Introduction
Human gliomas are the most prevalent malignant
neoplasms of the central nervous system, with an annual incidence
of ~5/100,000 worldwide (1). Despite
the use of aggressive surgery in combination with chemotherapy,
biological therapy and radiotherapy, gliomas continue to be
therapeutically challenging (2). For
patients with glioblastoma, the relative 5-year survival rate is
<5% (3). Novel therapies for the
treatment of glioma are warranted; recent advances in the
understanding of the molecular and biological nature of this
disease may facilitate the development of successful therapeutics
(4).
PDCD4 protein was initially determined to be
overexpressed during apoptosis, which subsequently suppresses
tumorigenesis (5,6). Loss of PDCD4 expression is closely
associated with the progression of a number of tumors, including
glioblastomas (7), and kidney,
ovarian and lung cancer (8–10). Low PDCD4 expression levels correlate
with poor outcomes in patients with glioblastoma multiforme
(11). The frequent loss of PDCD4 in
glioblastoma multiforme is partly due to epigenetic silencing
secondary to 5′ cytosine-phosphate-guanine island methylation
(12), in addition to overexpression
of microRNA (miRNA)-21, which targets PDCD4 mRNA for degradation
(13). Although several studies have
examined PDCD4 in glioma, the detailed molecular mechanisms
underlying the role of PDCD4 in glioma remain poorly
understood.
Long non-coding RNAs (lncRNAs) are non-protein
coding transcripts longer than 200 nucleotides, which are involved
in various important events, including transcriptional, epigenetic
and post-transcriptional regulation (14,15). A
previous study profiled the lncRNA homeobox transcript antisense
RNA (HOTAIR), and the results demonstrated that HOTAIR was closely
correlated with poor prognosis, molecular subtype and tumor grade
in patients with glioma (16).
However, the details of how HOTAIR regulates tumor suppressors,
including PDCD4, remain unclear.
The results of the present study demonstrated that
PDCD4 functions as a tumor suppressor in glioma cells, and its
downregulation is associated with a high level of histone 3 lysine
27 trimethylation (H3K27me3) at the PDCD4 promoter, a level that is
mediated by HOTAIR in a polycomb repressive complex 2
(PRC2)-dependent manner.
Materials and methods
Experimental subjects
A total of 24 brain glioma tissue samples and their
matched adjacent normal tissues from 24 patients obtained following
surgical resection were collected from the Department of
Neurosurgery, Yantai Yuhuangding Hospital Affiliated to Qingdao
University Medical College (Yantai, China) between August 2010 and
September 2012. Adjacent tissues were located 1 cm away from
lesions. All specimens were obtained under sterile conditions
during surgery, and immediately placed into Eppendorf tubes and
frozen at −80°C. The present study was approved by the ethics
committee of Shandong University (Jinan, China; approval no.
20130041). Written informed consent was obtained from all
patients.
Cell preparation and culture
The human astrocyte HA cell line was purchased from
ScienCell Research Laboratories (San Diego, CA, USA). The human
glioma cell lines U251, U87, LN-18 and H4 were all purchased from
the American Type Culture Collection (Manassas, VA, USA). All cell
lines were maintained in Dulbecco's modified Eagle's medium (DMEM;
GE Healthcare Life Sciences, Logan, UT, USA) supplemented with
heat-inactivated 10% fetal calf serum (GE Healthcare Life
Sciences), 2 mM L-glutamine and 100 U/ml penicillin/streptomycin.
Cells were incubated at 37°C in a humidified atmosphere with 5%
CO2.
Cell transfection and RNA
interference
The lentivirus for PDCD4 overexpression
(Lenti-PDCD4) and the control (Lenti-Empty) were commercially
constructed by Genechem Co., Ltd. (Shanghai, China). The lentivirus
was packaged in HEK-293T cells and collected from the supernatant
following the manufacturer's protocol. Glioma cells were infected
with lentiviral particles. Cell lines stably expressing PDCD4 were
established using puromycin as the selection marker.
Small interfering RNA (siRNA) targeting HOTAIR was
purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
Cells were transfected using Lipofectamine® 2000 (Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol.
Reverse transcriptase-quantitative
polymerase chain reaction (RT-qPCR)
RNA was isolated from the human glioma cell lines
using RNAzol® reagent (Vigorous Biotechnology Co., Ltd.,
Beijing, China), according to the manufacturer's protocol. The RNA
was treated with DNase H (Beyotime Institute of Biotechnology,
Haimen, China) to remove contaminating genomic DNA. cDNA was
synthesized in a 25 µl reaction mixture consisting of 2 µg total
RNA, 1 µl M-MLV Reverse Transcriptase 2 µl dNTPs, 5 µl 5X buffer, 1
µl random primers, 0.5 µl RNasin and diethylpyrocarbonate (all
Promega Corporation, Madison, WI, USA). qPCR was performed using
the ABI 7300 Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) with reagents from the SYBR®
Green Real-Time PCR Master Mix (Toyobo Co., Ltd., Osaka, Japan) and
the appropriate primers, which are presented in Table I. The PCR cycling conditions were as
follows: 95°C for 15 min, followed by 40 cycles of denaturation at
94°C for 15 sec, annealing at 60°C for 30 sec, and extension at
72°C for 30 sec. Relative mRNA expression levels were determined
following normalization to glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) using the 2−ΔΔCq method (17).
 | Table I.Primer sequences for each gene. |
Table I.
Primer sequences for each gene.
| Gene | Primer sequences,
5′-3′ | Product size, bp |
|---|
| GAPDH |
| 116 |
|
Forward |
TGTGGGCATCAATGGATTTGG |
|
|
Reverse |
ACACCATGTATTCCGGGTCAAT |
|
| PDCD4 |
| 108 |
|
Forward |
GGGAGTGACGCCCTTAGAAG |
|
|
Reverse |
ACCTTTCTTTGGTAGTCCCCTT |
|
| HOTAIR |
| 135 |
|
Forward |
GGCAGCACAGAGCAACTCTA |
|
|
Reverse |
GAGTGCAAAGTCCCGTTTG |
|
Cell proliferation
Cell Counting kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was used to determine cell
proliferation rate according to the manufacturer's protocol at the
indicated time points. Briefly, cells were seeded in 96-well plates
at a density of 2,000 cells/well. Cell proliferation reagent (10
µl) was added to each well, and the cells were incubated for 2 h at
37°C. Cell numbers were estimated by measuring the optical density
at 450 nm. The absorbance of cell-free wells containing medium was
set as zero. Data was obtained from three separate experiments and
three replications were performed each time.
Cell invasion assay
Transwell chambers (8.0 µm pore size; Corning
Incorporated, Corning, NY, USA) coated with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA) were used to measure the
invasiveness of glioma cells. In brief, 5×104 cells/well
were seeded in the upper chamber in DMEM without serum, and the
lower chamber contained DMEM supplemented with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.) to stimulate cell
invasion. Following 48 h of incubation at 37°C, cells that migrated
to the bottom of the chamber insert were fixed with 3%
paraformaldehyde, stained with 0.1% crystal violet, extracted with
33% acetic acid and finally detected quantitatively using a
standard microplate reader (at 570 nm). Data was obtained from
three separate experiments and three replications were performed
each time.
Western blot analysis
Protein extracts (10 µg) prepared with
radioimmunoprecipitation assay buffer were separated by 12%
SDS-PAGE and transferred to nitrocellulose membranes by
electroblotting. Subsequent to blocking with 5% non-fat milk, the
membranes were incubated overnight at 4°C with mouse anti-PDCD4
(#sc-376430) and mouse anti-GAPDH (#sc-25778) monoclonal antibodies
(dilutions, 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA). Blots were then incubated with peroxidase-conjugated goat
anti-mouse secondary antibodies (dilution, 1:1,000; #ZB2305 and
#ZB2307; Beijing Zhongshan Jinqiao Biotechnology, Co., Ltd.,
Beijing, China) for 1 h at room temperature and developed using a
SuperSignal™ West Pico Chemilumiscent substrate (Pierce
Biotechnology, Inc., Rockford, IL, USA). Immunoblots were scanned
using Image Lab™ software, version 1709690 (Bio-Rad Laboratories,
Inc., Hercules, CA, USA).
Chromatin immunoprecipitation (ChIP)
assay
Cells were fixed with 10% formaldehyde and sonicated
to prepare the chromatin sample. Chromatin samples were
immunoprecipitated with mouse anti-H3K27me3 monoclonal antibody
(#ab6002; Abcam, Cambridge, MA, USA), rabbit anti-enhancer of zeste
homolog 2 (EZH2) polyclonal antibody (#ab3748; Abcam) or rabbit
immunoglobulin G (IgG; #ab6785; Abcam) at 4°C for 3 h. Following
crosslink reversal, precipitated DNA was analyzed by PCR for
fragments of the PDCD4 promoter using the following primers:
Forward, 5′-GGGAGGAGGAATCGGACAG-3′; and reverse,
5′-TATGTTGGGAGGCGTGGC-3′ (141 bp). The PCR cycling conditions were
as follows: 95°C for 15 min, followed by 40 cycles of denaturation
at 94°C for 15 sec, annealing at 60°C for 30 sec, and extension at
72°C for 30 sec. The data obtained were normalized to those of
corresponding DNA precipitated by IgG.
Statistical analysis
All data are expressed as the mean ± standard
deviation. Comparisons between two groups were performed using
Student's t-test or among groups with one-way analysis of variance.
Statistical analyses were conducted using SPSS 13.0 software (SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
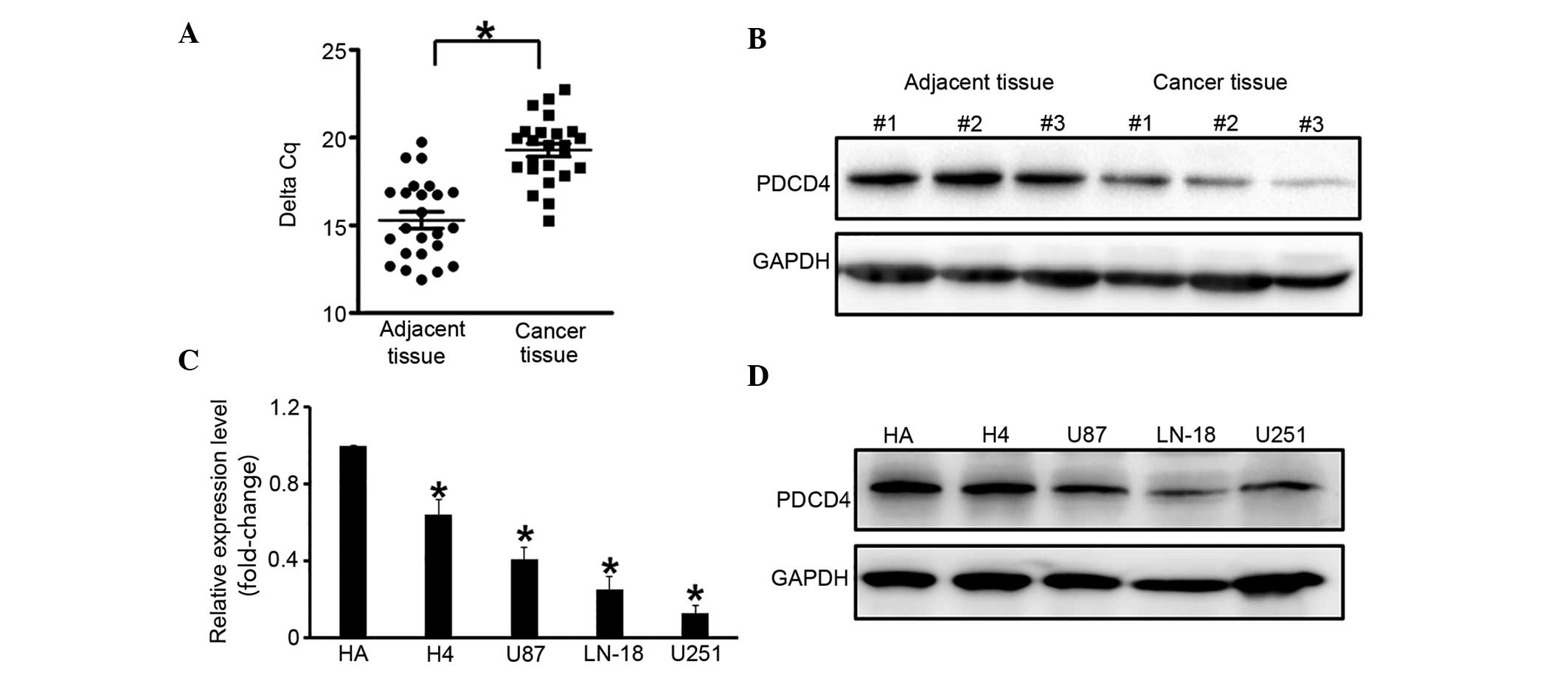
PDCD4 expression is downregulated in
glioma cells
Human glioma tissue samples (n=24) were analyzed to
detect the change in expression of PDCD4 in glioma. RT-qPCR
demonstrated that PDCD4 was significantly downregulated in glioma
tissues (by ~20%), as compared with adjacent normal tissues
(P=0.034; Fig. 1A), which was
supported by western blot analysis (Fig.
1B). In a few samples, no PDCD4 expression was detected.
This experiment was repeated with glioma cell lines
(U251, LN-18, U87 and H4), and the human astrocyte HA cell line was
used as the control. RT-qPCR and western blot analysis demonstrated
that PDCD4 expression was suppressed in all glioma cell lines
compared with the HA cells, and its expression was the lowest in
the U251 cells (Fig. 1C and D).
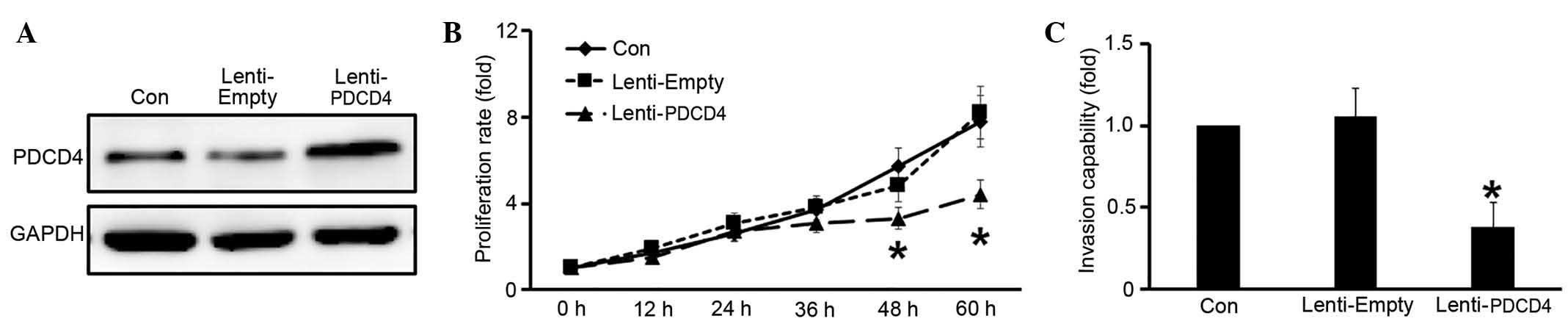
PDCD4 inhibits cell growth and
invasion in glioma cells
Next, the potential functions of PDCD4 in glioma
were investigated. A lentivirus was constructed, Lenti-PDCD4,
containing the full length PDCD4 cDNA, and PDCD4 was overexpressed
in U251 cells by infection with Lenti-PDCD4. Western blot analysis
indicated that PDCD4 was successfully overexpressed compared with
the Lenti-Empty construct (Fig. 2A).
CCK-8 assay was employed to determine whether PDCD4 affects the
proliferation of glioma cells. Glioma cells infected with
Lenti-PDCD4 exhibited a significantly lower proliferation rate than
the control at 48 and 60 h following transfection (P<0.05;
Fig. 2B). In addition, Transwell
migration assays were performed to verify invasive ability. The
results demonstrated that PDCD4 overexpression resulted in a
significant decrease in the invasion rate of U251 cells compared
with the control (P<0.05; Fig.
2C).
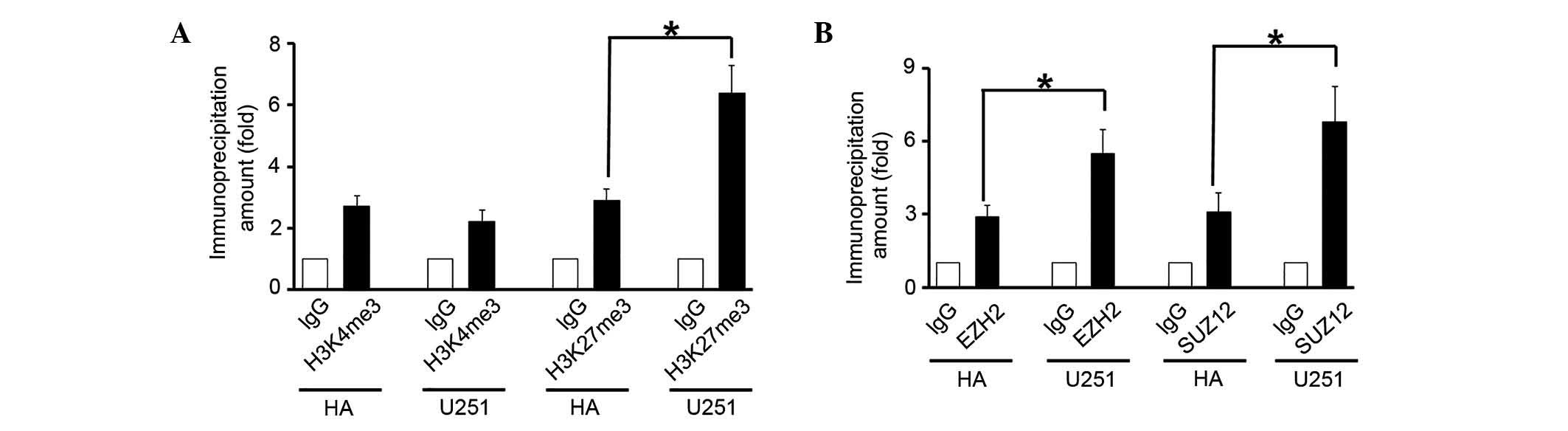
Histones at the PDCD4 promoter may be
methylated by the PRC2 complex
Next, mechanisms underlying PDCD4 downregulation in
glioma were investigated. Epigenetic modifications, particularly
methylation at specific histone sites, are important in gene
expression. ChIP assays demonstrated that the level of H3K27me3 at
the PDCD4 promoter region increased significantly in the U251
cells, as compared wih the HA cells (P=0.019; Fig. 3A), thus favoring transcriptional
silencing. Conversely, H3K4me3 exhibited little change between the
HA and U251 cells (P>0.05; Fig.
3A). The levels of EZH2 and suppressor of zeste 12 (SUZ12),
core components of the PRC2 complex, at the PDCD4 promoter region
were significantly increased in the U251 cells, as compared with
the HA cells (P<0.05; Fig. 3B).
These results indicated that the PRC2 complex was able to
downregulate PDCD4 expression by increasing the level of H3K27me3
at its promoter.
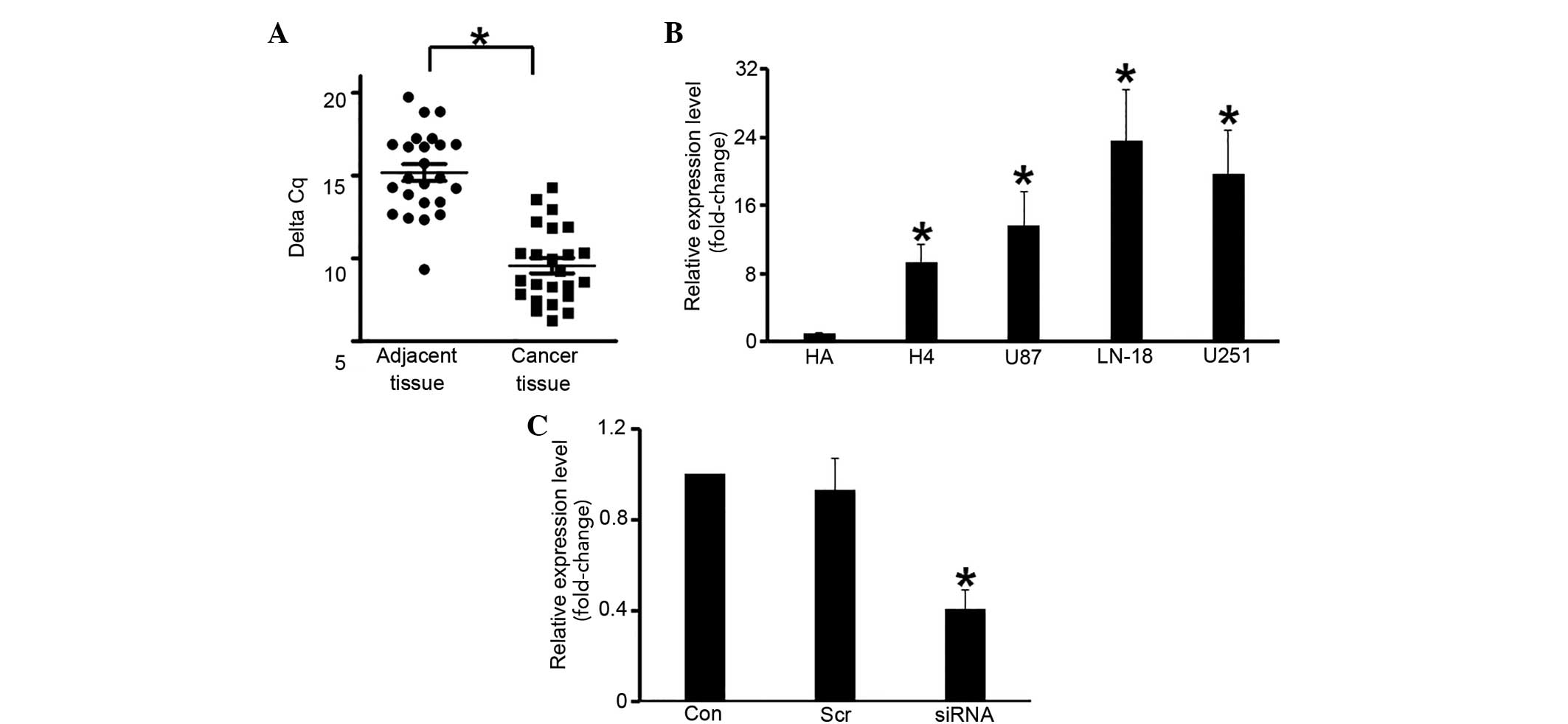
HOTAIR is upregulated in glioma
cells
The expression of HOTAIR was measured in human
glioma tissue samples. RT-qPCR demonstrated that HOTAIR expression
was significantly elevated (by ~30-fold) in glioma tissues, as
compared with normal tissues (P=0.039; Fig. 4A). A similar trend was observed in the
glioma cell lines, in which HOTAIR RNA levels were significantly
elevated compared with the HA cells (P<0.05; Fig. 4B). To investigate the function of
HOTAIR, the gene was knocked down in U251 cells using siRNA, which
significantly decreased its expression, as compared with the
scramble RNA (P<0.05; Fig.
4C).
HOTAIR participates in the silencing
of PDCD4 in glioma cells
The present study investigated how the expression of
PDCD4 was silenced in glioma cells. It was hypothesized that HOTAIR
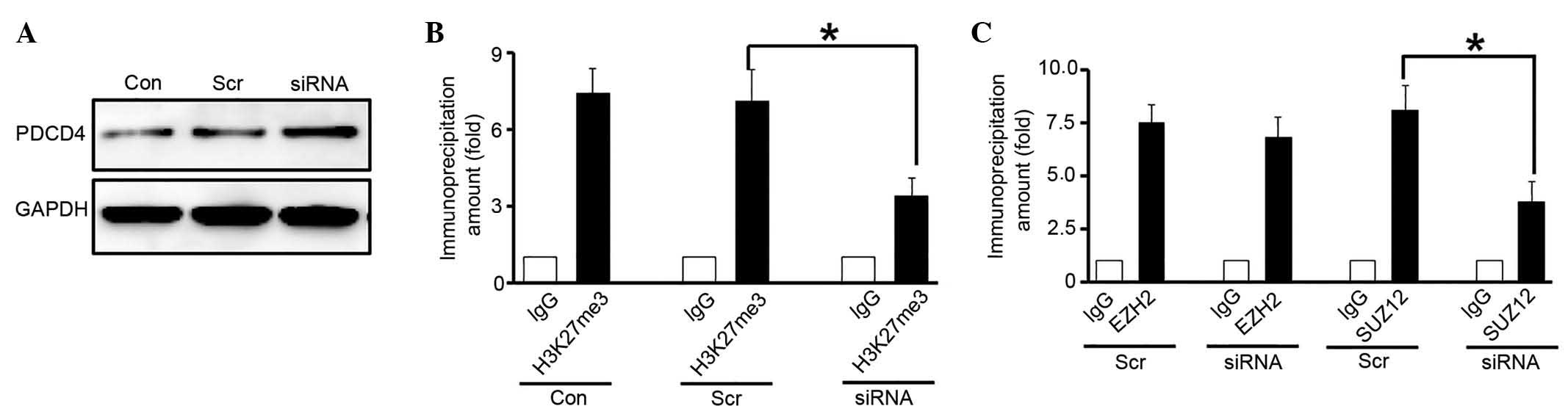
may induce the recruitment of PRC2 to the PDCD4 promoter. Western
blot analysis indicated that PDCD4 expression was upregulated when
HOTAIR was knocked down in glioma cells (Fig. 5A). Histone modifications at the PDCD4
promoter were measured by ChIP assays. The results demonstrated
that when HOTAIR was knocked down in glioma cells, the H3K27me3
level at the PDCD4 promoter was significantly reduced compared with
the control (P=0.031; Fig. 5B).
Furthermore, the recruitment of EZH2 and SUZ12 to the PDCD4
promoter was measured, and the results indicated that the level of
PRC2 components at the PDCD4 promoter was decreased in glioma cells
following HOTAIR-knockdown compared with the control (Fig. 5C). These results suggest that the
upregulation of HOTAIR results in PDCD4 silencing in glioma
cells.
Discussion
Glioma is the most aggressive form of tumor located
in the human brain, and despite advances in available therapies,
glioma remains incurable (18). The
tumors are particularly difficult to eradicate due to their highly
invasive and metastatic capabilities (19). In the present study, it was
demonstrated that PDCD4 expression was suppressed in glioma cells,
which suggested that PDCD4 may participate in glioma
tumorigenesis.
As a potential tumor suppressor, PDCD4 regulates the
expression of a variety of proteins, including p21 (20), urokinase receptor (21), hematopoietic progenitor kinase 1
(22), ornithine decarboxylase
(23), carbonic anhydrase II
(24) and c-Jun N-terminal
kinase/c-Jun/activator protein 1 (25). In glioma cells, the function of PDCD4
remains poorly understood. Liwak et al (26) reported that the loss of PDCD4
expression contributes to increased chemotherapy resistance in
glioblastoma multiforme by derepressing B-cell lymphoma-extra large
translation. Gaur et al (27)
reported that PDCD4 downregulation by miRNA-21 promotes
glioblastoma proliferation in vivo. In the current study,
when PDCD4 was overexpressed in glioma cells, the proliferation
rate and invasive capability significantly increased compared with
the control. However, no information regarding the regulation of
PDCD4 in glioma has been reported. The present study therefore
proposes that PDCD4 regulation depends on alterations in histone
modification at promoter region.
LncRNAs are generally defined as mRNA-like,
non-protein coding transcripts that are >200 nucleotides in
length (28,29). Using the most advanced sequencing
platforms and algorithms for assembling transcripts from deep
RNA-sequencing reads, it is estimated that there are ~20,000
distinct lncRNAs in humans (30).
LncRNAs demonstrate unique profiles in different forms of human
cancer, which serve as predictors of patient outcomes and reflect
disease progression (31,32). It was recently identified that lncRNAs
function in a number of aspects of cell biology and may aid the
development of tumors (33). HOTAIR,
a well-studied lncRNA, has emerged as an important regulator of
carcinogenesis and metastasis, and as a potential prognostic marker
(34,35). Therefore, an increasing amount of
research has focused on determining its functions, in addition to
identifying its target genes. The present study observed that the
expression of HOTAIR was dramatically upregulated in glioma cells
compared with normal human astrocyte cells.
Regarding the function of HOTAIR, previous studies
identified a possible role for HOTAIR in cancer. HOTAIR interacts
with PRC2, which increases the level of H3K27me3, and subsequently
decreases the expression of various genes, particularly
metastasis-suppressing genes (36,37).
Furthermore, the present study investigated the association between
HOTAIR and PDCD4. The results demonstrated that the elevated
expression of HOTAIR participated in PDCD4 regulation in a
PRC2-dependent manner.
In conclusion, to the best of our knowledge, the
current study investigated the association between the tumor
suppressor PDCD4 and the lncRNA HOTAIR in glioma cells for the
first time, with the results demonstrating that suppression of
PDCD4 mediated by HOTAIR inhibits glioma cell proliferation and
invasion in a PRC2-dependent manner. The results of the present
study may aid the understanding of the detailed molecular
mechanisms underlying glioma tumorigenesis, and support the notion
that understanding the regulation of PDCD4 expression via HOTAIR
intervention may contribute to the development of therapeutic
strategies for the treatment of gliomas.
References
|
1
|
Morgan LL: The epidemiology of glioma in
adults: A ‘state of the science’ review. Neuro-Oncol. 17:623–624.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grauer OM, Wesseling P and Adema GJ:
Immunotherapy of diffuse gliomas: Biological background, current
status and future developments. Brain Pathol. 19:674–693. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cloughesy TF, Cavenee WK and Mischel PS:
Glioblastoma: From molecular pathology to targeted treatment. Annu
Rev Pathol. 9:1–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gu JJ, Gao GZ and Zhang SM: miR-218
inhibits the migration and invasion of glioma U87 cells through the
Slit2-Robo1 pathway. Oncol Lett. 9:1561–1566. 2015.PubMed/NCBI
|
|
5
|
Cmarik JL, Min H, Hegamyer G, Zhan S,
Kulesz-Martin M, Yoshinaga H, Matsuhashi S and Colburn NH:
Differentially expressed protein Pdcd4 inhibits tumor
promoter-induced neoplastic transformation. Proc Natl Acad Sci USA.
96:14037–14042. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lankat-Buttgereit B and Göke R: The tumour
suppressor Pdcd4: Recent advances in the elucidation of function
and regulation. Biol Cell. 101:309–317. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gao F, Zhang P, Zhou C, Li J, Wang Q, Zhu
F, Ma C, Sun W and Zhang L: Frequent loss of PDCD4 expression in
human glioma: Possible role in the tumorigenesis of glioma. Oncol
Rep. 17:123–128. 2007.PubMed/NCBI
|
|
8
|
Chen Y, Knösel T, Kristiansen G, Pietas A,
Garber ME, Matsuhashi S, Ozaki I and Petersen I: Loss of PDCD4
expression in human lung cancer correlates with tumour progression
and prognosis. J Pathol. 200:640–646. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Y, Li W, Yang Y, Lu Y, He C, Hu G, Liu
H, Chen J, He J and Yu H: MicroRNA-21 targets LRRFIP1 and
contributes to VM-26 resistance in glioblastoma multiforme. Brain
Res. 1286:13–18. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei NA, Liu SS, Leung TH, Tam KF, Liao XY,
Cheung AN, Chan KK and Ngan HY: Loss of programmed cell death 4
(Pdcd4) associates with the progression of ovarian cancer. Mol
Cancer. 8:702009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liwak U, L E Jordan, Von-Holt SD, Singh P,
Hanson JE, Lorimer IA, Roncaroli F and Holcik M: Loss of PDCD4
contributes to enhanced chemoresistance in Glioblastoma multiforme
through de-repression of Bcl-xL translation. Oncotarget.
4:1365–1372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao F, Wang X, Zhu F, Wang Q, Zhang X, Guo
C, Zhou C, Ma C, Sun W, Zhang Y, et al: PDCD4 gene silencing in
gliomas is associated with 5′CpG island methylation and
unfavourable prognosis. J Cell Mol Med. 13:4257–4267. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen Y, Liu W, Chao T, Zhang Y, Yan X,
Gong Y, Qiang B, Yuan J, Sun M and Peng X: MicroRNA-21
down-regulates the expression of tumor suppressor PDCD4 in human
glioblastoma cell T98 G. Cancer Lett. 272:197–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Zhang A, Wang Y, Liu N, You Y,
Kang C and Pu P: New insights into the roles of ncRNA in the STAT3
pathway. Future Oncol. 8:723–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang JX, Han L, Bao ZS, Wang YY, Chen LY,
Yan W, Yu SZ, Pu PY, Liu N, You YP, et al: HOTAIR, a cell
cycle-associated long noncoding RNA and a strong predictor of
survival, is preferentially expressed in classical and mesenchymal
glioma. Neuro Oncol. 15:1595–1603. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real time quantitative PCR and
the 2(Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu Y, Gao H, Vo C, Ke C, Pan F, Yu L,
Siegel E, Hess KR, Linskey ME and Zhou YH: Anti-EGFR function of
EFEMP1 in glioma cells and patient prognosis. Oncoscience.
1:205–215. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang XP, Deng XL and Li LY: MicroRNA-584
functions as a tumor suppressor and targets PTTG1IP in glioma. Int
J Clin Exp Pathol. 7:8573–8582. 2014.PubMed/NCBI
|
|
20
|
Göke R, Barth P, Schmidt A, Samans B and
Lankat-Buttgereit B: Programmed cell death protein 4 suppresses
CDK1/cdc2 via induction of p21(Waf1/Cip1). Am J Physiol Cell
Physiol. 287:C1541–C1546. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Leupold JH, Yang HS, Colburn NH, Asangani
I, Post S and Allgayer H: Tumor suppressor Pdcd4 inhibits
invasion/intravasation and regulates urokinase receptor (u-PAR)
gene expression via Sp-transcription factors. Oncogene.
26:4550–4562. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang HS, Matthews CP, Clair T, Wang Q,
Baker AR, Li CC, Tan TH and Colburn NH: Tumorigenesis suppressor
Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase
kinase 1 expression to suppress colon carcinoma cell invasion. Mol
Cell Biol. 26:1297–1306. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jansen AP, Camalier CE and Colburn NH:
Epidermal expression of the translation inhibitor programmed cell
death 4 suppresses tumorigenesis. Cancer Res. 65:6034–6041. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lankat-Buttgereit B, Gregel C, Knolle A,
Hasilik A, Arnold R and Göke R: Pdcd4 inhibits growth of tumor
cells by suppression of carbonic anhydrase type II. Mol Cell
Endocrinol. 214:149–153. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bitomsky N, Böhm M and Klempnauer KH:
Transformation suppressor protein Pdcd4 interferes with
JNK-mediated phosphorylation of c-Jun and recruitment of the
coactivator p300 by c-Jun. Oncogene. 23:7484–7493. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liwak U, Jordan LE, Von-Holt SD, Singh P,
Hanson JE, Lorimer IA, Roncaroli F and Holcik M: Loss of PDCD4
contributes to enhanced chemoresistance in Glioblastoma multiforme
through de-repression of Bcl-xL translation. Oncotarget.
4:1365–1372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gaur AB, Holbeck SL, Colburn NH and Israel
MA: Downregulation of Pdcd4 by mir-21 facilitates glioblastoma
proliferation in vivo. Neuro Oncol. 13:580–590. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ernst C and Morton CC: Identification and
function of long non-coding RNA. Front Cell Neurosci. 7:1682013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Moran VA, Perera RJ and Khalil AM:
Emerging functional and mechanistic paradigms of mammalian long
non-coding RNAs. Nucleic Acids Res. 40:6391–6400. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang GY, Zhu YY and Zhang YQ: The
functional role of long non-coding RNA in digestive system
carcinomas. Bull Cancer. 101:E27–E31. 2014.PubMed/NCBI
|
|
32
|
Bhan A and Mandal SS: Long noncoding RNAs:
Emerging stars in gene regulation, epigenetics and human disease.
ChemMedChem. 9:1932–1956. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu MX, Chen X, Chen G, Cui QH and Yan GY:
A computational framework to infer human disease-associated long
noncoding RNAs. PLoS One. 9:e844082014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wan Y and Chang HY: HOTAIR: Flight of
noncoding RNAs in cancer metastasis. Cell Cycle. 9:3391–3392. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lv DW, Ge P, Zhang M, Cheng ZW, Li XH and
Yan YM: Integrative network analysis of the signaling cascades in
seedling leaves of bread wheat by large-scale phosphoproteomic
profiling. J Proteome Res. 13:2381–2395. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li L, Liu B, Wapinski OL, Tsai MC, Qu K,
Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, et al: Targeted
disruption of Hotair leads to homeotic transformation and gene
derepression. Cell Rep. 5:3–12. 2013. View Article : Google Scholar : PubMed/NCBI
|