Introduction
Thyroid cancer is the most prevalent type of
malignant endocrine system cancer (1). Thyroid cancer is histologically
classified into papillary, follicular, anaplastic and medullary
thyroid cancer, and these subtypes account for ~80, 15, 2 and 3% of
all thyroid malignancies, respectively (2). During recent decades, there has been an
increasing incidence rate of thyroid cancer observed in several
countries, including France (3), USA
(4), UK (5) and China (6). The mortality rate of thyroid cancer is
relatively low; well-differentiated thyroid carcinoma has a
mortality rate of 4% (7), and the
10-year relative survival rate for thyroid carcinoma in the USA has
improved from 95.4% in 1983 to 98.6% in 1999 (8). However, the rate of disease recurrence
of this type of cancer is high due to increasing incurability rates
(9). Recurrence is identified by
fine-needle aspiration biopsy, which is considered valuable for the
evaluation of single thyroid tumors (10). Currently, routine neck ultrasonography
and monitoring of serum thyroglobulin levels are widely used for
recurrence surveillance (11,12). Treatments for thyroid cancer include
surgical management, suppression therapy using levothyroxine and
adjuvant radioactive iodine (RAI) therapy. RAI ablation has been
used routinely, particularly in low-risk patients with thyroid
cancer, for >30 years (13).
However, previous studies have suggested that RAI confers only a
minor benefit in reducing the risk of recurrence or mortality
(14,15). Considerable studies concerning
oncogenic genetic alterations have been performed in thyroid cancer
(16–18). Additional knowledge is required
regarding the molecular mechanisms of thyroid tumorigenesis, so
that targeted treatment may be improved for patients with this
disease.
Lysine-specific demethylase 1 (LSD1) was first
identified in 2004 (19). LSD1 has
been found to specifically catalyze the demethylation of histone 3
lysine 4 (H3K4) or histone 3 lysine 9 (H3K9) (20), and to demethylate the mono- and
di-methyl groups of proteins (19).
It has been shown that LSD1 suppresses gene expression by targeting
histone H3K4 (21,22), while gene expression is activated by
targeting histone H3K9. Although LSD1 is highly expressed in a
number of cancer types, its mechanisms of action remain unknown.
The present study aimed to evaluate the effects of LSD1
downregulation, induced by small interfering RNA (siRNA)
transfection, on the proliferation, colony formation, migration and
invasion of the papillary thyroid carcinoma K1 cell line.
Material and methods
Cell culture
The human papillary thyroid carcinoma K1 cell line
was purchased from the European Collection of Authenticated Cell
Cultures (Salisbury, UK). The cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and supplemented with inactivated 10% fetal
bovine serum (FBS; Hyclone; GE Healthcare, Logan, UT, USA),
glutamine (2 Mm), penicillin (100 U/ml) and streptomycin (100
mg/ml) in a 37°C incubator with 5% CO2.
Transfection of siRNA
The sequences of the siRNA were as follows:
LSD1-siRNA sense, 5′-CACAAGGAAAGCUAGAAGATT-3′ and anti-sense,
5′-UCUUCUAGCUUUCCUUGUGTT-3′; and scrambled non-targeting siRNA
sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and anti-sense,
5′-ACGUGACACGUUCGGAGAATT-3′ (Shanghai GenePharma Co., Ltd.,
Shanghai, China). K1 cells (1.5×105 per well) were
seeded in 6-well plates and incubated for 20 h at 37°C. Using
HiperFect Transfection Reagent (Qiagen GmbH, Wetzlar, Germany), the
cells were transfected with either LSD1-siRNA or non-targeting
siRNA. Diluted HiperFect Transfection reagent was placed in tube 1,
while diluted siRNA was placed in tube 2. The two samples were
diluted in serum-free medium and incubated at room temperature for
5 min. Next, the reagents in tube 1 were mixed with the siRNA in
tube 2 gently by vortexing and incubation at room temperature for
30 min. The mixture was then added to the K1 cells in the 6-well
plates. After 6 h, complete medium with 10% serum was added to the
K1 cells. The transfection medium was replaced after 24 h with
fresh medium.
Immunocytochemistry staining
Immunocytochemistry staining was applied to observe
the expression of LSD1 on the seeded K1 cells on cover slips.
Sterilized cover slips were placed into 6-well plates. A total of
100 µl K1 cell suspension was added to each cover slip. After 2–4
h, the K1 cells had adhered in situ. Complete culture medium
(3 ml) was added to each well of the 6-well plates and the K1 cells
were cultured for another 24–36 h. Each cover slip was gently
rinsed three times in phosphate-buffered saline (PBS). Next, the K1
cells were fixed with 4% (v/v) paraformaldehyde in PBS for 20 min
at room temperature. A brown colored stain was counted as positive
expression, while blue staining was regarded as a negative result.
An eyepiece graticule facilitated cell counting and was used to
count the positive cells at a higher magnification (x400), for
which a minimum of 10 fields were used. Each experiment was
performed on three replicate samples.
Cell Counting kit-8 assay
Cell Counting kit-8 (CCK8; Dojindo Molecular
Technologies Inc., Kumamoto, Japan) was applied to determine the
effect of LSD1-siRNA on cell growth. K1 cells (3.6×103) transfected
with siRNA-LSD1 were seeded into each well of 96-well plates with
100 µl complete culture medium. At the same time, 3.6×103 K1 cells
transfected with negative siRNA were also seeded into wells. The K1
cells were detected using CCK8 after being cultured for another 24,
48 and 72 h, respectively. At the indicated time points, the
supernatant was removed and 100 µl DMEM containing 10 µl CCK8 was
added to each well for another 2 h at 37°C. The wells with DMEM (90
µl) and CCK8 (10 µl) were regarded as blank controls. The
absorbance was recorded at a wavelength of 450 nm with a microplate
reader (Model 550; Bio-Rad Laboratories Inc., Hercules, CA, USA).
Each experiment was performed in sextuplicate and repeated three
times.
Soft agar colony formation assay
The 0.7% soft agar was made in 6-well plates as a
base layer and the 0.35% soft agar was placed on top mixed with
suspended cells (1.0×103 cells per well). Each layer consisted of 3
ml soft agar containing complete growth medium. The cells were
cultured for nearly 14 days until formed colonies were visible. The
visible colonies were then counted and images were captured. Each
experiment was performed on three replicate samples and repeated
three times.
Invasion and migration assays
The invasion ability of the K1 cells was detected in
a 24-well Transwell chamber (Corning Inc., Corning, NY, USA)
covered with 50 µl of 2.0 mg/ml Matrigel (BD Biosciences, Franklin
Lakes, NJ, USA). Cells (1.5×104) in 200 µl of serum-free medium
were added to the upper chamber. DMEM (600 µl) supplemented with
15% FBS filled the lower chamber. Following incubation for 18 h at
37°C, the remaining cells on the upper surface of the membrane were
swabbed. Methanol was used to fix the invading cells on the lower
surface for nearly 20 min, and crystal violet (0.1%) was used as a
dye. An eyepiece graticule facilitated cell counting of the
Transwell cells at a higher magnification (x400), for which a
minimum of 10 fields were counted. A migration assay was performed
using the same steps without the Matrigel layer, and 3×104 cells
were added to the upper chamber. Each experiment was performed on
three replicate samples and repeated three times.
Total RNA isolation, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Transfected cells were harvested at the 48 h time
point after transfection. Total RNA was isolated using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific). RNA (1 µg) was used
for the first-strand cDNA synthesis with reverse transcriptase
(Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocol. Next, RT-qPCR was performed using a SYBR
Green qPCR kit (Roche Diagnostics, Indianapolis, IN, USA) according
to the manufacturer's protocol, and the LineGene 9600 PCR detection
system (Hangzhou Bioer Technology Co., Ltd, Hangzhou, China).
Relative mRNA levels of LSD1 were normalized to levels of β-actin
and normalized using the 2-ΔΔCq method (23). The cycling conditions were as follows:
95°C for 5 min for 1 cycle, then 95°C for 5 sec, 50°C (annealing)
for 30 sec and 72°C for 32 sec, for 35 cycles. Specific primer
pairs were used for LSD1 (forward, 5′-ATGTGTGAGGGAACTTGCCACC-3′ and
reverse, 5′-TTGGCACACTCCAGGGCTTTCA-3′) and β-actin (forward,
5′-GAGCAAGAGAGGCATCCTCA-3′ and reverse, 5′-AGCCTGGATAGCAACGTACA-3′)
(Shanghai GenePharma Co., Ltd.). The experiment was performed on 3
replicate samples and was repeated 3 times.
Western blot analysis
Western blot analysis was performed at the 48 h time
point after transfection. The cells were washed with PBS three
times and were lysed with radioimmunoprecipitation assay buffer
(Beyotime Institute of Biotechnology, Haimen, China). The
concentration of LSD1 protein was determined using a Bicinchoninic
Acid Protein Assay kit (Pierce™; Thermo Fisher Scientific, Inc.).
Subsequent to being separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis, the LSD1 protein was
transferred onto polyvinylidene difluoride membranes. After being
blocked with PBS with Tween 20 (PBST) and 5% dried skimmed milk for
1 h, the membranes were incubated with the monoclonal rabbit
anti-human LSD1 primary antibodies (cat. no. 2184; dilution 1:400;
Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C for 24 h.
The membranes were then washed three times with PBST and incubated
with goat anti-rabbit horseradish peroxidase-conjugated
immunoglobulin G secondary antibody (cat. no. BA1054; dilution
1:1,000; BosterBio, Wuhan, China). Proteins were detected using the
Enhanced Chemiluminescent Kit for Western (BosterBio).
Statistical analysis
Statistical comparisons of data were evaluated by
using Student's t-test (two groups) or an analysis of variance
(≥three groups), using SPSS 13.0 software (SPSS Inc., Chicago, IL,
USA). P<0.05 was used to indicate a statistically significant
difference.
Results
Transfection of LSD1-siRNA
downregulates the expression of LSD1 in K1 cells
LSD1-siRNA and negative control siRNA were
transfected into the papillary thyroid carcinoma K1 cells.
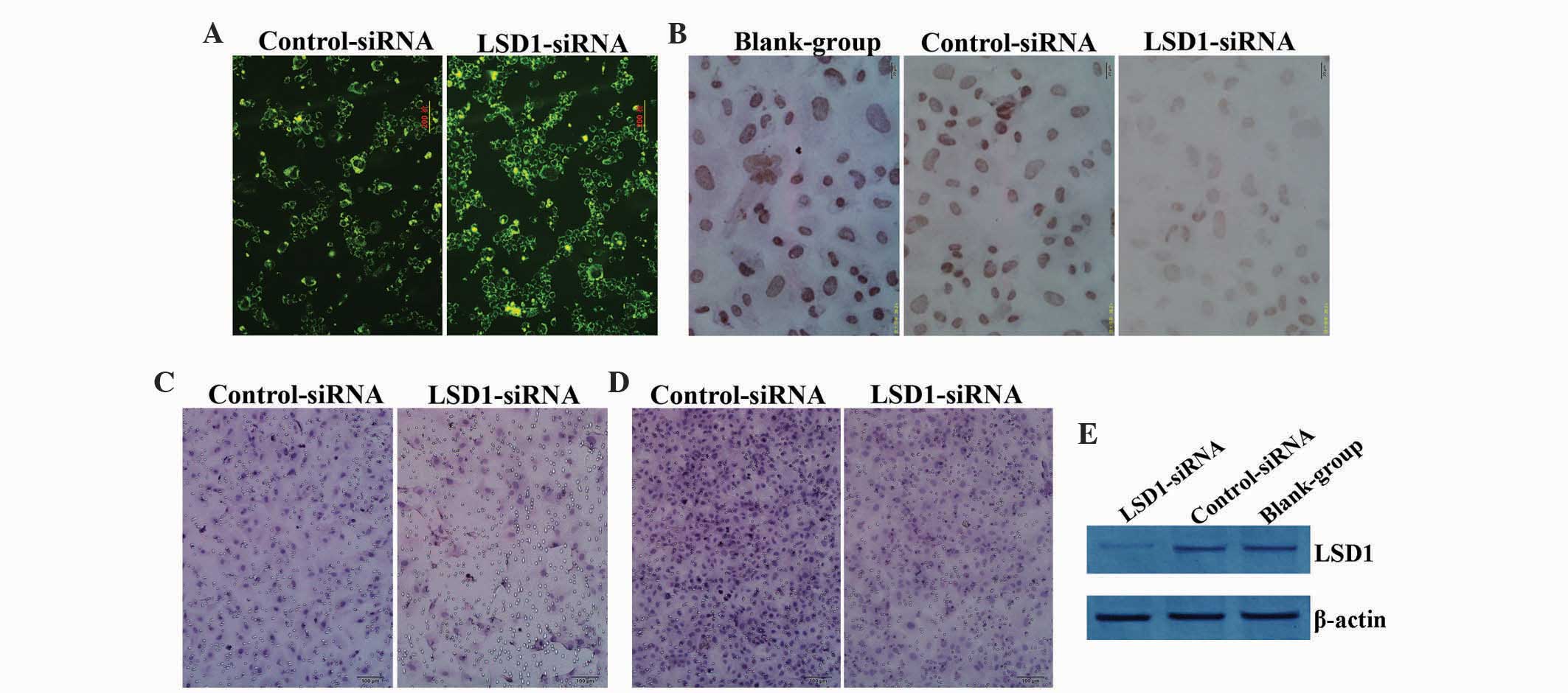
Fluorescent light was uniformly emitted in the cytoplasm, as
detected by fluorescence microscopy (Fig.
1A). Immunocytochemical (ICC) analysis was performed to detect
the expression of LSD1 in the three groups of K1 cells, namely the
blank, LSD1-siRNA and negative control siRNA groups. The results
demonstrated that the transfection of LSD1-siRNA downregulated the
expression of LSD1 (Fig. 1B). Higher
LSD1 expression was present in the nuclei of the blank and negative
control cells compared with the LSD1-siRNA K1 cells (F=15.192,
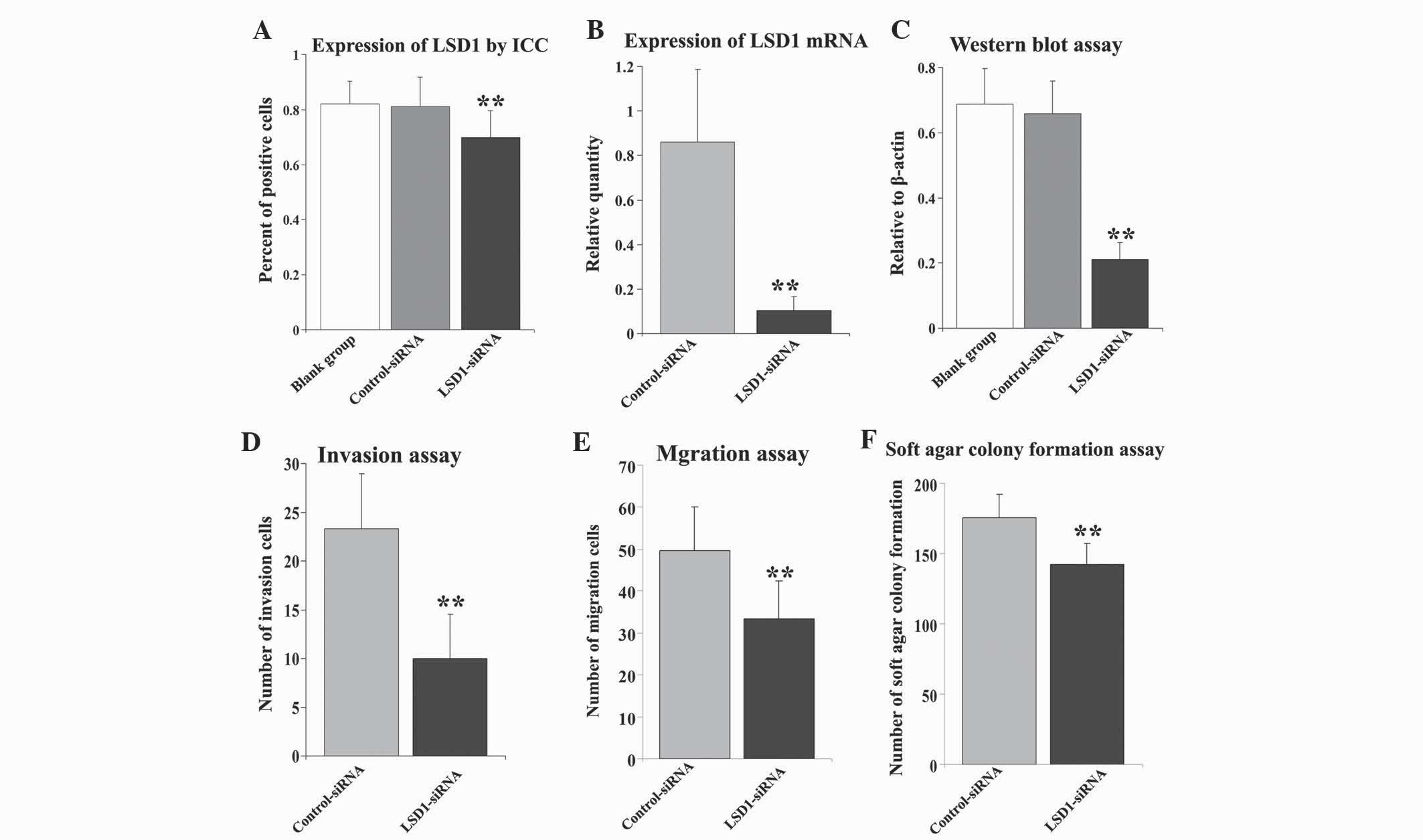
P<0.01) (Fig. 2A), with only weak
staining observed on the cover slips for cells with LSD1-siRNA.
There was no statistical difference between the blank and negative
control groups (P>0.05).
To verify the aforementioned data, the mRNA levels
of LSD1 were measured by RT-qPCR using the relative expression
value to β-actin. Transfection of siRNA targeting LSD1 resulted in
a significant decrease in the expression of LSD1. The relative
values revealed that compared with the control, transfection with
LSD1-siRNA statistically decreased the expression level of LSD1
mRNA (t=6.845, P<0.01) (Fig. 2B).
Western blot analysis was used to detect the expression level of
LSD1 protein using the β-actin gray value as a reference (Fig. 1E). The results indicated that compared
with the blank cells, the LSD1-siRNA transfected cells exhibited a
significantly reduced expression level of LSD1 protein (F=53.764,
P<0.01) (Fig. 2C), while no
statistical difference was found between the blank and scrambled
control groups (P>0.05).
Knockout of LSD1 reduces the
proliferation of K1 cells
CCK-8 was used to investigate the effect of
LSD1-siRNA on the proliferation of the K1 cells. CCK-8 detects the
number of viable cells in proliferation assays. In this process,
water-soluble tetrazolium-8 is reduced by dehydrogenase to produce
formazan, which exhibits an orange color, and is dissolved in the
culture medium. This means that the amount of formazan is directly
proportional to the number of viable cells. In the present study,
it was shown that compared with the control group, the optical
density of the K1 cells transfected with LSD1-siRNA at 24, 48 and
72 h was reduced notably to 0.419±0.047 (t=4.777, P<0.001),
0.796±0.132 (t=3.302, P=0.003) and 1.119±0.060 (t=3.017, P=0.006),
respectively (Table I).
 | Table I.OD value of the proliferation of the
K1 cells, as measured by Cell Counting kit-8 assay (mean ± standard
deviation). |
Table I.
OD value of the proliferation of the
K1 cells, as measured by Cell Counting kit-8 assay (mean ± standard
deviation).
|
| 24 h | 48 h | 72 h |
|---|
|
|
|
|
|
|---|
| Groups | OD | t-value | P-value | OD | t-value | P-value | OD | t-value | P-value |
|---|
| Control siRNA | 0.526±0.062 | 4.777 | <0.001 | 0.941±0.075 | 3.302 | 0.003 | 1.186±0.050 | 3.017 | 0.008 |
| LSD1-siRNA | 0.419±0.047 |
|
| 0.796±0.132 |
|
| 1.119±0.060 |
|
|
Absence of LSD1 inhibits the invasion
and migration of K1 cells in vitro
The cell invasion ability was investigated by
Transwell assay. Compared with the scrambled control group
(Fig. 2D), the amount of invasive
cells was reduced from 23.31±5.65 to 10.02±4.54 following
transfection with LSD1-siRNA (t=12.301, P<0.01; Fig. 1C). Similar results were observed
(Fig. 1D) showing that the number of
migrated cells was significantly higher for the negative control
cells than for the cells transfected with LSD1-siRNA (t=7.911,
P<0.01; Fig. 2E).
Loss of LSD1 downregulates the ability
of colony formation
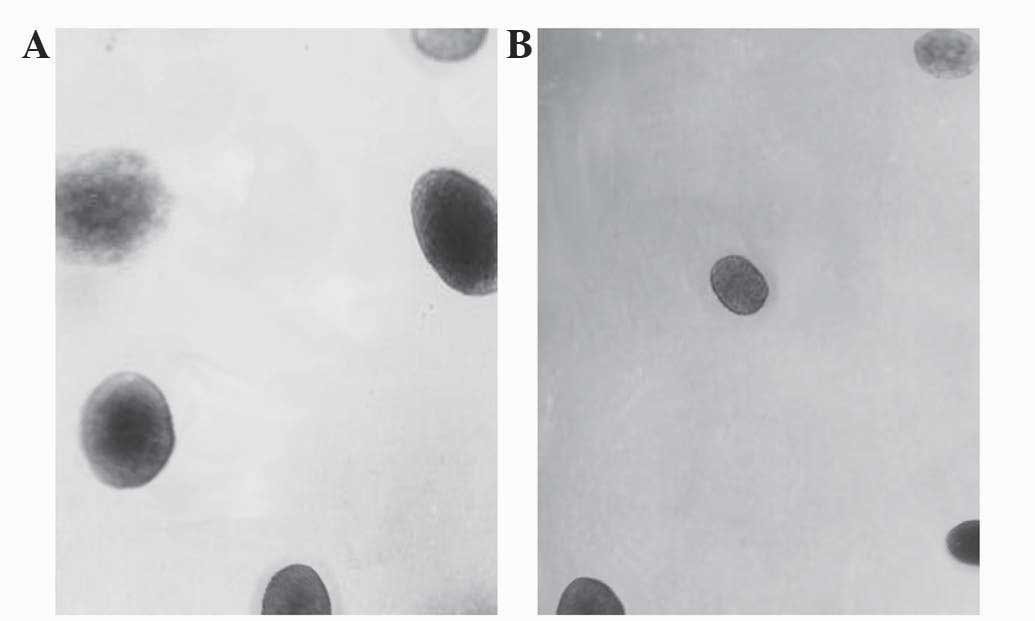
A soft agar colony formation assay was performed to
evaluate the effect of LSD1-siRNA on tumorigenicity in
vitro. Compared with in the control cells, fewer number of
colonies were observed (Fig. 3) in
the LSD1-siRNA group (t=3.612, P=0.005), as shown in Fig. 2F.
Discussion
Genetic and epigenetic modifications have been shown
to exhibit a significant role in the tumorigenesis and development
of cancer. Stable inheritance of epigenetic states is essential for
the maintenance of specific functions in cells and tissues
(24). Modification of epigenetic
states in gene regulation was suggested 40 years ago. Epigenetic
alterations are reversible, and significantly contribute to tumor
initiation and progression (24,25).
Histone methylation is a major determinant of chromatin structure
and function, and plays key roles in heterochromatin formation,
transcriptional regulation and DNA repair (26). Further study of the epigenetic
mechanisms may aid in the identification of a novel cancer-related
network, provide attractive targets for cancer and enable novel
insights into the biological characteristics of thyroid cancer to
be obtained.
LSD1 was first identified in 2004, and suggested
that methylation modifications may be reversible (19). Histone methylation shows a dynamic
process that is regulated through the alteration of methyl groups
by methylases and demethylases. As a member of the monoamine
oxidase family, LSD1 catalyzes the specific demethylation of
H3K4me1/2 and H3K9me1/2 via a process that requires flavin adenine
dinucleotide as an essential redox cofactor (19,21).
Recent studies have demonstrated that LSD1 exhibits an important
role in chromatin remodeling and transcriptional regulation in a
number of cancers (27), including
breast cancer (28), non-small cell
lung cancer (29), gastric cancer
(30), ovarian cancer (31), prostate cancer (32), osteosarcoma (33) and neuroblastoma (34). Epigenetic changes in LSD1 have been
shown to play a key role in carcinogenesis. LSD1 is regarded as a
predictive marker for malignant transformation as the
overexpression of LSD1 is associated with poor differentiation and
poor survival (31,34–36). LSD1
promotes the growth, invasion and migration of cancer cells
(28), while inhibiters of LSD1
suppress the proliferation, migration and invasion of cancer cells
(30,32,37).
Thyroid carcinoma is the most common malignancy of
the endocrine system and is rapidly increasing in incidence
(1). Therefore, clinical markers are
urgently required in order to form prognoses and diagnoses for
thyroid carcinoma. Our previous study showed that LSD1 plays an
important role in the occurrence, malignant transformation and
progression of thyroid tumors; the positive expression of LSD1 was
used to evaluate the diagnosis of thyroid carcinoma carcinogenesis
(38). Since LSD1 shows a strong
association with cancer, it may play an important role in the
molecular mechanism of thyroid carcinoma. Inhibiters of LSD1,
including miRNAs, are used to regulate histone modification
(39,40). Therefore, to continue our previous
study, the present study analyzed the effects of siRNA targeting
LSD1 in papillary thyroid carcinoma K1 cells. A focus was placed on
LSD1 in terms of its expression and the effects of this
downregulated expression on cell proliferation, colony formation,
migration and invasion in vitro. ICC data demonstrated that
siRNA targeting LSD1 lowered the expression of LSD1 in the
papillary thyroid carcinoma cells; the LSD1 mRNA and LSD1 protein
levels were significantly suppressed. siRNA also suppressed the
proliferation, and the invasion and migration abilities of the K1
cells. Tumorigenicity in vitro was evaluated by soft agar
colony formation assay, and the ability for colony formation was
reduced notably following transfection with siRNA.
Overall, the present data revealed that the
knockdown of LSD1 expression suppressed the proliferation,
migration, invasion and tumorigenicity in papillary thyroid
carcinoma cells. Due to the strong association with cancer, LSD1
could be a target for drug discovery, and efficient inhibiters of
LSD1 used in the regulation of histone modification could open up a
broad field of research in cancer therapy.
Acknowledgements
The current study was supported by the Natural
Science Foundation of Shandong (grant nos. ZR2011HM057 and
ZR2009CM070; Shandong, China).
References
|
1
|
Sherman SI: Thyroid carcinoma. Lancet.
361:501–511. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hundahl SA, Fleming ID, Fremgen AM and
Menck HR: A National Cancer Data Base report on 53,856 cases of
thyroid carcinoma treated in the U.S., 1985–1995. Cancer.
83:2638–2648. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Colonna M, Uhry Z, Guizard AV, Delafosse
P, Schvartz C, Belot A and Grosclaude P: FRANCIM network: Recent
trends in incidence, geographical distribution, and survival of
papillarythyroid cancer in France. Cancer Epidemiol. 39:511–518.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weir HK, Thompson TD, Soman A, Møller B
and Leadbetter S: The past, present, and future of cancer incidence
in the United States: 1975 through 2020. Cancer. 121:1827–1837.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McNally RJ, Blakey K, James PW, Pozo Gomez
B, Basta NO and Hale J: Increasing incidence of thyroid cancer in
Great Britain, 1976–2005: Age-period-cohort analysis. Eur J
Epidemiol. 27:615–622. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang BL, Sivasubramaniam PG, Zhang Q,
Wang J, Zhang B, Gao JD, Tang ZH, Chen GJ, Xie XM, Wang Z, et al:
Trends in radical surgical treatment methods for breast
malignancies in China: A multicenter 10-year retrospective study.
Oncologist. 20:1036–1043. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lei S, Ding Z, Ge J and Zhao D:
Association between prognostic factors and clinical outcome of
well-differentiated thyroid carcinoma: A retrospective 10-year
follow-up study. Oncol Lett. 10:1749–1754. 2015.PubMed/NCBI
|
|
8
|
Ho AS, Davies L, Nixon IJ, Palmer FL, Wang
LY, Patel SG, Ganly I, Wong RJ, Tuttle RM and Morris LG: Increasing
diagnosis of subclinical thyroid cancers leads to spurious
improvements in survival rates. Cancer. 121:1793–1799. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tuttle RM, Ball DW, Byrd D, Dilawari RA,
Doherty GM, Duh QY, Ehya H, Farrar WB, Haddad RI, Kandeel F, et al:
National Comprehensive Cancer Network: Thyroid carcinoma. J Natl
Compr Canc Netw. 8:1228–1274. 2010.PubMed/NCBI
|
|
10
|
Huang LY, Lee YL, Chou P, Chiu WY and Chu
D: Thyroid fine-needle aspiration biopsy and thyroid cancer
diagnosis: A nationwide population-based study. PLoS One.
10:e01273542015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wong CK and Wheeler MH: Thyroid nodules:
Rational management. World J Surg. 24:934–941. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Spencer C, Petrovic I, Fatemi S and
LoPresti J: Serum thyroglobulin (Tg) monitoring of patients with
differentiated thyroid cancer using sensitive (second-generation)
immunometric assays can be disrupted by false-negative and
false-positive serum thyroglobulin autoantibody misclassifications.
J Clin Endocrinol Metab. 99:4589–4599. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
American Thyroid Association (ATA)
Guidelines Taskforce on Thyroid Nodules and Differentiated Thyroid
Cancer. Cooper DS, Doherty GM, Haugen BR, Kloos RT, Lee SL, Mandel
SJ, Mazzaferri EL, McIver B, Pacini F, Schlumberger M, et al:
Revised American Thyroid Association management guidelines for
patients with thyroid nodules and differentiated thyroid cancer.
Thyroid. 19:1167–1214. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bourgeois P: A proposition for the use of
radioiodine in WDTC management. J Nucl Med. 50:328–329. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iyer NG, Morris LG, Tuttle RM, Shaha AR
and Ganly I: Rising incidence of second cancers in patients with
low-risk (T1N0) thyroid cancer who receive radioactive iodine
therapy. Cancer. 117:4439–4446. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Beaudenon-Huibregtse S, Alexander EK,
Guttler RB, Hershman JM, Babu V, Blevins TC, Moore P, Andruss B and
Labourier E: Centralized molecular testing for oncogenic gene
mutations complements the local cytopathologic diagnosis of thyroid
nodules. Thyroid. 24:1479–1487. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xing M: Molecular pathogenesis and
mechanisms of thyroid cancer. Nat Rev Cancer. 13:184–199. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saji M and Ringel MD: The PI3K-Akt-mTOR
pathway in initiation and progression of thyroid tumors. Mol Cell
Endocrinol. 321:20–28. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi Y, Lan F, Matson C, Mulligan P,
Whetstine JR, Cole PA, Casero RA and Shi Y: Histone demethylation
mediated by the nuclear amine oxidase homolog LSD1. Cell.
119:941–953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang L, He LL, Fu QT and Xu ZF: Selection
of reliable reference genes for gene expression studies in the
biofuel plant jatropha curcas using real-time quantitative PCR. Int
J Mol Sci. 14:24338–24354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Metzger E, Wissmann M, Yin N, Müller JM,
Schneider R, Peters AH, Günther T, Buettner R and Schüle R: LSD1
demethylates repressive histone marks to promote
androgen-receptor-dependent transcription. Nature. 437:436–439.
2005.PubMed/NCBI
|
|
22
|
Garcia-Bassets I, Kwon YS, Telese F,
Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J,
Escoubet-Lozach L, et al: Histone methylation-dependent mechanisms
impose ligand dependency for gene activation by nuclear receptors.
Cell. 128:505–518. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feinberg AP, Ohlsson R and Henikoff S: The
epigenetic progenitor origin of human cancer. Nature Rev Genet.
7:21–33. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Y and Reinberg D: Transcription
regulation by histone methylation: Interplay between different
covalent modifications of the core histone tails. Genes Dev.
15:2343–2360. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sorna V, Theisen ER, Stephens B, Warner
SL, Bearss DJ, Vankayalapati H and Sharma S: High-throughput
virtual screening identifies novel
N'-(1-phenylethylidene)-benzohydrazides as potent, specific and
reversible LSD1 inhibitors. J Med Chem. 56:9496–9508. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang X, Tanaka K, Yan J, Li J, Peng D,
Jiang Y, Yang Z, Barton MC, Wen H and Shi X: Regulation of estrogen
receptor α by histone methyltransferase SMYD2-mediated protein
methylation. Proc Natl Acad Sci USA. 110:17284–17289. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X
and Song Y: Over-expression of LSD1 promotes proliferation,
migration and invasion in non-small cell lung cancer. PLoS One.
7:e350652012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng YC, Duan YC, Ma JL, Xu RM, Zi X, Lv
WL, Wang MM, Ye XW, Zhu S, Mobley D, et al:
Triazole-dithiocarbamate based selective lysine specific
demethylase 1 (LSD1) inactivators inhibit gastric cancer cell
growth, invasion and migration. J Med Chem. 56:8543–8560. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Konovalov S and Garcia-Bassets I: Analysis
of the levels of lysine-specific demethylase 1 (LSD1) mRNA in human
ovarian tumors and the effects of chemical LSD1 inhibitors in
ovarian cancer cell lines. J Ovarian Res. 6:752013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rotili D, Tomassi S, Conte M, Benedetti R,
Tortorici M, Ciossani G, Valente S, Marrocco B, Labella D,
Novellino E, et al: Pan-histone demethylase inhibitors
simultaneously targeting jumonji C and lysine-specific demethylases
display high anticancer activities. J Med Chem. 57:42–55. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bennani-Baiti IM, Machado I,
Llombart-Bosch A and Kovar H: Lysine-specific demethylase 1
(LSD1/KDM1A/AOF2/BHC110) is expressed and is an epigenetic drug
target in chondrosarcoma, Ewing's sarcoma, osteosarcoma and
rhabdomyosarcoma. Human Pathol. 43:1300–1307. 2012. View Article : Google Scholar
|
|
34
|
Sakane C, Okitsu T, Wada A, Sagami H and
Shidoji Y: Inhibition of lysine-specific demethylase 1 by the
acyclic diterpenoid geranylgeranoic acid and its derivatives.
Biochem Biophys Res Commun. 444:24–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schulte JH, Lim S, Schramm A, Friedrichs
N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L,
Kuhfittig-Kulle S, et al: Lysine-specific demethylase 1 is strongly
expressed in poorly differentiated neuroblastoma: Implications for
therapy. Cancer Res. 69:2065–2071. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang Z, Li S, Song W, Li X, Li Q, Zhang
Z, Han Y, Zhang X, Miao S, Du R and Wang L: Lysine-specific
demethylase 1 (LSD1/KDM1A) contributes to colorectal tumorigenesis
via activation of the Wnt/β-catenin pathway by down-regulating
Dickkopf-1 (DKK1). PLoS One. 8:e700772013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ding D, Liu X and Guo SW: Overexpression
of lysine-specific demethylase 1 in ovarian endometriomas and its
inhibition reduces cellular proliferation, cell cycle progression
and invasiveness. Fertil Steril. 101:740–749. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kong L, Zhang G, Wang X, Zhou J, Hou S and
Cui W: Immunohistochemical expression of RBP2 and LSD1 in papillary
thyroid carcinoma. Rom J Morphol Embryol. 54:499–503.
2013.PubMed/NCBI
|
|
39
|
Cherblanc FL, Davidson RW, Di Fruscia P,
Srimongkolpithak N and Fuchter MJ: Perspectives on natural product
epigenetic modulators in chemical biology and medicine. Nat Prod
Rep. 30:605–624. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nana-Sinkam SP and Croce CM: Clinical
applications for microRNAs in cancer. Clin Pharmacol Ther.
93:98–104. 2013. View Article : Google Scholar : PubMed/NCBI
|