Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common malignancy and the third most common cause of cancer-related
mortality worldwide (1). Although
hepatectomy is one of the most effective options for HCC without
distant metastases (2–4), 80% of HCC patients experience
intrahepatic recurrence even after curative resection, and 50% die
within 5 years (5). The types of
intrahepatic recurrence are mainly divided into two types:
Intrahepatic metastasis (IM), which involves the development of HCC
foci from primary tumor cells and their spread to the remnant liver
via the portal vein before or during hepatectomy; and multicentric
occurrence (MO), which involves the development of new HCC foci due
to chronic active hepatitis or cirrhosis provoked by viruses,
alcohol, toxins or other HCC risk factors (6–9). In other
words, when the clinicopathological factors for HCC recurrence are
divided into two categories such as tumor factors and background
liver factors, IM tends to be related to tumor factors and MO is
rather related to background liver factors, since IM reflects the
characteristics of the primary tumor and MO exhibits different
genetic features from the primary lesions.
The clinical progression and outcomes of IM and MO
differ significantly, as determined by several studies (8–10).
Therefore, distinguishing between these conditions is important for
designing therapeutic strategies and predicting prognosis. Usually,
the pattern of HCC intrahepatic recurrence is determined
histologically, but it is sometimes difficult to distinguish them
(11). The present authors previously
examined mutations in the mitochondrial genome (12) and hypermethylation in tumor suppressor
gene promoters (13) in HCC, and
found distinctions between MO and IM. Our findings suggested that
MO was more common than IM. However, the study of another group
demonstrated that the proportion of two recurrence patterns was
almost the same (14). In any case,
MO recurrence pattern makes HCC totally different from other solid
carcinomas in view of recurrence.
Considering this unique MO pattern in HCC, simply
focusing on tumor tissue is insufficient; comparison of tumor
tissue and background non-tumorous liver tissue is also important.
When genetic and epigenetic changes related to HCC carcinogenesis
and recurrence are tried to be elucidated, many investigators have
attempted to evaluate only tumor tissue. However, we hypothesized
that molecular changes in the latter may directly cause MO or
indirectly affect the malignancy of the primary HCC. The present
study was designed to identify a unique molecular marker of HCC,
perhaps in background non-tumorous liver tissue, and to assess the
predictive value of the marker.
Materials and methods
Sample collection
For microarray analysis, non-tumorous liver tissue,
referred to as for corresponding normal (CN), was obtained from a
typical HCC patient during hepatectomy. The patient was a
58-year-old man; his HCC resulted from chronic hepatitis, and
recurred 3 years after resection at Nagoya University Hospital
(Nagoya, Japan). Pathology confirmed the absence of cancerous
regions from the CN sample. As controls, non-cancerous liver tissue
(not affected by hepatitis), referred to as super normal (SN), was
obtained from 11 patients with liver metastases who underwent
hepatectomy at Nagoya University Hospital. Their primary diseases
were colorectal cancer (n=5), gastrointestinal stromal tumor (n=2),
or gastric cancer, esophageal cancer, cervical cancer or tongue
cancer (n=1 each). The samples were collected between January 1998
and December 2011
For reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) assays, HCC and CN tissue was collected
from 158 consecutive patients who underwent curative primary
hepatectomy at Nagoya University Hospital between January 1998 and
December 2011. Median patient age was 65 years (range, 37–84
years), and the male:female ratio was 84:16. The median follow-up
duration was 48.5 months (range, 0.3–193.8 months). Patient
characteristics are summarized in Table
I. All tumor tissue samples were histologically confirmed as
HCC.
 | Table I.Characteristic of patients with
hepatocellular carcinoma (n=158). |
Table I.
Characteristic of patients with
hepatocellular carcinoma (n=158).
| Characteristic | Value |
|---|
| Age (years), median
(range) | 65 (37–84) |
| Sex (male:female), n
(%) | 132 (84):26 (16) |
| Viral infection
(HBV:HCV:non-HBV/HCV), n (%) | 41 (26):92 (58):28
(18) |
| Child-Pugh
classification (A:B), n (%) | 148 (94):9 (6) |
| Liver damage
classification (A:B:C), n (%) | 126 (83):25 (16):1
(1) |
| Albumin (mg/dl),
median (range) | 3.9 (2.3–4.9) |
| Total bilirubin,
(mg/dl), median (range) | 0.7 (0.2–7.3) |
| PT (%), median
(range) | 89.7
(46.9–138.0) |
| AFP (ng/ml), median
(range) | 17
(0.8–119,923.0) |
| Tumor size (cm),
median (range) | 3.50
(0.15–15.00) |
| Tumor number
(single:multiple), n (%) | 124 (78):34
(22) |
| ICG-R15 (%), median
(range) | 11.5
(1.6–35.2) |
| Japanese stage
(I:II:III:IV), n (%) | 17 (11):82 (52):40
(26):17 (11) |
All surgically obtained tissue samples were
immediately frozen in liquid nitrogen and stored at −80°C until
analysis. This study was approved by our institutional review board
of at Nagoya University (Nagoya, Japan), and all patients provided
written informed consent.
Microarray procedure
Total RNA was extracted from the CN and SN samples
using a miRNeasy Mini-kit (Qiagen, Inc., Valencia, CA, USA). The 11
SN samples were mixed to eliminate individual differences. RNA
integrity was assessed using an Agilent 2100 bioanalyzer (Agilent
Technologies, Inc., Santa Clara, CA, USA); an RNA integrity number
≥8 was indicative of good quality RNA. RNA was labeled with
cyanine-3 dye using a Quick Amp Labeling kit (Agilent Technologies,
Inc.) and hybridized to Agilent whole human genome (4×44 K)
microarrays (Agilent Technologies, Inc.) for 17 h in a rotating
SciGene model 700 oven (SciGene, Sunnyvale, CA, USA). The arrays
were scanned with a DNA microarray scanner (Agilent Technologies,
Inc.), and the data were feature-extracted using Feature Extraction
software 10.5.1.1 (Agilent Technologies, Inc.) and statistically
analyzed using the default settings for GeneSpring GX 11.0.1
software (Agilent Technologies, Inc.) (15).
RT-qPCR
PCR was performed using SYBR Premix Ex Taq II
(Takara Bio, Inc., Otsu, Japan) under the following conditions:
95°C for 10 sec, and 40 cycles at 95°C for 5 sec and 60°C for 30
sec. The SYBR Green signal was detected in real time using a
StepOne Plus Real-Time PCR system (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The PCR primers used to generate a 144-bp
fragment of AKR1B10 were 5′-GTGGGGGAAGCCATCCAAGA-3′ (sense,
exon 2) and 5′-CAGCTTCAGGTCCTTGAGGG-3′ (antisense, exon 3). The
primers used to generate an 85-bp fragment of Ras-related protein
Rab-25 (RAB25) were 5′-AAAGTGACCTCAGCCAGGCC-3′ (sense, exon
3) and 5′-GTCTCCAGGAAGAGCAGTCC-3′ (antisense, exon 4). The primers
used to generate a 95-bp fragment of erythrocyte membrane protein
band 4.1 like 4B (EPB41L4B) were 5′-AGCCTCTCACTGACCCTGGA-3′
(sense, exon 19) and 5′-GCAGGTGTTCCTGGACTCAG-3′ (antisense, exon
20). The primers used to generate a 145-bp fragment of lipoma HMGIC
fusion partner-like 1 (LHFPL1) were
5′-GGCTGCTGATAAGCTCAGGC-3′ (sense, exon 3) and
5′-GCTCCACCTCCAGCACAGTA-3′ (antisense, exon 4). GAPDH
expression was quantified in each sample for standardization
purposes. The primers used to generate a 226-bp fragment of GAPDH
were 5′-GAAGGTGAAGGTCGGAGTC-3′ (sense) and
5′-GAAGATGGTGATGGGATTTC-3′ (antisense). All RT-qPCR experiments
were performed at least three times, including negative controls
without a template. The absolute quantification method was used to
determine input copy number, which is based on a standard curve,
due to its advantages in studies with large sample numbers
(16). The expression of each gene
was calculated as follows: Value of the expressed gene/value of
GAPDH × 103.
Statistical analysis
Continuous variables were expressed as median and
range and compared using the Mann-Whitney U-test.
Categorical variables were compared using the χ2 or
Fisher's exact tests, as appropriate. Recurrence-free survival
(RFS) and overall survival (OS) rates were estimated using the
Kaplan-Meier method and compared using the log-rank test.
Univariate and multivariate Cox proportional hazards models were
used to determine the independent risk factors associated with RFS
and OS. All statistical analyses were performed using JMP Pro
software version 11.0.0 (SAS International Inc., Cary, NC, USA).
P-values are two-tailed, and P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression profiling via microarray
analysis
To identify novel tumor-related genes in the normal
liver tissue surrounding an HCC, the gene expression profiles of a
CN sample and the pooled SN samples were compared. Microarray
analysis revealed that AKR1B10, RAB25,
EPB41L4B and LHFPL1 were upregulated in the CN sample
(Table II). We focused on
AKR1B10 as a CN-expressed prognostic factor.
 | Table II.Hepatocellular carcinoma-related
genes identified in microarrays. |
Table II.
Hepatocellular carcinoma-related
genes identified in microarrays.
| Gene | RefSeq accession
#a | Fold change CN vs.
SN | Regulation CN vs.
SN | CN value | SN value | Flagsb CN | Flags SN |
|---|
| AKR1B10 | NM_020299 | 55.37 | Up | 14609.94 | 218.97 | Detected | Detected |
| RAB25 | NM_020387 | 43.96 | Up |
527.62 |
9.96 | Detected | Not detected |
|
EPB41L4B | NM_019114 | 36.51 | Up |
272.23 |
6.19 | Detected | Not detected |
| LHFPL1 | NM_178175 | 35.45 | Up |
290.66 |
6.80 | Detected | Not detected |
RT-qPCR analysis of HCC and CN
tissue
As determined via RT-qPCR, overall AKR1B10
expression (expression score/GAPDH × 1,000) was
significantly higher in HCC tissue (median, 9.2200; range,
0.0003–611.0200; n=158) than in CN tissue (median, 0.5461; range,
0.0018–69.0300; n=158) tissues (P<0.001). However, there was no
significant difference in expression between SN and CN tissue
(Fig. 1).
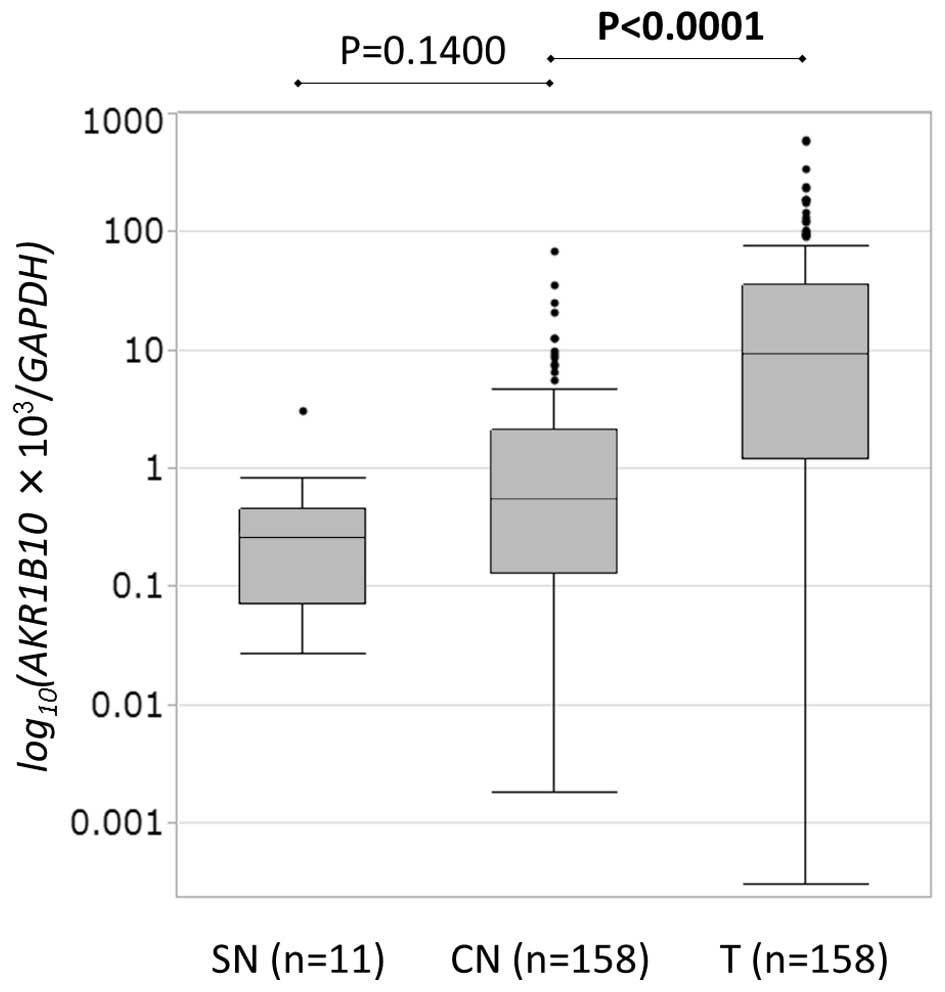 | Figure 1.AKR1B10 messenger RNA levels in
hepatocellular carcinoma and non-tumor tissue were quantified via
reverse transcription-quantitative polymerase chain reaction.
AKR1B10 expression (AKR1B10 score/GAPDH score ×
1,000) was significantly higher in T tissue (median, 9.2200; range,
0.0003–611.0200; n=158) than in CN tissue (median, 0.5461; range,
0.0018–69.0300; n=158) (P<0.001). However, there was no
significant difference between expression in CN tissue and SN
tissue (median, 0.2616; range, 0.0272–3.1250; n=11 vs. median,
0.5461; range, 0.0018–69.0300; n=158, respectively) (P=0.1400). CN,
corresponding normal; SN, super normal; T, tumor; AKR1B10,
aldo-keto reductase family 1, member B10. |
Correlation between AKR1B10 expression
and the clinicopathological characteristics of HCC
AKR1B10 expression significantly correlated
with liver damage (Child-Pugh score B or C vs. A) (P=0.035) and
capsule infiltration (P=0.0284) (Table
III). For example, 18 of 26 cases with liver damage scores of B
or C had significantly greater amounts of AKR1B10 messenger RNA
(mRNA) in CN tissue than in HCC tissue.
 | Table III.Association between the
clinicopathological characteristics of patients with hepatocellular
carcinoma and AKR1B10 expression. |
Table III.
Association between the
clinicopathological characteristics of patients with hepatocellular
carcinoma and AKR1B10 expression.
|
| AKR1B10
expression |
|---|
|
|
|
|---|
| Clinicopathological
factor | T<CN | T≥CN | P-value |
|---|
| Age (years) |
|
| 0.8321 |
|
≥65 | 13 | 69 |
|
|
<65 | 13 | 63 |
|
| Gender |
|
| 0.3191 |
|
Male | 20 | 112 |
|
|
Female | 6 | 20 |
|
| Virus
infection |
|
| 0.9517 |
|
HCV | 15 | 77 |
|
|
Others | 11 | 55 |
|
| Albumin
(mg/dl) |
|
| 0.1501 |
|
<3.5 | 8 | 24 |
|
|
≥3.5 | 18 | 107 |
|
| PT (%) |
|
| 0.3187 |
|
<70 | 5 | 14 |
|
|
≥70 | 21 | 117 |
|
| ICG-R15 (%) |
|
| 0.1183 |
|
≥15 | 7 | 22 |
|
|
<15 | 10 | 73 |
|
| Liver
cirrhosis |
|
| 0.1092 |
|
(+) | 5 | 50 |
|
|
(−) | 20 | 80 |
|
| Child-Pugh |
|
| 0.1705 |
| B | 3 |
6 |
|
| A | 23 | 125 |
|
| Liver damage |
|
| 0.0305 |
| B or
C | 8 | 18 |
|
| A | 17 | 109 |
|
| Tumor number |
|
| 0.6017 |
|
Multiple | 4 | 30 |
|
|
Solitary | 22 | 102 |
|
| Tumor size
(cm) |
|
| 0.1264 |
| ≥2 | 24 | 103 |
|
|
<2 | 1 | 22 |
|
| AFP (ng/ml) |
|
| 0.0556 |
|
≥20 | 16 | 53 |
|
|
<20 | 10 | 76 |
|
|
Differentiation |
|
| 1.0000 |
|
Poor | 2 | 10 |
|
|
Well/moderate | 23 | 119 |
|
| Growth form |
|
| 0.5454 |
|
Infiltrative | 5 | 18 |
|
|
Expansive | 21 | 111 |
|
| Formation of
capsule |
|
| 0.1036 |
|
(−) | 4 | 42 |
|
|
(+) | 22 | 90 |
|
| Infiltration to
capsule |
|
| 0.0284 |
|
(+) | 19 | 69 |
|
|
(−) | 6 | 63 |
|
| Septal
formation |
|
| 0.2419 |
|
(−) | 5 | 44 |
|
|
(+) | 19 | 86 |
|
| Serosal
invasion |
|
| 0.2108 |
|
(+) | 8 | 26 |
|
|
(−) | 16 | 95 |
|
| Portal vein or
hepatic vein invasion |
|
| 0.8830 |
|
(+) | 7 | 36 |
|
|
(−) | 19 | 91 |
|
| Surgical
margin |
|
| 0.7663 |
|
(+) | 3 | 21 |
|
|
(−) | 21 | 102 |
|
| Japanese stage |
|
| 0.7670 |
|
III/IV | 9 | 49 |
|
|
I/II | 17 | 81 |
|
Association between AKR1B10 expression
and prognosis in 158 HCC cases
The clinical relevance of AKR1B10 expression
was assessed in terms of its prognostic ability in HCC. The 158 HCC
cases were divided into two groups based on AKR1B10
expression levels in HCC or CN tissue. Analysis of several pairings
did not reveal any significant correlation between AKR1B10
expression and RFS or OS. The cases were also grouped as follows:
i) AKR1B10 expression in HCC tissue was higher than or equal
to AKR1B10 expression in CN tissue (HCC≥CN, n=132) and ii)
AKR1B10 expression was lower in HCC tissue than in CN tissue
(HCC<CN, n=26). The HCC<CN group had significantly worse RFS
(P=0.022) and OS (P<0.0001) than the HCC≥CN group (Fig. 2).
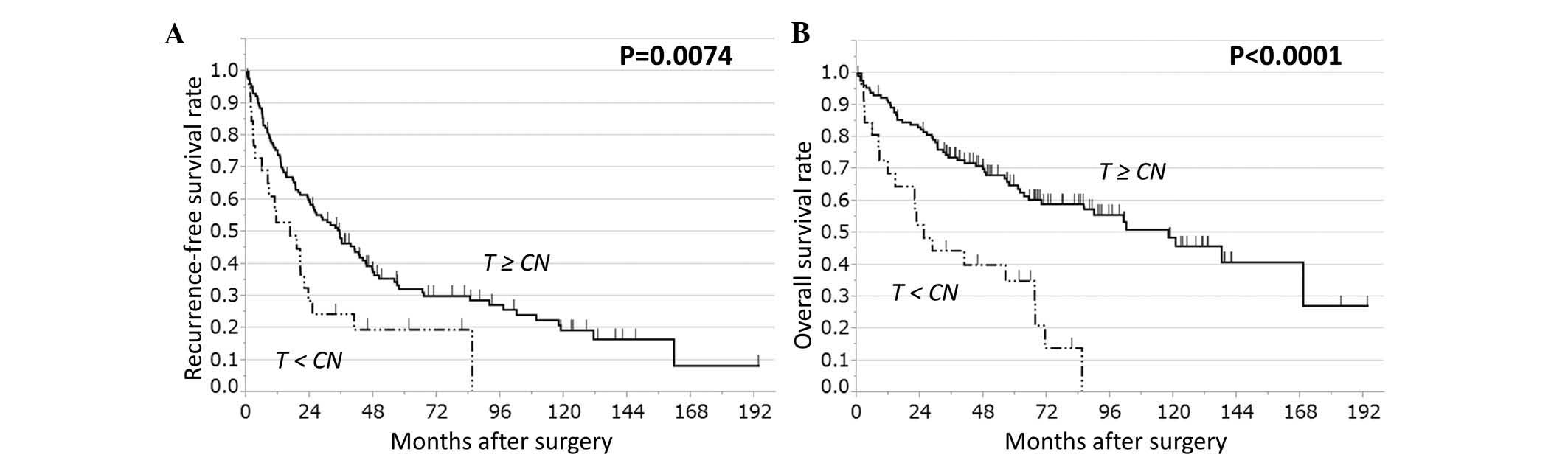 | Figure 2.RFS and OS rates in patients with HCC
based on AKR1B10 expression in T and CN tissue. HCC cases
(n=158) were stratified on the basis of AKR1B10 expression
in T tissue relative to CN tissue, either T≥CN (n=132) or T<CN
(n=26). T<CN cases have significantly worse (A) RFS and (B) OS
rates than T≥CN cases (log-rank test: RFS, P=0.0074; OS,
P<0.0001). CN, corresponding normal; T, tumor; RFS,
recurrence-free survival; OS, overall survival; AKR1B10,
aldo-keto reductase family 1, member B10; HCC, hepatocellular
carcinoma. |
A multivariate analysis with those factors that
displayed significant difference in an univariate analysis was next
performed. The risk associated with this approach is that certain
variables that were not significant in univariate analysis may have
the potential to be significant in multivariate analysis. However,
in situations when there is not enough information about the
importance of each factor, this approach seems to be feasible. In
the multivariate analysis, the finding for OS was confirmed
(P=0.0011) (Table IV), whereas the
finding for RFS was not (P=0.1884) (Table
V). Multivariate analysis also revealed significant
associations between survival (both OS and RFS) and serosal
invasion (P=0.0407), and between OS and vascular invasion
(P=0.0104) (Tables IV and V). Our findings suggest that the ratio of
AKR1B10 mRNA levels in HCC and CN tissues predicts prognosis after
curative hepatectomy, with low expression in HCC tissue relative to
normal liver tissue being indicative of poor prognosis.
 | Table IV.Univariate and multivariate analysis
of overall survival. |
Table IV.
Univariate and multivariate analysis
of overall survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinicopathological
factor | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) |
|
| ≥65 vs.
<65 | 1.55 | 0.98–2.48 | 0.0585 |
|
| Gender |
|
| Male
vs. female | 1.25 | 0.69–2.52 | 0.4709 |
|
| Virus
infection |
|
| HCV vs.
others | 1.50 | 0.94–2.46 | 0.0848 |
|
| Albumin
(mg/dl) |
|
| <3.5
vs. ≥3.5 | 1.65 | 0.95–2.75 | 0.0731 |
|
| PT (%) |
|
| <70
vs. ≥70 | 1.75 | 0.90–3.12 | 0.0914 |
|
| ICG-R15 (%) |
|
| ≥15 vs.
<15 | 1.74 | 0.92–3.19 | 0.0836 |
|
| Liver
cirrhosis |
|
| (+) vs.
(−) | 1.29 | 0.80–2.06 | 0.2876 |
|
| Child-Pugh |
|
| B vs.
A | 1.63 | 0.63–3.48 | 0.2824 |
|
| Liver damage |
|
| B or C
vs. A | 2.07 | 1.16–3.50 | 0.0149 |
|
| Tumor number |
|
|
Multiple vs. single | 1.68 | 0.99–2.75 | 0.0534 |
|
| Tumor size
(cm) |
|
| ≥2 vs.
<2 | 2.02 | 0.95–5.23 | 0.0681 |
|
| AFP (ng/ml) |
|
| ≥20 vs.
<20 | 2.06 | 1.30–3.30 | 0.0022 | 1.40 | 0.79–2.46 | 0.2368 |
|
Differentiation |
|
| Poor
vs. well/moderate | 2.29 | 1.05–4.38 | 0.0365 | 1.35 | 0.43–3.46 | 0.5732 |
| Growth form |
|
|
Infiltrative vs.
expansive | 1.57 | 0.86–2.71 | 0.1334 |
|
| Formation of
capsule |
|
| (−) vs.
(+) | 0.91 | 0.54–1.49 | 0.7282 |
|
| Infiltration to
capsule |
|
| (+) vs.
(−) | 0.97 | 0.61–1.54 | 0.9168 |
|
| Septal
formation |
|
| (−) vs.
(+) | 1.05 | 0.64–1.70 | 0.8221 |
|
| Serosal
invasion |
|
| (+) vs.
(−) | 2.51 | 1.48–4.17 | 0.0009 | 1.86 | 1.02–3.28 | 0.0407 |
| Portal vein or
hepatic vein invasion |
|
| (+) vs.
(−) | 2.25 | 1.38–3.62 | 0.0014 | 2.15 | 1.20–3.76 | 0.0104 |
| Surgical
margin |
|
| (+) vs.
(−) | 1.84 | 1.00–3.18 | 0.0498 | 1.37 | 0.63–2.71 | 0.4034 |
| Japanese stage |
|
| III/IV
vs. I/II | 1.56 | 0.97–2.47 | 0.0622 |
|
| AKR1B10
expression |
|
| T<CN
vs. T≥CN | 3.14 | 1.81–5.23 | <0.0001 | 3.06 | 1.58–5.71 | 0.0011 |
 | Table V.Univariate and multivariate analysis
of recurrence-free survival. |
Table V.
Univariate and multivariate analysis
of recurrence-free survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinicopathological
factor | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) |
|
| ≥65 vs.
<65 | 1.11 | 0.77–1.61 | 0.5539 |
|
| Gender |
|
| Male
vs. female | 1.57 | 0.95–2.78 | 0.0755 |
|
| Virus
infection |
|
| HCV vs.
others | 1.45 | 1.00–2.15 | 0.0496 | 1.14 | 0.65–2.02 | 0.6440 |
| Albumin
(mg/dl) |
|
| <3.5
vs. ≥3.5 | 1.74 | 1.10–2.68 | 0.0189 | 1.48 | 0.76–2.80 | 0.2373 |
| PT (%) |
|
| <70
vs. ≥70 | 1.33 | 0.75–2.20 | 0.3092 |
|
| ICG-R15 (%) |
|
| ≥15 vs.
<15 | 2.31 | 1.40–3.71 | 0.0012 | 1.72 | 0.90–3.19 | 0.0939 |
| Liver
cirrhosis |
|
| (+) vs.
(−) | 1.23 | 0.84–1.80 | 0.2742 |
|
| Child-Pugh |
|
| B vs.
A | 1.66 | 0.74–3.20 | 0.1981 |
|
| Liver damage |
|
| B or C
vs. A | 1.87 | 1.15–2.92 | 0.0121 |
|
| Tumor number |
|
|
Multiple vs. single | 1.65 | 1.05–2.49 | 0.0277 | 1.41 | 0.66–2.72 | 0.3432 |
| Tumor size
(cm) |
|
| ≥2 vs.
<2 | 1.81 | 1.03–3.49 | 0.0352 | 0.88 | 0.42–1.97 | 0.7536 |
| AFP (ng/ml) |
|
| ≥20 vs.
<20 | 1.43 | 0.98–2.08 | 0.0614 |
|
|
Differentiation |
|
| Poor
vs. well/moderate | 1.44 | 0.70–2.64 | 0.2899 |
|
| Growth form |
|
|
Infiltrative vs.
expansive | 1.18 | 0.68–1.93 | 0.5158 |
|
| Formation of
capsule |
|
| (−) vs.
(+) | 0.67 | 0.43–1.01 | 0.0577 |
|
| Infiltration to
capsule |
|
| (+) vs.
(−) | 1.18 | 0.82–1.72 | 0.3606 |
|
| Septal
formation |
|
| (−) vs.
(+) | 1.01 | 0.67–1.50 | 0.9240 |
|
| Serosal
invasion |
|
| (+) vs.
(−) | 2.75 | 1.76–4.19 | <0.0001 | 2.23 | 1.17–4.14 | 0.0151 |
| Portal vein or
hepatic vein invasion |
|
| (+) vs.
(−) | 1.94 | 1.28–2.88 | 0.0019 | 1.56 | 0.84–2.79 | 0.1481 |
| Surgical
margin |
|
| (+) vs.
(−) | 1.16 | 0.68–1.86 | 0.5608 |
|
| Japanese stage |
|
| III/IV
vs. I/II | 1.37 | 0.93–1.99 | 0.1012 |
|
| AKR1B10
expression |
|
| T<CN
vs. T≥CN | 1.90 | 1.14–3.01 | 0.0138 | 1.59 | 0.78–3.04 | 0.1884 |
Discussion
A major obstacle in HCC treatment is the high
frequency of tumor recurrence even after curative resection and
liver transplantation (17), and even
in cases of small, well-differentiated tumors (18). We previously reported that MO was more
common than IM in HCC (12,13). Accordingly, the detection of
metachronous multicentric recurrent carcinoma at an early stage and
the instigation of appropriate therapy may prolong survival
(14). Furthermore, evaluation of CN
liver tissue may provide useful information regarding MO risk along
with the evaluation of the cancer tissue.
In the present study, microarray analysis revealed
that four genes, including AKR1B10, were more highly
expressed in CN tissue than in SN tissue. We decided to further
investigate AKR1B10 as a potential non-tumor prognostic
predictor of HCC outcome.
NAD(P)H-dependent oxidoreductases catalyze the
reduction of a variety of carbonyl compounds, and AKR1B10, a member
of this superfamily, efficiently reduces aliphatic and aromatic
aldehydes (19). AKR1B10 is expressed
in the kidney, nasal epithelium, liver and cervical epithelium,
according to GeneCards (http://www.genecards.org/), and in several cancer cell
lines, including liver, kidney, lung, colon, brain, prostate,
cervix and breast (20). In non-small
cell lung cancers, especially squamous cell carcinomas, AKR1B10
expression highly correlates with smoking (21). As shown by Zhang et al
(22), AKR1B10 promotes pancreatic
carcinogenesis via modulation of the K-RAS/E-cadherin pathway.
Several studies demonstrated AKR1B10 expression in HCC via
immunohistochemistry (23–26). To the best of our knowledge, however,
there are no reports comparing AKR1B10 expression in HCC and
CN tissue or determining its association with HCC prognosis.
In this study, and as determined by RT-qPCR,
AKR1B10 expression was significantly higher in HCC than in
CN samples, but not significantly different in SN and CN samples.
Although the result of microarray analysis demonstrated higher
AKR1B10 expression in CN than in SN, it was only one typical
HCC case that was used to compare. Therefore, AKR1B10
expression is not an HCC marker specific to CN tissue. There was no
significant correlation between AKR1B10 expression and HCC
prognosis when these parameters were compared in 158 HCC surgical
samples. However, when samples were grouped according to
AKR1B10 expression in HCC tissue relative to CN tissue
(expression in HCC≥CN or expression in HCC<CN), it was observed
that HCC<CN was associated with significantly worse RFS and OS
rates, and that AKR1B10 was an independent risk factor for OS in a
multivariate analysis using a Cox hazard model.
Via immunohistochemistry, a previous study reported
that AKR1B10 levels were higher in HCC than in surrounding tissue,
and suggested that this enzyme may be useful for distinguishing HCC
from benign hepatic tumors (25).
Other studies found that HCCs with low AKR1B10 levels were highly
proliferative, poorly differentiated and had a poor prognosis
(23,24). In agreement with these studies, the
present study identified that AKR1B10 mRNA levels were elevated in
HCCs, and that prognosis (RFS and OS) was worse in cases in which
the amounts of AKR1B10 mRNA were lower in HCC tissue than in CN
tissue. AKR1B10 expression also correlated with capsule
infiltration and liver damage. Capsule infiltration may cause
genetic changes in non-tumorous liver tissue adjacent to HCCs and
may be related to cell invasiveness. Liver damage may increase
AKR1B10 expression in CN tissue. Sato et al (27) demonstrated that chronic hepatitis
C-mediated AKR1B10 upregulation correlated with serum
alpha-fetoprotein levels and HCC recurrence. Higher AKR1B10
expression in CN tissue may be associated with the oncogenic status
of the background liver tissue, while lower AKR1B10
expression in HCC tissue may indicate HCC malignancy.
Interestingly, factors considered to be prognostic such as vascular
invasion (28–30) were unrelated to AKR1B10
expression. Therefore, changes in AKR1B10 expression may be
worth considering as prognostic predictors, even if other
prognostic factors are not observed.
HCC-resected patients with a low tumor-to-CN ratio
of AKR1B10 mRNA could be offered a more intense follow-up program
consisting of frequent examinations with ultrasonography or
computed tomography, and adjuvant therapy should be considered if
possible in the future. Further study is required to elucidate how
genes such as AKR1B10 in tumor-adjacent normal liver tissue
respond to HCC development and recurrence. Such knowledge would
facilitate the design of novel approaches for prediction,
prevention and treatment of HCC.
In conclusion, our findings suggest that the ratio
of AKR1B10 expression in HCC tissue and background
non-tumorous liver tissue may be a prognostic indicator in patients
receiving curative hepatectomies. Its combination with other
prognostic factors may more accurately predict HCC prognosis.
Acknowledgements
This work was supported by the Japanese Society for
the Promotion of Science (Tokyo, Japan) through a KAKENHI
Grant-in-Aid for Scientific Research (C) (grant number
25461979).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rahbari NN, Mehrabi A, Mollberg NM, Müller
SA, Koch M, Büchler MW and Weitz J: Hepatocellular carcinoma:
Current management and perspectives for the future. Ann Surg.
253:453–469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kobayashi A, Kawasaki S, Miyagawa S, Miwa
S, Noike T, Takagi S, Iijima S and Miyagawa Y: Results of 404
hepatic resections including 80 repeat hepatectomies for
hepatocellular carcinoma. Hepatogastroenterology. 53:736–741.
2006.PubMed/NCBI
|
|
4
|
Chen MS, Li JQ, Zheng Y, Guo RP, Liang HH,
Zhang YQ, Lin XJ and Lau WY: A prospective randomized trial
comparing percutaneous local ablative therapy and partial
hepatectomy for small hepatocellular carcinoma. Ann Surg.
243:321–328. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Taura K, Ikai I, Hatano E, Fujii H, Uyama
N and Shimahara Y: Implication of frequent local ablation therapy
for intrahepatic recurrence in prolonged survival of patients with
hepatocellular carcinoma undergoing hepatic resection: An analysis
of 610 patients over 16 years old. Ann Surg. 244:265–273. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen PJ, Chen DS, Lai MY, Chang MH, Huang
GT, Yang PM, Sheu JC, Lee SC, Hsu HC and Sung JL: Clonal origin of
recurrent hepatocellular carcinomas. Gastroenterology. 96:527–529.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Imamura H, Matsuyama Y, Tanaka E, Ohkubo
T, Hasegawa K, Miyagawa S, Sugawara Y, Minagawa M, Takayama T,
Kawasaki S and Makuuchi M: Risk factors contributing to early and
late phase intrahepatic recurrence of hepatocellular carcinoma
after hepatectomy. J Hepatol. 38:200–207. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Portolani N, Coniglio A, Ghidoni S,
Giovanelli M, Benetti A, Tiberio GA and Giulini SM: Early and late
recurrence after liver resection for hepatocellular carcinoma:
Prognostic and therapeutic implications. Ann Surg. 243:229–235.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cucchetti A, Piscaglia F, Caturelli E,
Benvegnù L, Vivarelli M, Ercolani G, Cescon M, Ravaioli M, Grazi
GL, Bolondi L and Pinna AD: Comparison of recurrence of
hepatocellular carcinoma after resection in patients with cirrhosis
to its occurrence in a surveilled cirrhotic population. Ann Surg
Oncol. 16:413–422. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Matsuda M, Fujii H, Kono H and Matsumoto
Y: Surgical treatment of recurrent hepatocellular carcinoma based
on the mode of recurrence: Repeat hepatic resection or ablation are
good choices for patients with recurrent multicentric cancer. J
Hepatobiliary Pancreat Surg. 8:353–359. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morimoto O, Nagano H, Sakon M, Fujiwara Y,
Yamada T, Nakagawa H, Miyamoto A, Kondo M, Arai I, Yamamoto T, et
al: Diagnosis of intrahepatic metastasis and multicentric
carcinogenesis by microsatellite loss of heterozygosity in patients
with multiple and recurrent hepatocellular carcinomas. J Hepatol.
39:215–221. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nomoto S, Yamashita K, Koshikawa K, Nakao
A and Sidransky D: Mitochondrial D-loop mutations as clonal markers
in multicentric hepatocellular carcinoma and plasma. Clin Cancer
Res. 8:481–487. 2002.PubMed/NCBI
|
|
13
|
Nomoto S, Kinoshita T, Kato K, Otani S,
Kasuya H, Takeda S, Kanazumi N, Sugimoto H and Nakao A:
Hypermethylation of multiple genes as clonal markers in
multicentric hepatocellular carcinoma. Br J Cancer. 97:1260–1265.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kumada T, Nakano S, Takeda I, Sugiyama K,
Osada T, Kiriyama S, Sone Y, Toyoda H, Shimada S, Takahashi M and
Sassa T: Patterns of recurrence after initial treatment in patients
with small hepatocellular carcinoma. Hepatology. 25:87–92. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stangegaard M: Gene expression analysis
using agilent DNA microarrays. Methods Mol Biol. 529:133–145. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsai SJ and Wiltbank MC: Quantification of
mRNA using competitive RT-PCR with standard-curve methodology.
Biotechniques. 21:862–866. 1996.PubMed/NCBI
|
|
17
|
Wang Z, Zhang G, Wu J and Jia M: Adjuvant
therapy for hepatocellular carcinoma: Current situation and
prospect. Drug Discov Ther. 7:137–143. 2013.PubMed/NCBI
|
|
18
|
Takenaka K, Adachi E, Nishizaki T,
Hiroshige K, Ikeda T, Tsuneyoshi M and Sugimachi K: Possible
multicentric occurrence of hepatocellular carcinoma: A
clinicopathological study. Hepatology. 19:889–894. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao D, Fan ST and Chung SS: Identification
and characterization of a novel human aldose reductase-like gene. J
Biol Chem. 273:11429–11435. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Endo H, Shiroki T, Nakagawa T, Yokoyama M,
Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, et
al: Enhanced expression of long non-coding RNA HOTAIR is associated
with the development of gastric cancer. PLoS One. 8:e770702013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kang MW, Lee ES, Yoon SY, Jo J, Lee J, Kim
HK, Choi YS, Kim K, Shim YM, Kim J and Kim H: AKR1B10 is associated
with smoking and smoking-related non-small-cell lung cancer. J Int
Med Res. 39:78–85. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang W, Li H, Yang Y, Liao J and Yang GY:
Knockdown or inhibition of aldo-keto reductase 1B10 inhibits
pancreatic carcinoma growth via modulating Kras-E-cadherin pathway.
Cancer Lett. 355:273–280. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heringlake S, Hofdmann M, Fiebeler A,
Manns MP, Schmiegel W and Tannapfel A: Identification and
expression analysis of the aldo-ketoreductase1-B10 gene in primary
malignant liver tumours. J Hepatol. 52:220–227. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schmitz KJ, Sotiropoulos GC, Baba HA,
Schmid KW, Müller D, Paul A, Auer T, Gamerith G and Loeffler-Ragg
J: AKR1B10 expression is associated with less aggressive
hepatocellular carcinoma: A clinicopathological study of 168 cases.
Liver Int. 31:810–816. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Matkowskyj KA, Bai H, Liao J, Zhang W, Li
H, Rao S, Omary R and Yang GY: Aldoketoreductase family 1B10
(AKR1B10) as a biomarker to distinguish hepatocellular carcinoma
from benign liver lesions. Hum Pathol. 45:834–843. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsuzura H, Genda T, Sato S, Murata A,
Kanemitsu Y, Narita Y, Ishikawa S, Kikuchi T, Mori M, Hirano K, et
al: Expression of aldo-keto reductase family 1 member b10 in the
early stages of human hepatocarcinogenesis. Int J Mol Sci.
15:6556–6568. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sato S, Genda T, Hirano K, Tsuzura H,
Narita Y, Kanemitsu Y, Kikuchi T, Iijima K, Wada R and Ichida T:
Up-regulated aldo-keto reductase family 1 member B10 in chronic
hepatitis C: Association with serum alpha-fetoprotein and
hepatocellular carcinoma. Liver Int. 32:1382–1390. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kosuge T, Makuuchi M, Takayama T, Yamamoto
J, Shimada K and Yamasaki S: Long-term results after resection of
hepatocellular carcinoma: Experience of 480 cases.
Hepatogastroenterology. 40:328–332. 1993.PubMed/NCBI
|
|
29
|
Izumi R, Shimizu K, Ii T, Yagi M, Matsui
O, Nonomura A and Miyazaki I: Prognostic factors of hepatocellular
carcinoma in patients undergoing hepatic resection.
Gastroenterology. 106:720–727. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hanazaki K, Kajikawa S, Koide N, Adachi W
and Amano J: Prognostic factors after hepatic resection for
hepatocellular carcinoma with hepatitis C viral infection:
Univariate and multivariate analysis. Am J Gastroenterol.
96:1243–1250. 2001. View Article : Google Scholar : PubMed/NCBI
|
















