Introduction
Colorectal cancer is one of the most common types of
cancer in males and females. It is estimated that there are
~140,000 novel cases of colorectal cancer expected in the USA and
it remains the third leading cause of cancer-associated mortality
in 2014 (1). In China, colorectal
cancer was the fifth most common type of cancer and the fifth
leading cause of cancer-associated mortality in 2011 (2). Although the 5-year mortality rate of
colorectal cancer has slightly decreased in the last decade, novel
prognostic factors and potential therapeutic targets for this
disease are required. Furthermore, the underlying
pathophysiological mechanisms of the development of colorectal
cancer remain unclear (3,4).
Long non-coding RNAs (lncRNAs), >200 nucleotides
in length, have been identified as novel gene expression regulators
in the last decade (5,6). A previous study suggested that a number
of lncRNAs may be involved in the development and metastasis of
cancer (7). LncRNA-imprinted
maternally expressed transcript (non-protein coding) (H19),
initially reported in 1991 by Bartolomei et al (8), was identified to be expressed at
increased levels in extraembryonic tissues, embryonic tissues and
the majority of fetal tissues; however, the expression level of
lncRNA H19 is markedly decreased following birth (9). As the first imprinting lncRNA to be
identified, H19, which is expressed by the maternal allele rather
than the paternal, is transcribed from the H19/insulin-like growth
factor 2 gene cluster located on human chromosome 11p15.5 (10). Previous studies have revealed that H19
serves an important function in the progression of a number of
types of cancer (11–13). Additionally, H19 overexpression was
identified to be markedly associated with a poor prognosis in
bladder and gastric cancer (14,15).
Currently, the expression levels and the function of H19 in
colorectal cancer remain unclear.
In the present study, the expression level of H19
and its association with clinicopathological variables and disease
outcome was examined using colorectal tumor samples and paired
adjacent normal tissues. In addition, the effects of H19
overexpression on the viability, migration and EMT process of colon
cancer cells were investigated in HCT-116 and SW-480 cells, using
an H19-expressing plasmid. The results suggested that H19
overexpression was significantly associated with pre-treatment
metastasis (P=0.020), poor differentiation level (P=0.022) and
advanced tumor-node-metastasis (TNM) stages (P=0.046). Furthermore,
H19 overexpression was identified to be an independent risk factor
for decreased recurrence-free survival (RFS) [hazard ratio (HR),
3.037; 95% confidence interval (CI), 1.107–8.329; P=0.031] and
overall survival (OS) time (HR, 4.028; 95% CI, 1.332–12.183;
P=0.014) in patients with colorectal cancer. H19 overexpression was
identified to be associated with increased viability and migratory
potential of HCT-116 and SW-480 cells. Additionally, H19
overexpression decreased the expression level of epithelial
(E-)cadherin and increased the expression levels of vimentin and
snail family transcriptional repressor 1 (Snail) in colon cancer
cells. Immunofluorescence suggested that H19 overexpression
increased the expression of filamentous (F-) actin in SW-480 cells.
The results of the present study suggested that H19 overexpression
is associated with decreased RFS and OS rates in patients with
colorectal cancer, and increased viability and migration of colon
cancer cells.
Materials and methods
Patients and samples
Between April 2006 and October 2009, 96 patients
with colorectal cancer, who underwent surgery at Peking University
First Hospital (Beijing, China), were included in the present
study. Baseline characteristics of all patients, including sex,
age, tumor size, tumor histological differentiation status, the
status of mucin and TNM stages, according to the American Joint
Committee on Cancer staging (7th edition), were collected from the
medical records and surgical pathology reports (Table I) (16).
Patient follow-up was carried out every 3 months for the first 2
years, every 6 months for the following 3 years and annually
thereafter. The present study was approved by the Institutional
Review Board at Peking University First Hospital, conducted in
accordance with the principles of The Declaration of Helsinki and
written informed consent was obtained from all patients. Samples of
colorectal tumors and paired adjacent normal tissues were collected
immediately following resection, and stored at −80°C until use.
 | Table I.Association between H19 expression and
clinicopathological variables. |
Table I.
Association between H19 expression and
clinicopathological variables.
|
| Total, n=96 | H19 |
|
|---|
|
|
|
|
|
|---|
| Characteristic | No. | High | Low | P-value |
|---|
| Sex |
|
|
| 0.365 |
| Male | 42 | 21 | 21 |
|
|
Female | 54 | 32 | 22 |
|
| Age, years |
|
|
| 0.845 |
| ≤60 | 39 | 22 | 17 |
|
|
>60 | 57 | 31 | 26 |
|
| T-stage |
|
|
| 0.646 |
| T1-2 | 16 | 8 | 8 |
|
| T3-4 | 80 | 45 | 35 |
|
| N-stage |
|
|
| 0.874 |
| N0-1 | 84 | 45 | 36 |
|
| N2 | 12 | 8 | 7 |
|
| M-stage |
|
|
| 0.020 |
| M0 | 77 | 38 | 39 |
|
| M1 | 19 | 15 | 4 |
|
| Tumor size, cm |
|
|
|
|
| ≤5 | 75 | 40 | 35 | 0.485 |
|
>5 | 21 | 13 | 8 |
|
|
Differentiation |
|
|
| 0.022 |
|
Well/moderate | 83 | 42 | 41 |
|
|
Poor | 13 | 11 | 2 |
|
| Mucin
production |
|
|
| 0.315 |
|
Absent | 82 | 47 | 35 |
|
|
Present | 14 | 6 | 8 |
|
| TNM stage |
|
|
| 0.046 |
|
I–II | 54 | 25 | 29 |
|
|
III–IV | 42 | 28 | 14 |
|
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) to detect H19
Total RNA was extracted using the TRIzol one-step
extraction method (TRIzol reagent; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and reverse-transcribed into
cDNA using a Reverse Transcription System kit (cat. no. A3500;
Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocol. qPCR analysis was carried out using a
final reaction volume of 25 µl and the TaqMan® Universal PCR Master
Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.), according
to the manufacturer's protocol. All reactions were performed in
triplicate using a 7500 Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The thermocycling conditions
comprised the following steps: initial denaturation for 15 min at
95°C, then 40 cycles of denaturation at 94°C for 15 sec, then
annealing for 30 sec at 55°C and extension for 30 sec at 72°C.
Relative RNA expression was calculated as a fold-change using the
comparative quantification (Cq) method (2−ΔΔCq), with
GAPDH as the internal control gene (17). The primers and probes used, listed
5′-3′, were as follows: GAPDH forward, CAGTCAGCCGCATCTTCTTTT; GAPDH
reverse, GTGACCAGGCGCCCAATAC; GAPDH probe, tetramethylrhodamine
(TAMRA)-CGTCGCCAGCCG AGCCACA-Black Hole Quencher (BHQ)2; H19
forward, AATCGGCTCTGGAAGGTGAA; H19 reverse, CTGCTG TTCCGATGGTGTCTT;
H19 probe, TAMRA-CTAGAG GAACCAGACCTCATCAGCCCAAC-BHQ1.
Cell culture
Human colon cancer cell lines HCT-116 and SW-480
were purchased from the American Type Culture Collection (Manassas,
VA, USA). HCT-116 cells were cultured in McCoy's 5A medium (Thermo
Fisher Scientific, Inc.) with 10% fetal bovine serum (FBS; Thermo
Fisher Scientific, Inc.). SW-480 cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) with
10% FBS. H19 expressing plasmid pcDNA 3.1 (+)-H19 (cat. no. C05003)
or pcDNA 3.1(+) empty vector as control were purchased from Suzhou
GenePharma Co., Ltd. (Suzhou, China). Transfection was conducted
using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. A total of
1×106 cells were seeded for transfection.
MTT viability assays
A total of 1,000 cells/well were seeded in a 96-well
plate for 24 h. The cells were subsequently transfected with 0.2 µg
H19 or control plasmid for 24 h and incubated at 37°C for 5 days.
The cells were incubated in 50 µl 0.1 mg/ml solution of MTT
(Sigma-Aldrich, USA) at 37°C for 4 h and subsequently lysed in 150
µl dimethyl sulfoxide at room temperature for 30 min. The
absorbance in each well was measured at 580 nm using a microplate
reader. Experiments were performed in triplicate and repeated 3
times.
Migration assays
Cell migration assays were analyzed using Transwell
chambers (8.0 µm pore size; Corning Incorporated, Corning, NY,
USA). At 24 h post-transfection, with H19 or control plasmid, cells
were incubated in serum-free medium for 12 h at 37°C. A total of
5×104 cells, in 200 µl DMEM or MacCoy's 5A medium
without serum, were added to the upper chamber and 600 µl DMEM or
MacCoy's 5A medium with 20% FBS was placed in the bottom of wells.
The plates were incubated at 37°C in an atmosphere containing 5%
CO2 for 24 h. The cells that migrated to the opposite
side of the membrane were fixed and stained with hematoxylin and
eosin, and the number of invading cells was determined using a
microscope (Olympus Corporation, Tokyo, Japan) using a previously
described method (18). Experiments
were performed in triplicate and repeated 3 times.
Western blot analysis
Total protein was extracted from cells 72 h after
transfection with H19 or control plasmid using a Cell Total Protein
Extraction kit, (Beyotime Institute of Biotechnology, Haimen,
China). The concentrations of protein were determined using the
bicinchoninic acid assay method and equal quantities of 25 µg
protein/lane were separated by SDS-PAGE (10% gel). Subsequently,
the separated proteins were transferred onto a polyvinylidene
fluoride membrane. The membrane was blocked for non-specific
binding for 1 h [5% bovine serum albumin (BSA) in TBS-Tween-20
buffer] at room temperature, and subsequently incubated overnight
at 4°C with the following rabbit monoclonal antibodies:
anti-E-cadherin (cat. no. 3195; dilution, 1:1,000; Cell Signaling
Technology, Inc., Danvers, MA, USA), rabbit anti-vimentin (cat. no.
5741; dilution, 1:1,000), rabbit anti-Snail (cat. no. 3879;
dilution, 1:1,000) and rabbit anti-GAPDH (cat. no. 2118; dilution,
1:1,000) (all from Cell Signaling Technology, Inc.). The membrane
was subsequently incubated at room temperature for 1 h with
anti-rabbit immunoglobulin G horseradish peroxidase-linked
secondary antibodies (cat. no. 7074; dilution, 1:5,000; Cell
Signaling Technology, Inc.). Blots were developed with enhanced
chemiluminescence detection reagents (Merck KGaA, Darmstadt,
Germany).
Immunofluorescence
The immunofluorescence of F-actin was carried out
according to a previously described protocol (19), with certain modifications. Following
transfection with H19 or control plasmid for 72 h, SW-480 cells, on
glass slides, were fixed with ice-cold acetone for 5 min at −20°C.
Subsequently, slides were placed at the bottom of the wells of the
6-well plates with the cells facing upwards. Slides were rinsed
with PBS, followed by blocking with 1% BSA for 1 h at room
temperature. F-actin was stained following incubation with
rhodamine-phalloidin (Thermo Fisher Scientific, Inc.) and nuclei
were stained using DAPI (Thermo Fisher Scientific, Inc.). Following
washing with PBS, slides were mounted, using the ProLong Gold
Antifade reagent (Molecular Probes, USA), and stored at 4°C in the
dark until analyzed. Fluorescence was visualized using a Fluoview
1000 confocal microscope (Olympus Corporation).
Statistical analysis
The results for each tumor were evaluated by
determining the relative ratio of tumor to normal tissue in the
same patient. Data were tested for normal distribution using the
Kolmogorov-Smirnov test and the means of the two groups were
compared using Wilcoxon signed-rank test. A relative ratio of tumor
to normal tissue ≥4 and <4 was classified as high expression and
low expression, respectively. Pearson's χ2 test was used
to analyze the association between H19 expression and
clinicopathological variables. Kaplan-Meier estimator and Cox's
proportional hazard tests were used for survival analysis. The
results were expressed as the mean ± standard error of the mean and
analyzed using a Student's t-test for unpaired data. P<0.05
indicated a statistically significant difference. All statistical
analyses were carried out using SPSS software (version 23.0; IBM
Corp., Armonk, NY, USA).
Results
Patient characteristics
The male to female ratio of the study population was
0.78:1 and the median age was 65 years (range, 25–84 years). A
total of 40 patients (42%) exhibited lymphoid node metastasis and
19 patients (20%) exhibited distant metastasis at the time of
surgery. The mean follow-up period was 45 months (range, 3–60
months). In total, 34 patients (35%) experienced tumor recurrence
and distant recurrence occurred in 15 patients (16%; Table I).
Relative H19 expression in tumor and
paired adjacent normal tissues
The Kolmogorov-Smirnov test indicated that the
expression of H19 in tumor tissues exhibited a non-normal
distribution (P<0.001). Analysis of 96 paired rectal tumors and
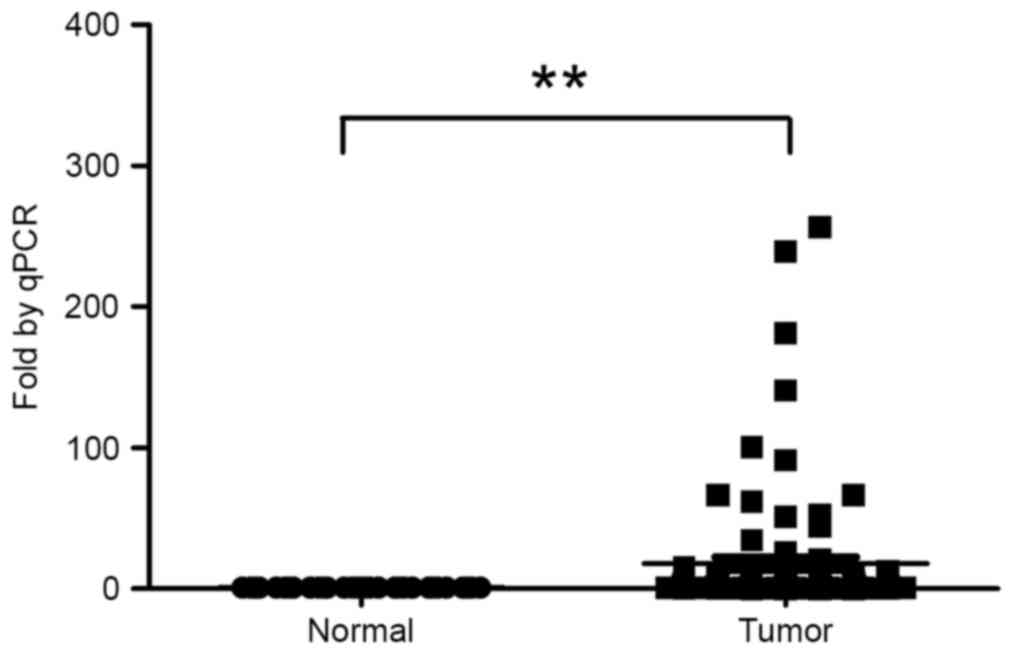
paired adjacent normal tissues revealed a significant upregulation
(median, 4.19-fold) of H19 expression in tumor tissues compared
with matched normal tissues (Fig. 1).
The 96 patients were subsequently divided into two groups on the
basis of the fold change of H19 expression [H19-high (≥4-fold
upregulation, n=53) and H19-low (<4-fold upregulation,
n=43)].
Association between H19 expression
with clinicopathological variables
Table I demonstrates
the association between H19 expression in colorectal tumor tissues
and clinicopathological variables. H19 overexpression in colorectal
tumor tissues was identified to be associated with pre-treatment
metastasis (P=0.020), poor differentiation level (P=0.022) and
advanced TNM stages (P=0.046).
Expression of H19 in association with
prognosis of rectal cancer
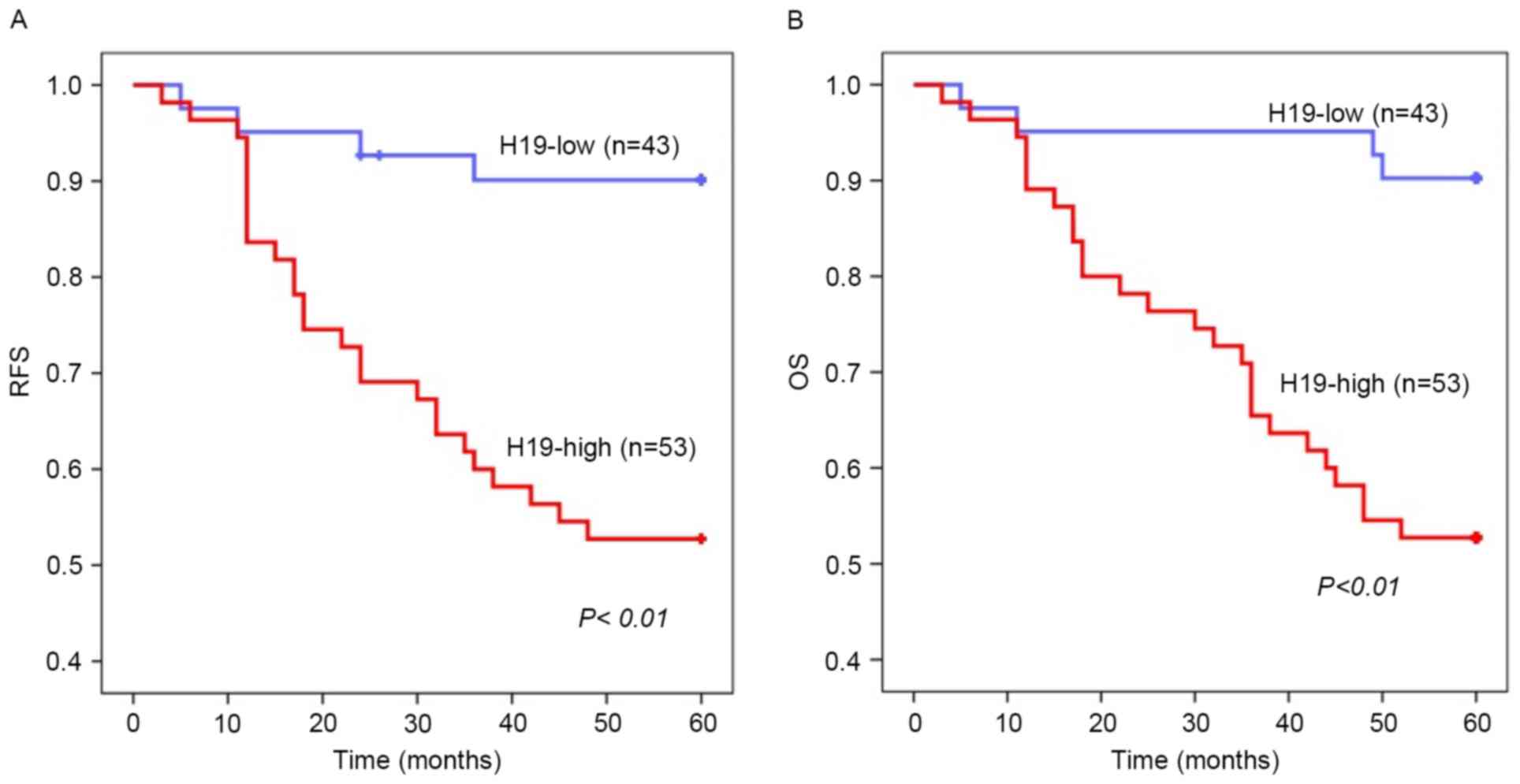
Kaplan-Meier estimator survival analysis identified
that overexpression of H19 in colorectal tumor tissues was
significantly associated with poor RFS (P<0.01) and OS
(P<0.01) rates (Fig. 2A and B).
Cox's univariate proportional hazard analysis of RFS revealed that
lymph node invasion, metastasis, poor differentiation level,
advanced pre-surgery TNM stage and H19 overexpression in tumors
were significantly associated with tumor recurrence (P=0.036,
P=0.001, P=<0.01, P<0.01 and P=0.002, respectively). Advanced
TNM stage and increased H19 expression were identified as
independent risk factors for recurrence in multivariate analysis
(P=0.023 and P=0.031, respectively; Table IIA). Metastasis, poor differentiation
level, advanced pre-surgery TNM stage and increased H19 expression
in tumors were significantly associated with decreased OS rates
using Cox's univariate proportional hazard analysis (P<0.01,
P<0.01, P<0.01 and P=0.001, respectively). Increased H19
expression and advanced TNM stage were identified using Cox's
multivariate analysis as independent risk factors for decreased OS
rates (P=0.014 and P=0.007, respectively; Table IIB). These results suggested that
overexpression of H19 is associated with unfavorable prognosis in
patients with colorectal cancer.
 | Table II.Cox's proportional hazard
analysis. |
Table II.
Cox's proportional hazard
analysis.
| A, Cox's
proportional hazard analysis of recurrence-free survival |
|---|
|
|---|
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Sex, female | 1.163 | 0.581–2.329 | 0.670 |
|
|
|
| Age, >60
years | 1.379 | 0.665–2.862 | 0.388 |
|
|
|
| T-stage, 3/4 | 2.125 | 0.647–6.977 | 0.214 |
|
|
|
| N-stage, 2 | 2.461 | 1.062–5.705 | 0.036 | 1.311 | 0.503–3.418 | 0.580 |
| M-stage, 1 | 3.464 | 1.704–7.041 | 0.001 | 1.471 | 0.621–3.487 | 0.380 |
| Tumor size, >5
cm | 1.007 | 0.435–2.329 | 0.987 |
|
|
|
| Differentiation,
poor | 5.416 | 2.583–11.356 | 0.000 | 1.667 | 0.670–4.418 | 0.272 |
| Mucin, present | 2.756 | 0.658–11.535 | 0.165 |
|
|
|
| TNM-stage,
III/IV | 5.223 | 2.339–11.666 | 0.000 | 3.138 | 1.171–8.410 | 0.023 |
| H19 expression,
high | 4.514 | 1.737–11.733 | 0.002 | 3.037 | 1.107–8.329 | 0.031 |
|
| B, Cox's
proportional hazard analysis of overall survival rate |
|
|
| Univariate | Multivariate |
|
|
|
|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|
| Sex, female | 0.722 | 0.353–1.476 | 0.372 |
|
|
|
| Age, >60
years | 1.486 | 0.695–3.176 | 0.307 |
|
|
|
| T-stage, 3/4 | 3.089 | 0.736–12.973 | 0.123 |
|
|
|
| N-stage, 2 | 1.960 | 0.800–4.800 | 0.141 |
|
|
|
| M-stage, 1 | 4.096 | 1.960–8.395 | 0.000 | 1.127 | 0.463–2.743 | 0.792 |
| Tumor size, >5
cm | 1.105 | 0.474–2.574 | 0.818 |
|
|
|
| Differentiation,
poor | 5.772 | 2.702–12.333 | 0.000 | 2.261 | 0.950–5.378 | 0.065 |
| Mucin, present | 2.584 | 0.615–10.848 | 0.195 |
|
|
|
| TNM-stage,
III/IV | 5.570 | 2.384–13.016 | 0.000 | 3.841 | 1.454–10.146 | 0.007 |
| H19 expression,
high | 7.64 | 2.315–25.216 | 0.001 | 4.028 | 1.332–12.183 | 0.014 |
H19 increases proliferation and
migration of colon cancer cells
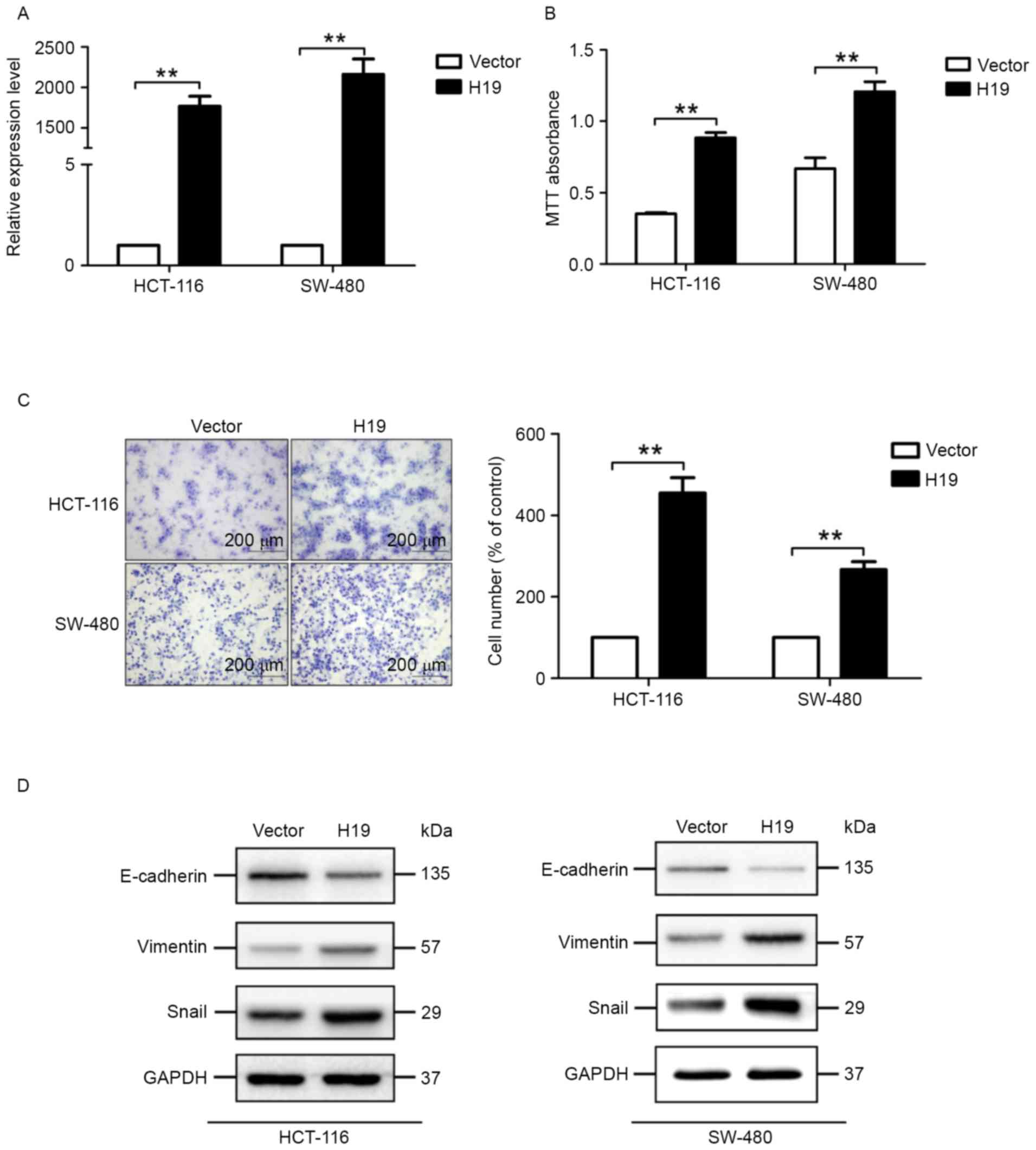
The expression level of H19 in cells transfected
with the H19-expressing plasmid is significantly increased,
compared with cells transfected with the control plasmid (Fig. 3A). MTT assays suggested that colon
cancer cells transfected with the H19-expressing plasmid exhibited
significantly increased viability potential (Fig. 3B). In addition, migration assays
suggested that H19 overexpression significantly increased the
migration potential of HCT-116 and SW-480 cells (Fig. 3C).
H19 induces EMT in colon cancer
cells
Since decreased expression of E-cadherin and
increased expression of vimentin- and EMT-associated transcription
factors, including Snail, are well-known markers of the EMT
process, the expression level of E-cadherin, vimentin and Snail was
determined. H19 overexpression markedly decreased the expression of
E-cadherin and increased the expression of vimentin and Snail,
determined by western blot analysis (Fig.
3D). These results suggested that H19 overexpression induced
the EMT process in colon cancer cells.
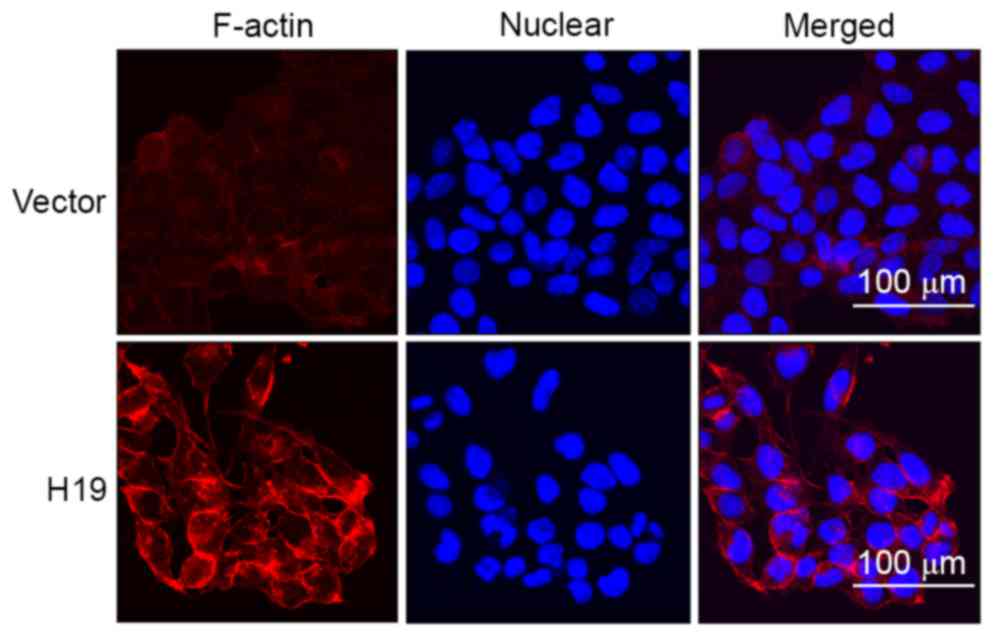
H19 induces increased expression of
F-actin in SW-480 cells
Previous studies identified that increased
expression of F-actin is associated with increased migratory
potential of cancer cells and is considered as a marker of EMT
process, which prompted the determination of the expression of
F-actin in SW-480 cells using immunofluorescence in the present
study. The results suggested that H19 overexpression induced
increased expression of F-actin and the increased expression was
primarily located at the cell edge (Fig.
4).
Discussion
Although the mortality rate of colorectal cancer has
decreased slightly in the last decade, it remains a
life-threatening disease. The identification of novel factors
involved in the development of colorectal cancer as novel
therapeutic targets is required. H19, an imprinted and maternally
expressed lncRNA, is abundant in embryonic tissues of endodermal
and mesodermal origin, but is thought to be repressed following
birth in the majority of tissues (8,9). In
addition, the expression level of H19 has been investigated in a
number of types of cancer. Song et al (20) identified that there was an >8-fold
increase in H19 expression in gastric tumor tissues, compared with
paired normal tissues. Furthermore, increased expression of H19 was
markedly associated with an early recurrence of bladder cancer and
may be considered a predictive marker for poor prognosis (14). Zhou et al (21) revealed that plasma H19 expression
enabled the differentiation of early stage gastric cancer from
controls with an area under the curve of 0.877, sensitivity of
85.5% and specificity of 80.1% (18).
However, the underlying molecular mechanisms of the
oncogenic function of H19 are complex. Previous studies have
identified that H19 is able to regulate the expression of a number
of types of cancer-associated protein, including calneuron 1, the
tumor suppressor retinoblastoma and ubiquitin ligase E3 family, by
acting as the precursor of microRNA (miR)-675 (15,22,23).
Additionally, a previous study suggested that H19 may function as a
competing endogenous RNA for miR-138 and miR-200a, antagonizing
their functions and thus leading to the derepression of their
endogenous targets vimentin, zinc finger E-box-binding homeobox
(ZEB) OL-9705-NPS 1 and ZEB2, all of which were associated with the
EMT process (24). Further studies
are required to determine the underlying molecular mechanisms of
the role of H19 in distinct types of cancer.
In the present study, the expression level of H19
was identified to be markedly increased in colorectal tumor
tissues, compared with adjacent normal tissues, which suggested
that H19 serves a function in the tumorigenesis of colorectal
cancer. Subsequently, the association between the expression level
of H19 and the clinicopathological variables and patient survival
was analyzed. Overexpression of H19 was identified to be associated
with pre-treatment metastasis, poor differentiation level and
advanced TNM stages. In addition, overexpression of H19 was
revealed to be an independent risk factor for decreased RFS and OS.
These results were consistent with previous studies, which suggests
that H19 exhibits a tumorigenesis function in the development of a
number of types of cancer (12–15).
Notably, overexpression of H19 was identified to be associated with
pre-treatment metastasis in the present study. Considering the
important role of metastasis in the development of cancer, the
effect of H19 on the metastasis potential of colon cancer cells,
and the underlying molecular mechanism, was investigated.
Previous studies have suggested that H19 may be
involved in the EMT process in a number of types of cancer cells
(25,26). EMT is considered to serve an important
function in the process of metastasis and is characterized by
‘cadherin switch’, increased expression of vimentin and relevant
transcription factors, including Snail (27,28). In
the present study, the expression levels of E-cadherin, vimentin
and Snail were determined in HCT-116 and SW-480 cells. The results
suggested that H19 overexpression markedly decreased the expression
of E-cadherin, and increased the expression of vimentin and Snail,
indicating that H19 overexpression induced the EMT process in colon
cancer cells.
Since increased expression of F-actin is associated
with increased migration potential of cancer cells and considered a
marker of EMT process (29), the
expression level of F-actin in SW-480 cells was determined using
immunofluorescence. The results suggested that H19 overexpression
induced increased expression of F-actin and this increased
expression was primarily located at cell edges.
The expression level of H19 is markedly increased in
colorectal tumor tissues, compared with adjacent normal tissues,
and overexpression of H19 is an independent risk factor for
decreased RFS and OS rates in patients with colorectal cancer.
Overexpression of H19 is additionally associated with increased
viability and migration of colon cancer cells. In addition, it is
hypothesized that the induction of the EMT process may be one of
the underlying molecular mechanisms of the effect of H19 on the
metastatic potential of colon cancer cells.
Acknowledgements
The authors thank Professor Ding-fang Bu for his
technical support and recommendations with the present study, and
all the staff of the Division of General Surgery (Peking University
First Hospital, Beijing, China) for their assistance with the
collection of samples and in the process of follow-up.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zeng H and Zhang S: The
updated incidences and mortalities of major cancers in China, 2011.
Chin J Cancer. 34:502–507. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fearnhead NS, Wilding JL and Bodmer WF:
Genetics of colorectal cancer: Hereditary aspects and overview of
colorectal tumorigenesis. Br Med Bull. 64:27–43. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wolpin BM and Mayer RJ: Systemic treatment
of colorectal cancer. Gastroenterology. 134:1296–1310. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Whitehead J, Pandey GK and Kanduri C:
Regulation of the mammalian epigenome by long noncoding RNAs.
Biochim Biophys Acta. 1790:936–947. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li L, Feng T, Lian Y, Zhang G, Garen A and
Song X: Role of human noncoding RNAs in the control of
tumorigenesis. Proc Natl Acad Sci. 106:pp. 12956–12961. 2009;
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartolomei MS, Zemel S and Tilghman SM:
Parental imprinting of the mouse H19 gene. Nature. 351:153–155.
1991. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tabano S, Colapietro P, Cetin I, Grati FR,
Zanutto S, Mandò C, Antonazzo P, Pileri P, Rossella F, Larizza L,
et al: Epigenetic modulation of the IGF2/H19 imprinted domain in
human embryonic and extra-embryonic compartments and its possible
role in fetal growth restriction. Epigenetics. 5:313–324. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Poirier F, Chan CT, Timmons PM, Robertson
EJ, Evans MJ and Rigby PW: The murine H19 gene is activated during
embryonic stem cell differentiation in vitro and at the time of
implantation in the developing embryo. Development. 113:1105–1114.
1991.PubMed/NCBI
|
|
11
|
Byun HM, Wong HL, Birnstein EA, Wolff EM,
Liang G and Yang AS: Examination of IGF2 and H19 loss of imprinting
in bladder cancer. Cancer Res. 67:10753–10758. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Berteaux N, Lottin S, Monté D, Pinte S,
Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T and
Adriaenssens E: H19 mRNA-like noncoding RNA promotes breast cancer
cell proliferation through positive control by E2F1. J Biol Chem.
280:29625–29636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim SJ, Park SE, Lee C, Lee SY, Jo JH, Kim
JM and Oh YK: Alterations in promoter usage and expression levels
of insulin-like growth factor-II and H19 genes in cervical
carcinoma exhibiting biallelic expression of IGF-II. Biochim
Biophys Acta. 1586:307–315. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ariel I, Sughayer M, Fellig Y, Pizov G,
Ayesh S, Podeh D, Libdeh BA, Levy C, Birman T and Tykocinski ML:
The imprinted H19 gene is a marker of early recurrence in human
bladder carcinoma. Mol Pathol. 53:320–323. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kiernan JA: Histological and Histochemical
Methods: Theory and Practice. 4th. Bloxham, UK: Scion; 2008
|
|
19
|
Chen S, Zhu J, Zuo S, Ma J, Zhang J, Chen
G, Wang X, Pan Y, Liu Y and Wang P: 1,25(OH)2D3 attenuates
TGF-β1/β2-induced increased migration and invasion via inhibiting
epithelial-mesenchymal transition in colon cancer cells. Biochem
Biophys Res Commun. 468:130–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song H, Sun W, Ye G, Ding X, Liu Z, Zhang
S, Xia T, Xiao B, Xi Y and Guo J: Long non-coding RNA expression
profile in human gastric cancer and its clinical significances. J
Transl Med. 11:2252013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou X, Yin C, Dang Y, Ye F and Zhang G:
Identification of the long non-coding RNA H19 in plasma as a novel
biomarker for diagnosis of gastric cancer. Sci Rep. 5:115162015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung
JJ and Kwok TT: Oncofetal H19-derived miR-675 regulates tumor
suppressor RB in human colorectal cancer. Carcinogenesis.
31:350–358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vennin C, Spruyt N, Dahmani F, Julien S,
Bertucci F, Finetti P, Chassat T, Bourette RP, Le Bourhis X and
Adriaenssens E: H19 non coding RNA-derived miR-675 enhances
tumorigenesis and metastasis of breast cancer cells by
downregulating c-Cbl and Cbl-b. Oncotarget. 6:29209–29223.
2015.PubMed/NCBI
|
|
24
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan
Z and Ai K: H19 promotes pancreatic cancer metastasis by
derepressing let-7's suppression on its target HMGA2-mediated EMT.
Tumour Biol. 35:9163–9169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matouk IJ, Raveh E, Abu-lail R, Mezan S,
Gilon M, Gershtain E, Birman T, Gallula J, Schneider T, Barkali M,
et al: Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys
Acta. 1843:1414–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tsanou E, Peschos D, Batistatou A,
Charalabopoulos A and Charalabopoulos K: The E-cadherin adhesion
molecule and colorectal cancer. A global literature approach.
Anticancer Res. 28:3815–3826. 2008.PubMed/NCBI
|
|
28
|
Costa LC, Leite CF, Cardoso SV, Loyola AM,
Faria PR, Souza PE and Horta MC: Expression of
epithelial-mesenchymal transition markers at the invasive front of
oral squamous cell carcinoma. J Appl Oral Sci. 23:169–178. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ouderkirk JL and Krendel M: Non-muscle
myosins in tumor progression, cancer cell invasion and metastasis.
Cytoskeleton (Hoboken). 71:447–463. 2014. View Article : Google Scholar : PubMed/NCBI
|