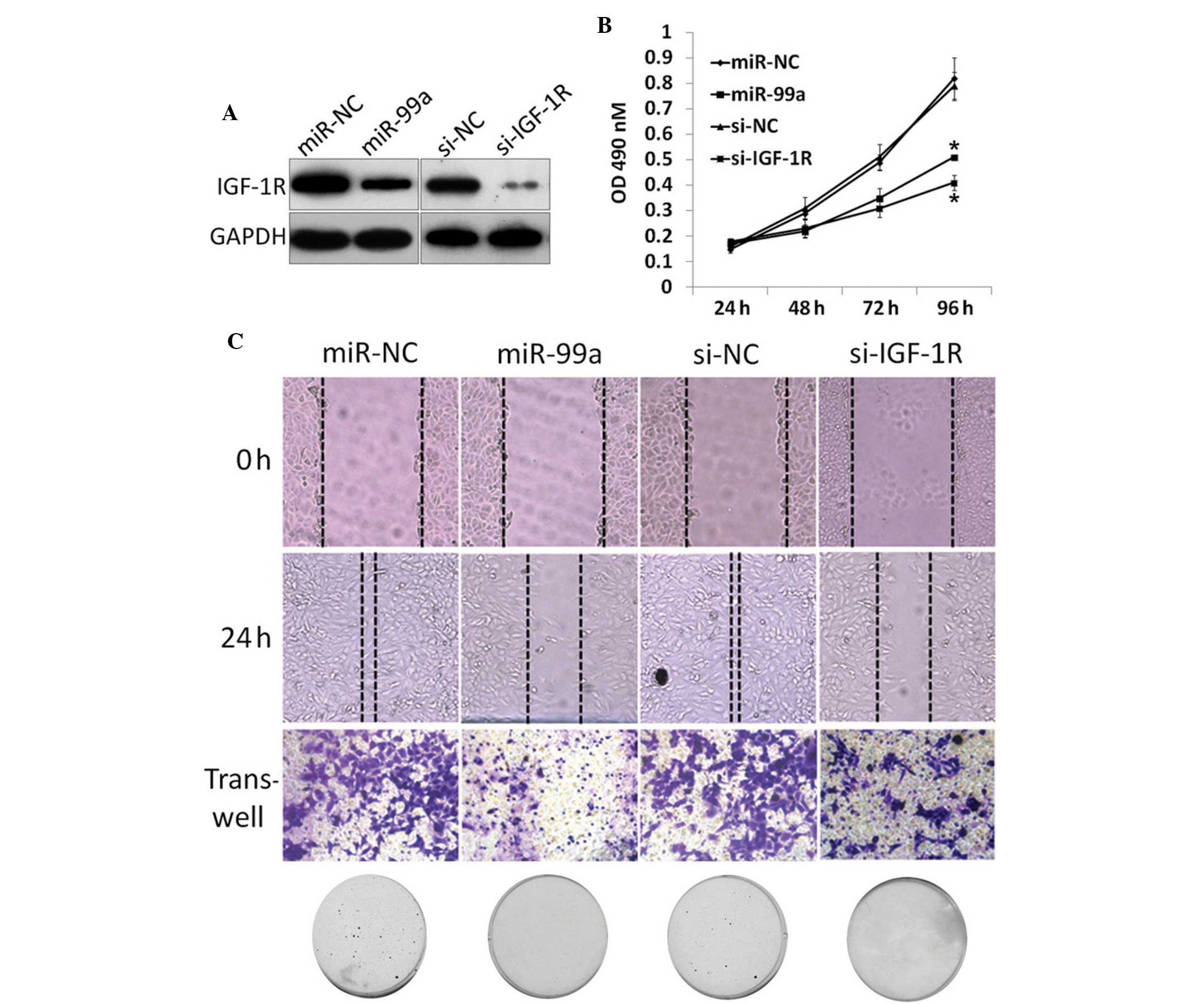
To further reveal the mechanisms underlying this
tumor suppressor effect of miR-99a, IGF-1R, a target mRNA of
miR-99a, was knocked-down in NSCLC cells. The A549 cells were
transfected with IGF-1R siRNA or negative control (NC), followed by
functional assays (Fig. 4A).
Proliferation and colony formation results revealed that knockdown
of IGF-1R decreases proliferation (Fig.
4B) and colony formation (Fig.
4C), similar to the phenotype observed upon miR-99a restoration
in the A549 and H1299 cells. Scratch and Transwell assays also
revealed a similar phenotype compared with miR-99a restoration
(Fig. 4C). Collectively, it was
concluded that the tumor suppressor role of miR-99a is associated
with IGF-1R pathway regulation.
Discussion
Lung cancer is considered to be the most dangerous
cancer worldwide and is a leading cause of cancer mortality.
Currently, a model for lung cancer pathogenesis hypothesizes that
the pathogenesis is due to the combination of genetic risk and
environmental factors (20). As
aforementioned, NSCLC accounts for ~80% of all lung cancer subtypes
(3). Understanding the carcinogenic
mechanisms may aid in the identification of a biomarker or
therapeutic target for lung cancer, and this promotes the
elucidation of the underlying mechanism of NSCLC development.
Although the carcinogenesis and pathophysiology of NSCLC have been
intensively investigated in the past several decades (1,2), the
underlying mechanism of NSCLC development remains poorly understood
and no efficient therapeutic strategies have emerged from the
extensive number of studies that have been performed.
As a prevailing area for cancer biology studies in
previous years, miRNA has been revealed to play important
regulatory roles in tumorigenesis and cancer development, not only
in cell proliferation and development (21–23).
The regulatory roles of miRNA include the upregulation of mir574-3p
in prostate cancer (24) and the
extreme downregulation of miR-23a in gastric cancer (25). The miRNAs that have been identified
as associated with lung cancer include miR-210 (26), miR-365 (27) and miR-449 (28), indicating an association between
miRNA and NSCLC.
In the present study, it was found that miR-99a was
significantly downregulated in NSCLC adenocarcinoma tissues and
cell lines, which was consistent with previous findings that
miR-99a is reduced in several human tumors and cancer cell lines
compared with normal adjacent lung tissues and normal cell lines
(19). The expression of miR-99a
was low in 13 out of 15 NSCLC tissues, although no correlation was
observed between the reduction in miR-99a expression and the
clinical characteristics of NSCLC in the 15 pairs of tissues. This
indicates that low expression of miR-99a may be involved in NSCLC
carcinogenesis. In addition, it was demonstrated that miR-99a
expression is elevated in NSCLC cell lines compared with the HBE
normal human bronchial epithelial cell line, with the exception of
A549 and H1299, and these findings also indicate the role of
miR-99a in NSCLC adenocarcinoma carcinogenesis.
To assess the role of miR-99a in NSCLC, the present
study investigated the effect of miR-99a gain of function on
various aspects of NSCLC. First, it was demonstrated that the
overexpression of miR-99a in A549 and H1299 cells, two
adenocarcinoma cell lines, led to a significant inhibition of cell
proliferation, colony formation, migration and invasion.
IGF-1R mRNA is an identified miR-99a target that is
known to be involved in the pathogenesis of psoriasis (29,30).
Although the mechanisms of IGF-1R in EMT are unclear, the
downregulation of IGF-1R is sufficient to drive tumor cell
migration, indicating that the presence of IGF-1R is critical for
inducing an EMT-like phenotype. The present study hypothesized that
miR-99a-mediated downregulation of IGF-1R induced an EMT-like
phenotype. As expected, siRNA-mediated IGF-1R knockdown caused a
morphological change in cells to a flatter shape. A corresponding
decrease in the levels of E-cadherin and γ-catenin, and an increase
of N-cadherin was also observed, which was similar to the effects
of miR-99a overexpression, indicating that overexpression of
miR-99a alters the epithelial phenotype of the cell in an
IGF-1R-dependent manner. In addition to EMT, there is usually an
increase in cell mobility. Consistent with this phenomenon, IDL
single-stranded RNA expression markedly increased the cell
migration of A549 cells, as examined by scratch and Transwell
assays.
In conclusion, the present study identified miR-99a
as an effector of the IGF-1R pathway during EMT in NSCLC cells. The
data revealed that miR-99a is significantly downregulated in NSCLC
adenocarcinoma cells and it can be hypothesized that miR-99a may
play a key role in NSCLC development and progression by modulating
IGF-1R signaling. The identification of the downregulation of
miR-99a in NSCLC adenocarcinoma highlights the possibility of
therapeutic applications for miR-99a in cancer.
Acknowledgements
This study was supported by the National Science
Foundation of China (grant no. 81330016 and 31171020), the Major
State Basic Research Development Program (grant no. 2013CB967404),
grants from the Ministry of Education of China (grant no. 313037,
20110181130002 and IRT0935), a grant from the State Commission of
Science Technology of China (grant no. 2012BAI04B04) and grants
from the Science and Technology Bureau of Sichuan Province (grant
no. 2010SZ0280 and 2011JTD0005).
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang L, Parkin DM, Li L and Chen Y: Time
trends in cancer mortality in China: 1987–1999. Int J Cancer.
106:771–783. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ramalingam SS, Owonikoko TK and Khuri FR:
Lung cancer: new biological insights and recent therapeutic
advances. CA Cancer J Clin. 61:91–112. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Toyooka S, Mitsudomi T, Soh J, et al:
Molecular oncology of lung cancer. Gen Thorac Cardiovasc Surg.
59:527–537. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Engels BM and Hutvagner G: Principles and
effects of microRNA-mediated post-transcriptional gene regulation.
Oncogene. 25:6163–6169. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Van Rooij E, Purcell AL and Levin AA:
Developing microRNA therapeutics. Circ Res. 110:496–507. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takamizawa J, Konishi H, Yanagisawa K, et
al: Reduced expression of the let-7 microRNAs in human lung cancers
in association with shortened postoperative survival. Cancer Res.
64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kosaka N, Takeshita F, Yoshioka Y and
Ochiya T: Therapeutic application of microRNAs in cancer. RNA
Interference from Biology to Therapeutics. Howard KA: Springer; New
York, NY: pp. 299–314. 2013, View Article : Google Scholar
|
|
16
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li D, Liu X, Lin L, et al: MicroRNA-99a
inhibits hepatocellular carcinoma growth and correlates with
prognosis of patients with hepatocellular carcinoma. J Biol Chem.
286:36677–36685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cui L, Zhou H, Zhao H, et al: MicroRNA-99a
induces G1-phase cell cycle arrest and suppresses tumorigenicity in
renal cell carcinoma. BMC Cancer. 12:5462012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao W, Shen H, Liu L, et al: MiR-21
overexpression in human primary squamous cell lung carcinoma is
associated with poor patient prognosis. J Cancer Res Clin Oncol.
137:557–566. 2011. View Article : Google Scholar
|
|
20
|
Fong KM, Sekido Y, Gazdar AF and Minna JD:
Lung cancer 9; Molecular biology of lung cancer: clinical
implications. Thorax. 58:892–900. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Raveche ES, Salerno E, Scaglione BJ, et
al: Abnormal microRNA-16 locus with synteny to human 13q14 linked
to CLL in NZB mice. Blood. 109:5079–5086. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hatley ME, Patrick DM, Garcia MR, et al:
Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21.
Cancer Cell. 18:282–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang Z, Huang S, Wang Q, et al:
MicroRNA-95 promotes cell proliferation and targets sorting Nexin 1
in human colorectal carcinoma. Cancer Res. 71:2582–2589. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chiyomaru T, Yamamura S, Fukuhara S, et
al: Genistein upregulates tumor suppressor microRNA-574-3p in
prostate cancer. PLoS One. 8:e589292013. View Article : Google Scholar
|
|
25
|
An J, Pan Y, Yan Z, et al: MiR-23a in
Amplified 19p13.13 loci targets metallothionein 2A and promotes
growth in gastric cancer cells. J Cell Biochem. 114:2160–2169.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grosso S, Doyen J, Parks SK, et al:
MiR-210 promotes a hypoxic phenotype and increases radioresistance
in human lung cancer cell lines. Cell Death Dis. 4:e5442013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kang SM, Lee HJ and Cho JY: MicroRNA-365
regulates NKX2-1, a key mediator of lung cancer. Cancer Lett.
335:487–494. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Miao LJ, Huang SF, Sun ZT, et al: MiR-449c
targets c-Myc and inhibits NSCLC cell progression. FEBS Lett.
587:1359–1365. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hodak E, Gottlieb AB, Anzilotti M and
Krueger JG: The insulin-like growth factor 1 receptor is expressed
by epithelial cells with proliferative potential in human epidermis
and skin appendages: correlation of increased expression with
epidermal hyperplasia. J Invest Dermatol. 106:564–570. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wraight CJ, White PJ, McKean SC, et al:
Reversal of epidermal hyperproliferation in psoriasis by
insulin-like growth factor I receptor antisense oligonucleotides.
Nat Biotechnol. 18:521–526. 2000. View
Article : Google Scholar : PubMed/NCBI
|