Introduction
Previous comprehensive genetic analyses of
colorectal cancer by microarrays identified translocase of the
outer mitochondrial membrane 34 (TOMM34) and ring finger protein 43
(RNF43) as oncogenes expressed in colorectal cancer (1,2). TOMM34
and RNF43 are considered to be associated with cell proliferation,
and clinical studies of vaccine therapy with an artificially
synthesized cancer peptide based on the amino acid sequence of
tumor antigens such as TOMM34 and RNF43 are ongoing (3–6). A tumor
antigen inoculated by vaccine is used for antigen presentation from
dendritic cells to lymphocytes, and cytotoxic T-lymphocytes (CTLs)
are induced in vivo. The induced CTLs are expected to
recognize the tumor antigen and show an antitumor effect against
cancer cells expressing the tumor antigen. Since it is important
that vaccine therapy should be performed based on the expression
levels of tumor antigens in an individual for the effective
treatment, analysis of oncogene expression is essential.
Formalin-fixed, paraffin-embedded (FFPE) tissue samples are the
most available material for pathological studies. However, the
quality of mRNA extracted from FFPE specimens is generally
considered to be insufficient for gene analyses due to nucleic acid
degradation (7). The present study
examined methodologies for the quantification of TOMM34 and RNF43
in tissue samples extracted from FFPE colorectal cancer specimens.
The samples used in the present study were specimens from 19
patients with colorectal cancer and liver metastasis, and the
tumors were surgically removed in the Department of
Gastroenterological Surgery of Tokyo Women's Medical University
(Tokyo, Japan) between December 2004 and October 2008. The FFPE
samples were prepared from the resected tumors and stored at room
temperature until present examination. The tissue slides were
prepared from the FFPE samples and total RNA was extracted from the
slides. Subsequent to synthesizing cDNA from the extracted total
RNA, quantitative polymerase chain reaction was performed using the
Universal ProbeLibrary as a PCR probe to measure the gene
expression levels of TOMM34 and RNF43 in the several-year-old
colorectal FFPE samples.
Materials and methods
Subjects
Samples were obtained from 19 cases of colorectal
cancer with liver metastasis that were removed surgically at the
Department of Gastroenterological Surgery of Tokyo Women's Medical
University between December 2004 and October 2008. The tissue
samples were paraffin embedded upon formalin fixation and stored at
room temperature until examination. As a negative control for the
expression of TOMM34 and RNF43, peripheral blood mononuclear cells
(PBMCs) were obtained from healthy 35-year-old male. The present
study was approved by the Institutional Review Board of Tokyo
Women's Medical University. Informed consent was obtained from the
subjects for the use of their tissues.
Patient characteristics
As shown in Table I,
the patients' ages ranged from 52 to 78 years (average, 65.1
years). There were 14 males and 5 females. The histological type of
the primary tumors was well differentiated adenocarcinoma in 15
patients and moderately differentiated adenocarcinoma in 4
patients. The site of the primary tumor was the sigmoid colon in 8
(42%) cases, the ascending colon in 6 (32%) cases, the cecum in 3
(16%) cases, the rectum in 1 (5%) case and the transverse colon in
1 (5%) case.
 | Table I.Characteristics of 19 patients with
colorectal cancer and histological type of primary tumors. |
Table I.
Characteristics of 19 patients with
colorectal cancer and histological type of primary tumors.
| Patient no. | Age, years | Sex | Tumor site | Histological type of
primary tumor |
|---|
| 1 | 65 | Male | R | Moderately
differentiated adenocarcinoma |
| 2 | 70 | Male | S | Well differentiated
adenocarcinoma |
| 3 | 64 | Male | A | Well differentiated
adenocarcinoma |
| 4 | 64 | Male | S | Well differentiated
adenocarcinoma |
| 5 | 54 | Male | A | Well differentiated
adenocarcinoma |
| 6 | 69 | Male | S | Well differentiated
adenocarcinoma |
| 7 | 78 | Male | S | Well differentiated
adenocarcinoma |
| 8 | 59 | Female | A | Well differentiated
adenocarcinoma |
| 9 | 69 | Male | S | Well differentiated
adenocarcinoma |
| 10 | 75 | Male | C | Well differentiated
adenocarcinoma |
| 11 | 72 | Female | A | Moderately
differentiated adenocarcinoma |
| 12 | 56 | Female | C | Moderately
differentiated adenocarcinoma |
| 13 | 52 | Female | S | Well differentiated
adenocarcinoma |
| 14 | 55 | Male | A | Moderately
differentiated adenocarcinoma |
| 15 | 64 | Male | C | Well differentiated
adenocarcinoma |
| 16 | 70 | Female | A | Well differentiated
adenocarcinoma |
| 17 | 62 | Male | T | Well differentiated
adenocarcinoma |
| 18 | 72 | Male | S | Well differentiated
adenocarcinoma |
| 19 | 68 | Male | S | Well differentiated
adenocarcinoma |
Colon cancer CW2 cell line
The human colon cancer cell line CW2 (RIKEN
BioResource Center, Tsukuba, Japan) was selected as a positive
control for the expression of TOMM34 and RNF43. CW2 cells were
obtained from a 60-year-old female who underwent the surgical
removal of colon cancer, and whose case was described (8). The cell line was maintained in medium
consisting of RPMI-1640 (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) containing 10% heat-inactivated fetal bovine serum (Thermo
Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin. The cell line was cultured at 37°C in a 95% humidity
atmosphere containing 5% CO2.
Tissue sampling and extraction of
RNA
For the extraction of total RNA from CW2 and PBMCs
of a healthy 35-year-old male donor (mentioned in Subjects
paragraph), the RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA)
was used. From surgically resected specimens, separate slides of
normal and cancer tissue were prepared. The normal mucosa was
obtained from surgical edge of the resected colon which site was at
least 10 cm away from the tumor. Hematoxylin and eosin
(H&E)-stained slides were examined, and the normal and cancer
tissue areas in the slides were confirmed prior to all experimental
procedures. No cancer tissue contaminated the normal tissue slides,
which was confirmed by pathologists. For the extraction of total
RNA from paraffin-embedded specimens, cancer and normal mucosa
(mucosa without tumor) specimens were obtained from slides of the
paraffin blocks previously examined under H&E staining, and
total RNA was extracted upon deparaffinization using an RNeasy FFPE
kit (Qiagen, Inc.). All steps were performed according to the
manufacturer's protocol.
Reverse transcription-polymerase chain
reaction (RT-PCR)
Qualitative RT-PCR using 50 ng total RNA from CW2 or
PBMCs of a 35-year-old male healthy donor (mentioned in Subjects
paragraph) was performed with Ready-To-Go RT-PCR Beads (GE
Healthcare Life Sciences, Little Chalfont, UK) following the
manufacturer's protocol. The 5′-3′ sequences of the primers used
for the reaction were as follows: TOMM34-forward (F), TTG CAG ACA
TCA GCA ACC TC; TOMM34-reverse (R), ACC TTT CTG GTG CAA CAA CC;
RNF43-F, AAA GGA CCA CAG CAA ACA CC; RNF43-R, CTG AAC CCA CTG GCT
GGT AT; GAPDH-F, CGA CCA CTT TGT CAA GCT CA; and GAPDH-R, TGT GAG
GAG GGG AGA TTC AG. The conditions of the PCR were as follows: 95°C
for 5 min, 35 cycles of 95°C for 30 sec, 56°C for 30 sec and 72°C
for 30 sec, and 72°C for 5 min. The PCR products were evaluated by
1% agarose (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) gel
electrophoresis and ethidium bromide (Sigma-Aldrich; Merck KGaA)
staining.
Probes and primers for quantitative
PCR (qPCR) and PCR conditions
The Universal ProbeLibrary (Roche Diagnostics,
Basel, Switzerland) was selected for qPCR analysis, and various
primers and probes were selected using ProbeFinder software version
2.43 from the human set of the Universal ProbeLibrary Assay Design
Center (https://qpcr.probefinder.com/organism.jsp). The
sequences (5′-3′) of the primers for TOMM34 were as follows:
TOMM34-F, CAA ATC CAA AGA AAC CAC AGC; and TOMM34-R, AGA ACT CTG
GCT TTC TCC ACA, and the probe number was 3. The sequences (5′-3′)
of the primers for RNF43 were as follows: RNF43-F, TTA TCC GCA CTG
CCA GGT; and RNF43-R, CAC AGC TCC TCG AGT TCC TC, and the probe
number was 64. The sequences (5′-3′) of the primers for GAPDH were
as follows: GAPDH-F, AGC CAC ATC GCT CAG ACA C; and GAPDH-R, GCC
CAA TAC GAC CAA ATC C, and the probe number was 60. Complementary
DNA (cDNA) was obtained using SuperScript III First-Strand
Synthesis SuperMix for qRT-PCR (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with total RNA (50 ng)
extracted from the paraffin-embedded specimens. qPCR was performed
with this cDNA using LightCycler Taqman Master in a LightCycler
Instrument (both from Roche Diagnostics). All reactions were
conducted following the manufacturer's protocol. The PCR conditions
were as follows: 95°C for 10 min, followed by 45 cycles of 95°C for
10 sec, 60°C for 10 sec and 72°C for 10 sec.
Genetic sequences, relative
quantification of gene expression and statistical analysis
The genetic sequences for TOMM34, RNA43 and GAPDH
were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The accession
numbers were as follows: TOMM34 (NM_006809.4), RNF43 (NM_017763.4)
and GAPDH (NM_002046.3).
For the relative quantification of target gene
expression, a previously reported calculation method was used
(9). The point at which the
fluorescence of a sample exceeded that of the background was
identified as a crossing point (CP) in the amplification process of
the PCR. The CP value of the reference gene GAPDH was subtracted
from the CP value of each sample, and this value was defined as
ΔCP. The ΔCP of the colon cancer cell line CW2 used as a control
was subtracted from the ΔCP of each sample, and the resulting value
was defined as ΔΔCP. The relative gene expression level of each
sample was calculated as 2−ΔΔCP, and the data obtained
were used for further evaluation of gene expression. The gene
expression data was collected using Microsoft Excel 2013
(Microsoft, Redmond, WA, USA) and was statistical analyzed using
IBM SPSS Statistics 21 (IBM SPSS, Armonk, NY, USA). The Student's
t-test was performed to compare the results between two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
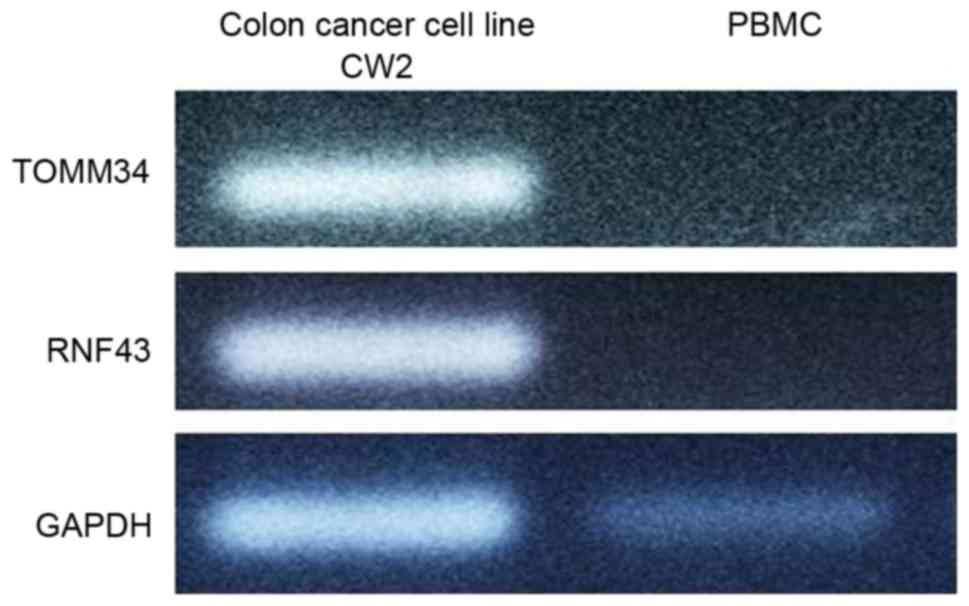
Qualitative PCR of the colon cancer
cell line CW2
Total RNA was extracted from CW2 cells and used in
qualitative PCR (Fig. 1). TOMM34 and
RNF43 expression was detected in CW2 cells but no in PBMCs. Based
on these results, CW2 cells were selected as a positive
control.
Relative gene expression level of each
sample
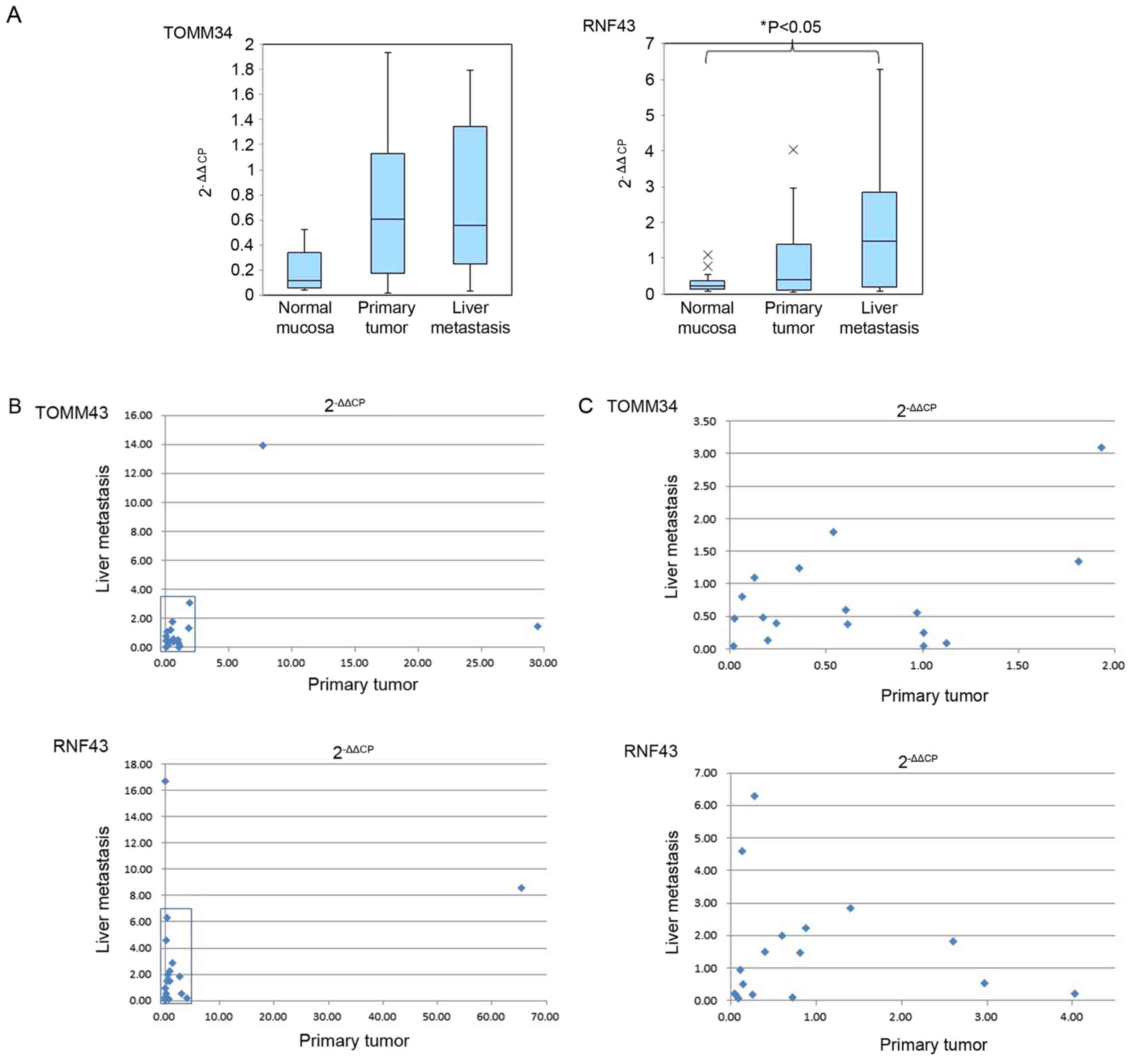
The 2−ΔΔCP data of the normal mucosa,
primary tumor and liver metastasis are summarized in Table II. TOMM34 expression was elevated,
compared with that in the normal mucosa, in 15 of the 19 primary
tumors (78.9%) and in 14 of the 19 samples of liver metastasis
(73.7%). The graph depicted in Fig.
2A indicates mean and SD of TOMM34 expression in normal mucosa,
primary tumor and liver metastasis obtained from all patients
showed higher expression in the primary tumors and liver metastases
compared with the gene expression in the normal mucosa, but the
difference was not significant. Similarly, RNF43 exhibited higher
expression, compared with that in the normal mucosa, in 12 of the
19 primary tumors (63.2%) and in 14 of the 19 liver metastases
(73.7%). The 2−ΔΔCP values of RNF43 in the primary
tumors and liver metastases were compared with those in the normal
mucosa. The expression of RNF43 in the liver metastases was
significantly higher (P<0.05) than that in the normal mucosa
(Fig. 2A). TOMM34 and RNF43 did not
show significant differences in relative gene expression level
(Fig. 2A) between their expression in
primary tumors and liver metastases. The gene expression level of
all patients' primary tumors and liver metastases were plotted in
Fig. 2B. Fig. 2C showed enlarged figure of squared
area of Fig. 2B. There was no
correlation of measured gene expression levels between primary
tumors and liver metastases in both TOMM34 and RNF43 data.
 | Table II.Relative gene expression levels
(2−ΔΔCP value) of TOMM34 and RNF43 in 19 patients with
colorectal cancer. |
Table II.
Relative gene expression levels
(2−ΔΔCP value) of TOMM34 and RNF43 in 19 patients with
colorectal cancer.
| A, TOMM34 |
|---|
|
|---|
|
|
Patient |
|---|
|
|
|
|---|
| Tissue | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 |
|---|
| Normal mucosa | 0.21 | 0.16 | 0.52 | 0.12 | 0.48 | 0.09 | 0.04 | 0.17 | 0.11 | 0.34 | 4.23 | 0.41 | 0.10 | 0.04 | 0.09 | 0.06 | 0.06 | 0.11 | 0.04 |
| Primary tumor | 0.54 | 1.82 | 0.02 | 0.36 | 1.93 | 0.97 | 1.13 | 0.60 | 0.61 | 1.01 | 7.73 | 0.24 | 0.06 | 0.17 | 0.02 | 1.01 | 0.19 | 0.13 | 29.45 |
| Liver metastasis | 1.79 | 1.35 | 0.46 | 1.24 | 3.10 | 0.55 | 0.09 | 0.59 | 0.37 | 0.25 | 13.93 | 0.39 | 0.80 | 0.48 | 0.04 | 0.04 | 0.12 | 1.09 |
1.48 |
|
| B, RNF43 |
|
|
| Patient |
|
|
|
| Tissue | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 |
|
| Normal mucosa | 0.78 | 0.13 | 0.27 | 0.11 | 0.09 | 0.30 | 0.16 | 0.15 | 0.14 | 0.56 | 0.32 | 0.08 | 0.36 |
0.13 | 0.33 | 1.09 | 0.23 | 0.42 |
0.16 |
| Primary tumor | 0.81 | 0.40 | 0.05 | 1.40 | 2.60 | 0.60 | 0.11 | 0.25 | 0.88 | 0.73 | 2.97 | 0.09 | 0.14 |
0.05 | 0.04 | 4.03 | 0.14 | 0.28 | 65.34 |
| Liver
metastasis | 1.46 | 1.51 | 0.18 | 2.85 | 1.82 | 1.99 | 0.95 | 0.20 | 2.23 | 0.11 | 0.54 | 0.07 | 4.59 | 16.68 | 0.22 | 0.21 | 0.51 | 6.28 |
8.57 |
Discussion
TOMM34 and RNF43 have been identified as oncogenes
by comprehensive gene studies such as microarrays, and
investigations of these genes may lead to the development of novel
and effective molecular target drugs (1,2). TOMM34
protein is present inside the nucleus and cytoplasm of tumor cells,
whereas no expression is observed in normal organs, with the
exception of the testes and ovaries (10,11). High
expression of TOMM34 is observed in colorectal cancer, which is
considered to be associated with tumor growth, since cell
proliferation is inhibited when the function of this gene is
blocked by small interfering RNA (1).
TOMM34 is also present in tumors such as hepatocellular carcinoma,
lung cancer, bladder cancer, acute myeloid leukemia and soft tissue
sarcoma (1). RNF43 is present at the
endoplasmic reticulum and nucleus membrane; it is associated with
cell proliferation and has ubiquitin ligase activity. High
expression of RNF43 is observed in colorectal cancer and colorectal
adenoma (2,12–14). In
addition, RNF43 mutation has been reported in hepatocellular
carcinoma, lung cancer, pancreas cystic tumor and
cholangiocarcinoma (15–18).
Dendritic cells are antigen-presenting cells, and it
is considered that these cells present cancer antigens to
lymphocytes and induce tumor antigen-specific lymphocytes in
vivo (19). A clinical trial of
cancer vaccine therapy with dendritic cells pulsed with tumor
lysate (used as an adjuvant postoperative treatment for patients
with cholangiocarcinoma) demonstrated improvement of overall
survival and relapse-free survival (20). Therefore, further development of such
cancer vaccine therapies is expected.
Clinical trials of vaccine therapy using
artificially synthesized cancer peptides based on the amino acid
sequence of the tumor antigens TOMM34 and RNF43 are ongoing
(3–6).
A tumor antigen inoculated by vaccine is used for antigen
presentation from dendritic cells to lymphocytes, and cytotoxic
T-lymphocytes (CTLs) are induced in vivo. CTLs that
recognize the tumor antigen show an anti-tumor effect against
cancer cells expressing the tumor antigen. In fact, higher CTL
induction was confirmed in proportion to the peptide concentration,
and patients with a strong skin reaction at the vaccination site or
a greater number of CTL responses to the peptide exhibited
significantly longer overall survival (4,6).
In light of these results, cancer peptide vaccine
therapy appears to be a promising strategy by which treatment can
be customized to the tumor antigen expression levels on each tumor.
RT-qPCR can be conducted to quantify the gene expression of the
tumor, and even a small amount of mRNA can be evaluated by this
method. Using this assay, the present study was able to quantify
the tumor antigen expression of an individual cancer, and a cancer
peptide vaccine therapy could be tailored to the results.
Furthermore, the correlation between the effectiveness of the
vaccine and the gene expression level can be evaluated by measuring
the gene expression in a stored specimen. However, mRNA from an old
sample or a sample upon formalin fixation and paraffin embedding
may be degraded by these procedures, and the quality of the mRNA
may not be suitable for RT-qPCR.
In fact, TOMM34 and RNF43 expression could not be
detected in samples derived from a paraffin block by qualitative
PCR in our preliminary studies (data not shown). To the best of our
knowledge, no study has attempted a quantification of TOMM34 and
RNF43 expression in paraffin block samples. The present study was
able to quantify the expression levels of TOMM34 and RNF43 mRNA
upon extracting total RNA from primary colorectal cancer, liver
metastasis and normal large intestine mucous membrane using the
Universal ProbeLibrary assay. In this assay, the pair of probes
created from 8–9 bases and the primers exhibited the specificity of
the transcription product. Since the length of the PCR product was
<150 bp, it may be possible to perform this type of assay
without the influence of RNA destruction.
The present study observed that the mRNA expression
of TOMM34 and RNF43 in the primary tumor and liver metastasis
tissue samples tended to be higher than that in the normal mucosa,
but a significant difference was observed only for RNF43 between
the liver metastases and normal mucosa, possibly due to an
insufficient number of subjects. However, these oncogenes were
confirmed to be present in the canceration process of the
large-intestine mucous membrane, and effectiveness of a cancer
vaccine therapy against a cancer that expresses those tumor
antigens is expected. The gene quantitative methodology used in the
present study may be useful for the refinement of cancer vaccine
therapies based on tumor antigen expression. In future studies, the
correlation between tumor antigen levels in stored samples and
therapeutic outcomes will be evaluated.
In conclusion, although qualitative RT-PCR to
confirm the expression of TOMM34 and RNF43 in paraffin-embedded
samples was difficult to perform, the present study was able to
quantify the expression levels of these genes using a Universal
ProbeLibrary assay. This methodology may be useful for both
customized cancer vaccine therapy based on tumor antigen expression
levels and for the development of molecular targeting drugs. The
present results confirmed that TOMM34 and RNF43 tended to increase
in the canceration process of colorectal cancer in several-year-old
samples stored in paraffin blocks. However, there was no
significant difference in gene expression between the primary tumor
and liver metastasis tissues in the canceration process. The
present findings should be assessed in further studies with
additional cases.
References
|
1
|
Shimokawa T, Matsushima S, Tsunoda T,
Tahara H, Nakamura Y and Furukawa Y: Identification of TOMM34,
which shows elevated expression in the majority of human colon
cancers, as a novel drug target. Int J Oncol. 29:381–386.
2006.PubMed/NCBI
|
|
2
|
Uchida N, Tsunoda T, Wada S, Furukawa Y,
Nakamura Y and Tahara H: Ring finger protein 43 as a new target for
cancer immunotherapy. Clin Cancer Res. 10:8577–8586. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Matsushita N, Aruga A, Inoue Y, Kotera Y,
Takeda K and Yamamoto M: Phase I clinical trial of a peptide
vaccine combined with tegafur-uracil plus leucovorin for treatment
of advanced or recurrent colorectal cancer. Oncol Rep. 29:951–959.
2013.PubMed/NCBI
|
|
4
|
Hazama S, Nakamura Y, Takenouchi H, Suzuki
N, Tsunedomi R, Inoue Y, Tokuhisa Y, Iizuka N, Yoshino S, Takeda K,
et al: A phase I study of combination vaccine treatment of five
therapeutic epitope-peptides for metastatic colorectal cancer;
safety, immunological response, and clinical outcome. J Transl Med.
12:632014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yasuda S, Tsuchiya I, Okada K, Tanaka A,
Suzuki T, Sadahiro S, Takeda K, Yamamoto S and Nakui M: Significant
clinical response of advanced colon cancer to peptide vaccine
therapy: A case report. Tokai J Exp Clin Med. 37:57–61.
2012.PubMed/NCBI
|
|
6
|
Okuno K, Sugiura F, Hida JI, Tokoro T,
Ishimaru E, Sukegawa Y and Ueda K: Phase I clinical trial of a
novel peptide vaccine in combination with UFT/LV for metastatic
colorectal cancer. Exp Ther Med. 2:73–79. 2011.PubMed/NCBI
|
|
7
|
Srinivasan M, Sedmak D and Jewell S:
Effect of fixatives and tissue processing on the content and
integrity of nucleic acids. Am J Pathol. 161:1961–1971. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miyazono Y, Kamogawa Y, Ryo K, Furukawa T,
Mitsuhashi M, Yamauchi K, Kameoka T and Hayashi N: Effect of
B7.1-transfected human colon cancer cells on the induction of
autologous tumour-specific cytotoxic T cells. J Gastroenterol
Hepatol. 14:997–1003. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Terada KJ, Ueno S, Yomogida K, Imai T,
Kiyonari H, Takeda N, Yano M, Abe S, Aizawa S and Mori M:
Expression of Tom34 splicing isoforms in mouse testis and knockout
of Tom34 in mice. J Biochem. 133:625–631. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chewawiwat N, Yano M, Terada K, Hoogenraad
NJ and Mori M: Characterization of the novel mitochondrial protein
import component, Tom34, in mammalian cells. J Biochem.
125:721–727. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sugiura T, Yamaguchi A and Miyamoto K: A
cancer-associated RING finger protein, RNF43, is a ubiquitin ligase
that interacts with a nuclear protein, HAP95. Exp Cell Res.
314:1519–1528. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takahashi N, Yamaguchi K, Ikenoue T, Fujii
T and Furukawa Y: Identification of two Wnt-responsive elements in
the intron of RING finger protein 43 (RNF43) gene. PLoS One.
9:e865822014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shinada K, Tsukiyama T, Sho T, Okumura F,
Asaka M and Hatakeyama S: RNF43 interacts with NEDL1 and regulates
p53-mediated transcription. Biochem Biophys Res Commun.
404:143–147. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yagyu R, Furukawa Y, Lin YM, Shimokawa T,
Yamamura T and Nakamura Y: A novel oncoprotein RNF43 functions in
an autocrine manner in colorectal cancer. Int J Oncol.
25:1343–1348. 2004.PubMed/NCBI
|
|
16
|
Koo BK, Spit M, Jordens I, Low TY, Stange
DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM
and Clevers H: Tumour suppressor RNF43 is a stem-cell E3 ligase
that induces endocytosis of Wnt receptors. Nature. 488:665–669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu J, Jiao Y, Dal Molin M, Maitra A, de
Wilde RF, Wood LD, Eshleman JR, Goggins MG, Wolfgang CL, Canto MI,
et al: Whole-exome sequencing of neoplastic cysts of the pancreas
reveals recurrent mutations in components of ubiquitin-dependent
pathways. Proc Natl Acad Sci USA. 108:pp. 21188–21193. 2011;
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ong CK, Subimerb C, Pairojkul C, Wongkham
S, Cutcutache I, Yu W, McPherson JR, Allen GE, Ng CC, Wong BH, et
al: Exome sequencing of liver fluke-associated cholangiocarcinoma.
Nat Genet. 44:690–693. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sadanaga N, Nagashima H, Mashino K, Tahara
K, Yamaguchi H, Ohta M, Fujie T, Tanaka F, Inoue H, Takesako K, et
al: Dendritic cell vaccination with MAGE peptide is a novel
therapeutic approach for gastrointestinal carcinomas. Clin Cancer
Res. 7:2277–2284. 2001.PubMed/NCBI
|
|
20
|
Shimizu K, Kotera Y, Aruga A, Takeshita N,
Takasaki K and Yamamoto M: Clinical utilization of postoperative
dendritic cell vaccine plus activated T-cell transfer in patients
with intrahepatic cholangiocarcinoma. J Hepatobiliary Pancreat Sci.
19:171–178. 2012. View Article : Google Scholar : PubMed/NCBI
|