Introduction
Folinic acid, leucovorin, 5-fluorouracil and
oxaliplatin (FOLFOX)-based chemotherapy (CT) is widely used in the
treatment of colorectal adenocarcinoma (CRC). The addition of the
platinum-containing compound oxaliplatin to the standard adjuvant
treatment with 5-fluorouracil plus leucovorin enhances the efficacy
of CT, doubling the response rate and prolonging progression-free
survival of patients with stage II–III CRC (1,2). Despite
positive results obtained from this chemotherapeutic regimen,
unwanted side effects are significant and require monitoring, as
they affect, to various degrees, the quality of patient.
In an attempt to identify biomarkers that may
predict the onset of these secondary effects, a recent study
investigated the differential gene expression of peripheral
leukocytes in patients with CRC prior to and following 3 cycles of
oxaliplatin-based CT (3). The study
identified 502 differentially-expressed genes that were
significantly up- or downregulated in the peripheral white cells
(PWCs) of patients following CT treatment (3).
To determine whether the changes in gene expression
observed in PWCs may be detected in tumors following the
administration of adjuvant CT, the present study performed
immunohistochemical analysis of a selected number of genes that had
previously been identified in the differential transcriptome
profile (4). The study observed that
one gene among four, whose transcriptional expression levels varied
from ‘off’ to ‘on’, was that coding for IQ-motif containing GTPase
activating protein 1 (IQGAP1) (3).
IQGAP1 is a ubiquitously expressed scaffold protein,
which contains a number of protein interaction domains (5). The protein interacts with cell adhesion
molecules, components of the cytoskeleton, and various signaling
molecules to regulate cell motility and morphology, cell cycle and
other cellular functions (5,6). By regulating its binding partners,
IQGAP1 integrates a number of signaling pathways, several of which
contribute to tumorigenesis. For example, IQGAP1 is associated with
actin dynamics by direct binding to actin or indirect regulation
via cell division cycle 42/Rac1 (7).
Through its polyproline protein-protein domain (WW domain), the
protein modulates the mitogen-activated protein kinase (MAPK)
pathway, which is associated with cell cycle control (8). Therefore, IQGAP1 links MAPK signaling
(for example, decisions regarding cell fate) to the cytoskeleton or
cellular adhesion, with important implications for cancer.
Furthermore, interactions of IQGAP1 with
extracellular signal-regulated kinases 1/2 and MEK1/2 may lead to
activation of the MAPK signaling pathway, thus modulating cell
differentiation and proliferation (8). Therefore, IQGAP1 serves pivotal roles in
several cellular functions, including control of polarization, cell
adhesion, migration, proliferation and angiogenesis.
Numerous immunohistochemical studies have
demonstrated that IQGAP1 is overexpressed in several forms of
cancer and an aberrant membrane accumulation is observed,
particularly at the invasive front (9,10). Higher
expression and altered localization of IQGAP1 from the cytoplasm to
the membrane correlates with tumor grade and poor prognosis
(11). The presence of IQGAP1 in the
cell membrane may decrease adherens junction (AJ) function,
favoring dissociation of the tumor cells (12).
The present study reports the alteration of IQGAP1
expression in CRC and liver metastases following CT administration
at the protein level by performing an immunohistochemical
analysis.
Patients and methods
Patients
The study was approved by the Ethics Committee of La
Laguna University (La Laguna, Canary Islands, Spain) and the
Ethical Committee of Nuestra Señora de Candelaria University
Hospital (HUNSC; Santa Cruz de Tenerife, Canary Islands, Spain).
All patients were treated at University Hospital of the Nuestra
Señora de Candelaria, Santa Cruz de Tenerife, Spain between May
2007 and September 2015 and provided informed consent for the
diagnosis and research of tissue specimens prior to enrollment in
the study. All patients had colonic cancer and liver metastasis,
which had been treated with FOLFOX-CT, (day 1, oxaliplatin 100
mg/m2 iv over 2 h; leucovorin calcium 400
mg/m2 iv over 2 h; followed by 5-fluorouracil 400
mg/m2 iv bolus and by 5-fluorouracil 2400
mg/m2 iv over 46 h; every 14 days). All patients
received CT following resection of the primary tumor. Therefore,
the primary tumors were CT-naïve, while the liver metastases were
CT-treated.
Tumor tissue
Paraffin-embedded tissue samples from 15 patients (7
males and 8 females), ensuring patient's anonymity, and the
corresponding clinical data were obtained from the reference
medical areas of HUNSC.
Antibodies
The following antibodies were used: Rabbit
anti-human polyclonal antibody against IQGAP1 (dilutions, 1:500 for
immunohistochemistry and 1:250 for immunofluorescence; cat. no.
ABT186; EMD Millipore, Billerica, MA, USA); mouse monoclonal
antibody clone PC10 against anti-proliferating cell nuclear antigen
(PCNA; dilution, 1:100; cat. no. 1486772; Roche Diagnostics GmbH,
Mannheim, Germany); mouse monoclonal anti-human cluster of
differentiation (CD)34 class II clone QBEnd 10 (ready-to-use; cat.
no. IR632; Dako; Agilent Technologies GmbH, Waldbronn, Germany);
and mouse monoclonal anti-β tubulin (dilution, 1:150; cat. no.
sc-101527; Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
Secondary antibodies: Biotin-conjugated anti-rabbit secondary
antibody (dilution, 1:300; cat. no. 31820; Pierce; Thermo Fisher
Scientific, Inc., Waltham, MA, USA); fluorescein isothiocyanate
(FITC)-conjugated goat polyclonal antibody against rabbit IgG (cat.
no. F9887; Sigma-Aldrich, Merck Merck Millipore, Darmstadt,
Germany; dilution, 1:200); goat polyclonal antibody against mouse
IgG (DyLight® 650; cat. no. ab97018; Abcam, Cambridge, UK;
dilution, 1:100).
Immunohistochemistry
Immunoperoxidase staining of paraffin-embedded
tissue sections (fixed in 10% formalin, for 48–72 h at 4°C) was
performed using the avidin-biotin reaction. Briefly, 5-µm-thick
tissue sections were deparaffinized in xylene and hydrated in a
graded series of alcohol baths. Heat-induced epitope retrieval was
achieved by heating samples in sodium citrate buffer (pH 6.0) at
120°C for 10 min in an autoclave. Once non-specific sites were
blocked with 5% non-fat dry milk in TBS for 1 h at room
temperature, endogenous biotin was blocked using the Avidin/Biotin
Blocking kit (Vector Laboratories Inc., Burlingame, CA, USA). The
primary antibody against IQGAP1 was applied (dilution, 1:500) to
slides over night at 4°C. To block endogenous peroxidase activity,
the slides were incubated with 3% hydrogen peroxidase in methanol
for 15 min. Biotin-conjugated anti-rabbit secondary antibody
(dilution, 1:300; Pierce; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) was incubated for 2 h at 37°C, and ABC Peroxidase Staining
kit (Thermo Fisher Scientific, Inc.) was used to amplify the
specific antibody staining. 3,3′-diaminobenzidine substrate
concentrate (no. IHC-101F; Bethyl Laboratories Inc., Montgomery,
TX, USA) was used to visualize immunohistochemical reactions.
Samples incubated without primary antibodies were used as a
negative control. Slides were counterstained with Harris
hematoxylin solution DC (Panreac Química SLU, Barcelona, Spain) to
visualize cell nuclei and mounted with Eukitt mounting medium
(Panreac Química SLU). An optical light microscope (BX50; Olympus
Corporation, Tokyo, Japan) was used to visualize the results of the
immunostaining.
Image analysis and statistical
analysis
To compile tables, two independent observers
evaluated the specimens blindly. Staining intensities were graded
as absent (−), weak (+), moderate (++) or strong (+++). These
cut-offs were established by consensus between each investigator
following an initial survey of the entire blind-coded material. In
cases where scorings differed by more than one unit, the observers
re-evaluated the specimens to reach a consensus. In other cases,
means of the scorings were calculated.
Double immunofluorescence simultaneous
staining
Following deparaffinization, hydration and a
heat-induced epitope retrieval procedure (as described above),
tissue sections were incubated simultaneously with a mixture of two
distinct primary antibodies (rabbit anti-human polyclonal antibody
against IQGAP1 and mouse monoclonal antibody anti-PCNA, anti-CD34
or anti-β-tubulin) overnight at 4°C. Slides were then incubated for
1 h at room temperature in the dark with a mixture of two secondary
antibodies raised in different species and conjugated to two
different fluorochromes [anti-rabbit fluorescein
isothiocyanate-conjugated antibody (Sigma-Aldrich; Merck Millipore)
and anti-mouse DyLight®650-conjugated antibody (Abcam)]. Slides
were mounted with ProLong® Diamond Anti-fade Mountant with DAPI
(Molecular Probes®; Thermo Fisher Scientific, Inc.) to visualize
cell nuclei. Slides were analyzed using a confocal microscope
(FV1000; Olympus Corporation).
Results
IQGAP1 expression and localization in
healthy colon and CRC tissue sections
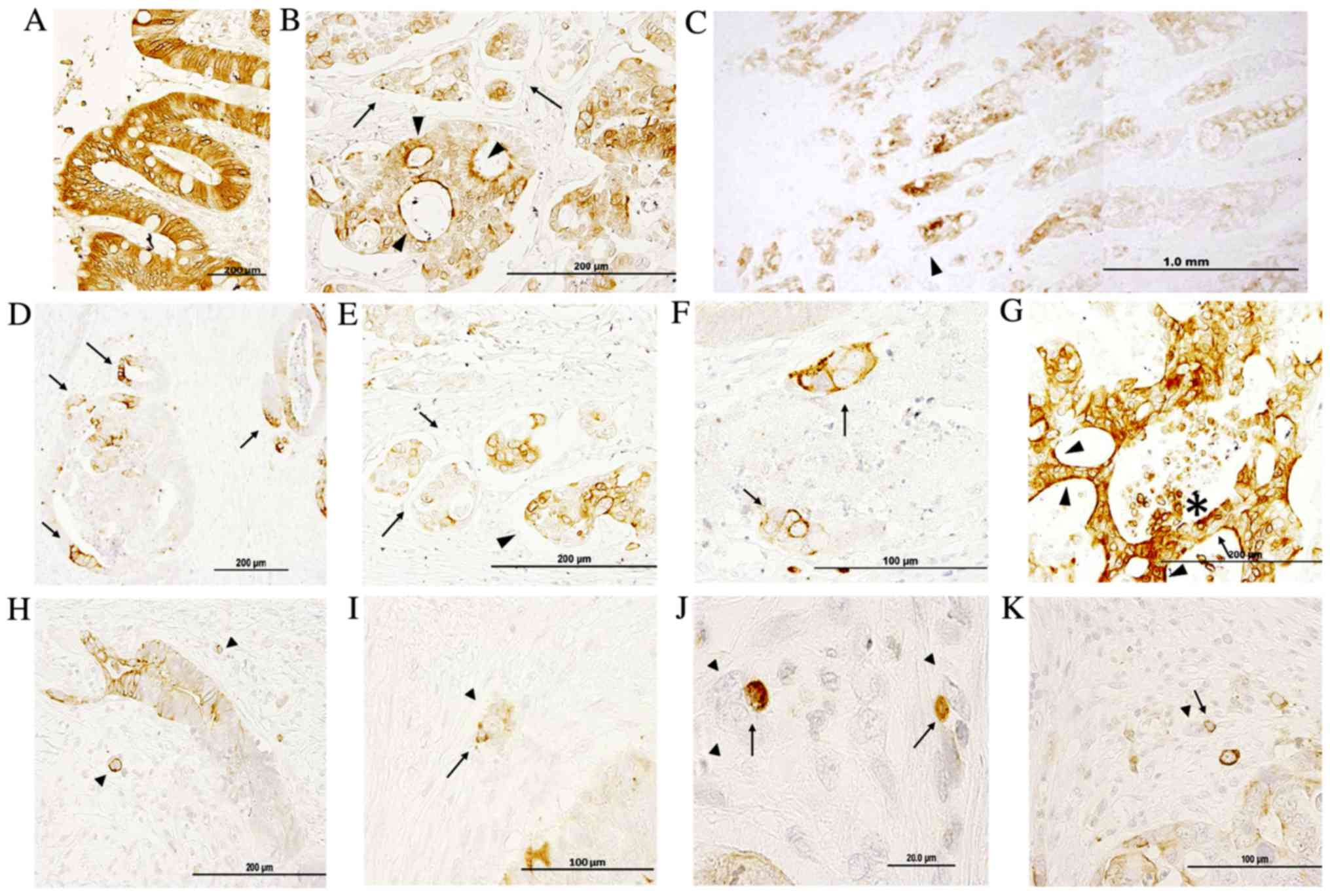
Fig. 1 demonstrates
IQGAP1 immunoreactivity in paraffin-embedded sections of normal
colon and in healthy areas of adenocarcinoma sections. The signal
revealed positive staining in enterocytes (nuclear membrane,
cytoplasm, apical and lateral cell membrane) (Fig. 1A); the majority of cells were stained
and only a few exhibited a lighter level of staining. IQGAP1
protein was localized in cytoplasm, nuclear envelope, cell
junctions, plasma membrane and apical membrane, and this variable
localization could be observed in the same structure concurrently
(Fig. 1B and D-F). Within the tumor,
tumor cells exhibited a diffuse pattern with a variable level of
staining (from no staining to high intensity) in the nuclear
envelope and in the cytoplasm. The intensity of staining was higher
at the invasive front (Fig. 1C).
Cancer cell nests exhibited variable positive perinuclear and
cytoplasmic staining in the open-lumen lymphatic ducts located in
the submucosa (Fig. 1B and E, black
arrows). In Fig. 1D, the intense
membranous IQGAP1 staining appeared polarized in several tumor
glands, while the other cells of the lesion were negative or
exhibited only weak reactivity. Thus, the expression pattern and
localization of IQGAP1 protein in the CRC tissue sections was
heterogeneous, both in the normal glandular epithelium and in tumor
glands and nests. Strongly IQGAP1-positive cells were intermixed
with unstained tumor cells within tumor lesions (Fig. 1G, black arrow); in several carcinoma
cell clusters located in close proximity to tumor glands, a strong
and diffuse membranous immunolocalization was observed, while
nuclei were negative (Fig. 1F, black
arrows). In several lesions, strong apical cell membrane staining
was observed (arrowhead in Fig. 1G);
in addition, strong IQGAP1 expression was observed in areas of the
lesion where cells were detaching into the lumen (asterisk in
Fig. 1G).
Fig. 1H-K shows
representative images of budding tumor cells (arrowheads) either as
small clusters of cells (<5 cells) or as single cells. IQGAP1
expression was variable, with certain cells exhibiting a strong
positive signal all along the cell membrane, while others were
completely negative. The presence of IQGAP1+ immune
cells (arrows) was associated with budding cells.
IQGAP1 expression and localization in
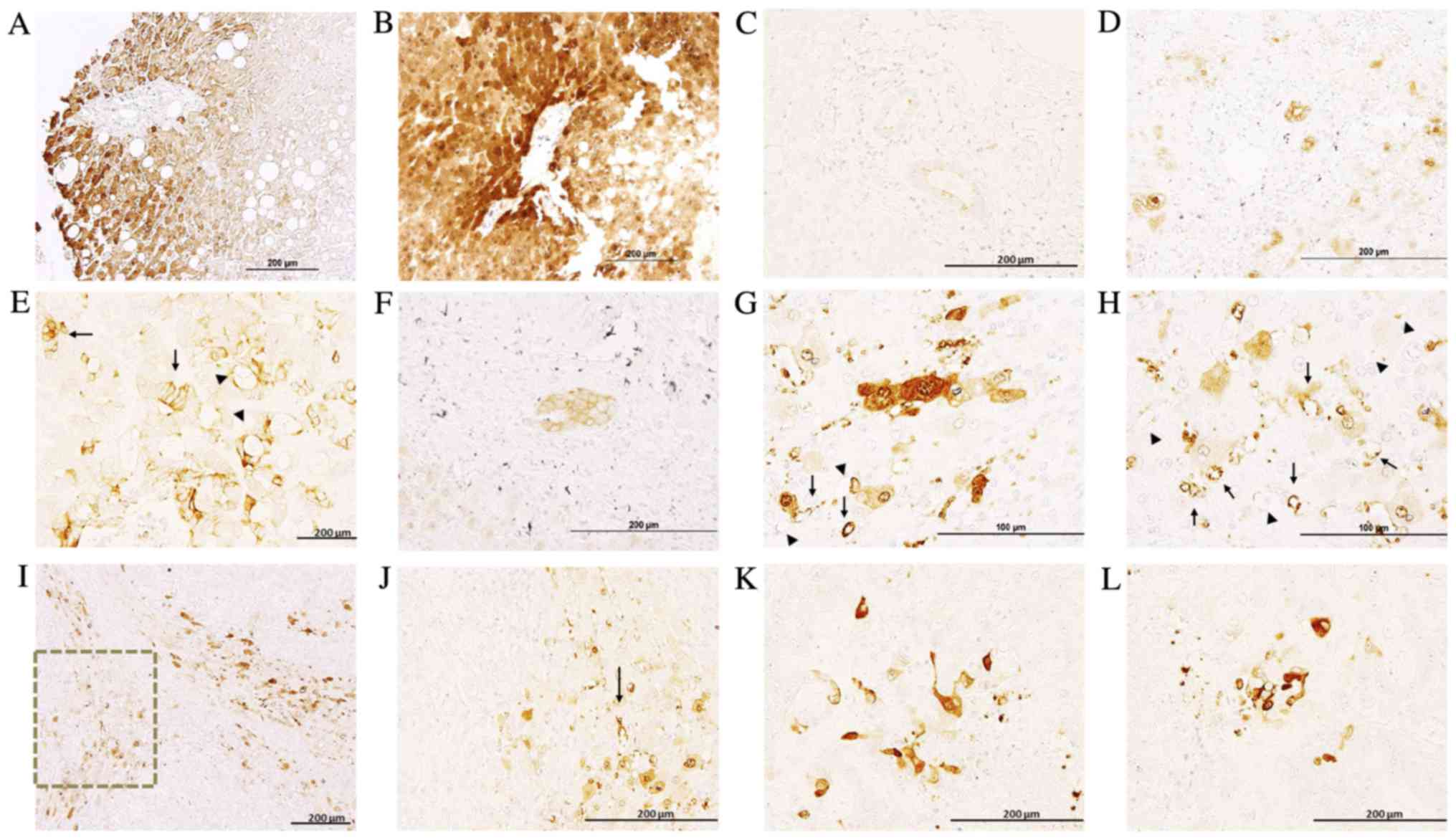
healthy and metastasized liver tissue sections
In healthy liver and the healthy region of
metastasized liver tissues, hepatocytes exhibited a clearly
delimitated labeling in the plasma membrane, cytoplasm and nuclear
envelope, exhibiting non-homogenous IQGAP1+
immunostaining, with neighboring hepatocytes demonstrating variable
levels of expression (Fig. 2). Along
the tissue sections, there were areas with no signal and areas
exhibiting variable positive staining. The intensity of the signal
was higher in the metastasized tissue sections compared with normal
liver sections (Fig. 2A and B). Bile
ducts exhibited weak apical staining in normal liver sections
(Fig. 2C), while in metastasized
liver sections they demonstrated intense membranous positive
staining (Fig. 2D).
Metastases exhibited variable levels of IQGAP1
expression, and were heterogeneous in terms of intensity and
localization (Fig. 2E-G). Positive
immunostaining was observed in several stromal cells (Fig. 2H-L) and in microvessels present in
areas adjacent to the metastasis (Fig. 2G
and H). To assess the nature of these IQGAP1+
vessels, the vascular endothelium marker CD34 was used. Confocal
analyses of tissue sections double immune-labeled for CD34 and
IQGAP1 revealed co-localization of each protein in several vessels
(Fig. 3A-E).
Tables I and II specify the IQGAP1 expression signatures
observed in the tissue specimens.
 | Table I.IQGAP1 expression and localization in
healthy colon and CRC tissues. |
Table I.
IQGAP1 expression and localization in
healthy colon and CRC tissues.
| Cell type | Healthy colon | CRC |
|---|
| Epithelial cells
(Mucosae) | +++ | +++/− |
|
| lm, am, n, c | pm, lm, c, n |
| Stromal cells
(Mucosae) | − | ++/− |
| Tumor cells | − | +++/− |
|
|
| pm, c, n |
| Tumor associated
stromal cells | − | ++/− |
| Immune cells | ? | +++ |
| Endothelial
cells | ++ | ++ |
|
| n | n, c |
| Smooth muscle
cells | ++ | − |
|
| n |
|
| Neurons
(Myenteric plexus) | ++ | − |
| Glial cells
(Myenteric plexus) | ++ | − |
 | Table II.IQGAP1 expression and localization in
healthy and colorectal adenocarcinoma metastasized liver. |
Table II.
IQGAP1 expression and localization in
healthy and colorectal adenocarcinoma metastasized liver.
| Cell type | Healthy liver | Metastasized
liver |
|---|
| Tumor cells | − | ++/− |
|
|
| pm, c |
| Hepatocytes | +++/− | +++/− |
|
| n, c | n, c |
| Epithelial
cells | + | ++ |
| (bile ducts) | am | pm |
| Tumor associated
stromal cells | − | ++ |
To evaluate the proliferative activity of tumor
cells, the proliferation marker PCNA was used in double
immunofluorescence experiments on formalin-fixed, paraffin-embedded
tissue sections. In several malignant cells forming tumor nests,
IQGAP1 and PCNA protein exhibited partial co-localization. The
distribution of intranuclear PCNA and IQGAP1 surrounding the outer
membrane of the nucleus suggests that these cells were in the early
S phase (Fig. 3F-I).
In Fig. 3J-M,
IQGAP1+ staining was observed along the cell membrane of
several tumor cells, and in tumor-associated blood and
microvessels. Within the lesion, PCNA intensity and localization
varied depending on the stage of the cell cycle in which the cells
were. Notably, IQGAP1+ staining was frequently observed
in cells in G1/S phase when PCNA begins to translocate from the
cytoplasm to the nucleus (Fig. 3F-I),
or in cells in early S phase when PCNA expression in the nucleus is
weaker (Fig. 3J-M, yellow arrowhead).
High levels of PCNA in the nucleus helps to identify late S phase
cells; within these cells, no IQGAP1+ signal was
observed (Fig. 3J-M, white
arrowhead), except occasionally in the nucleolus (Fig. 3Q).
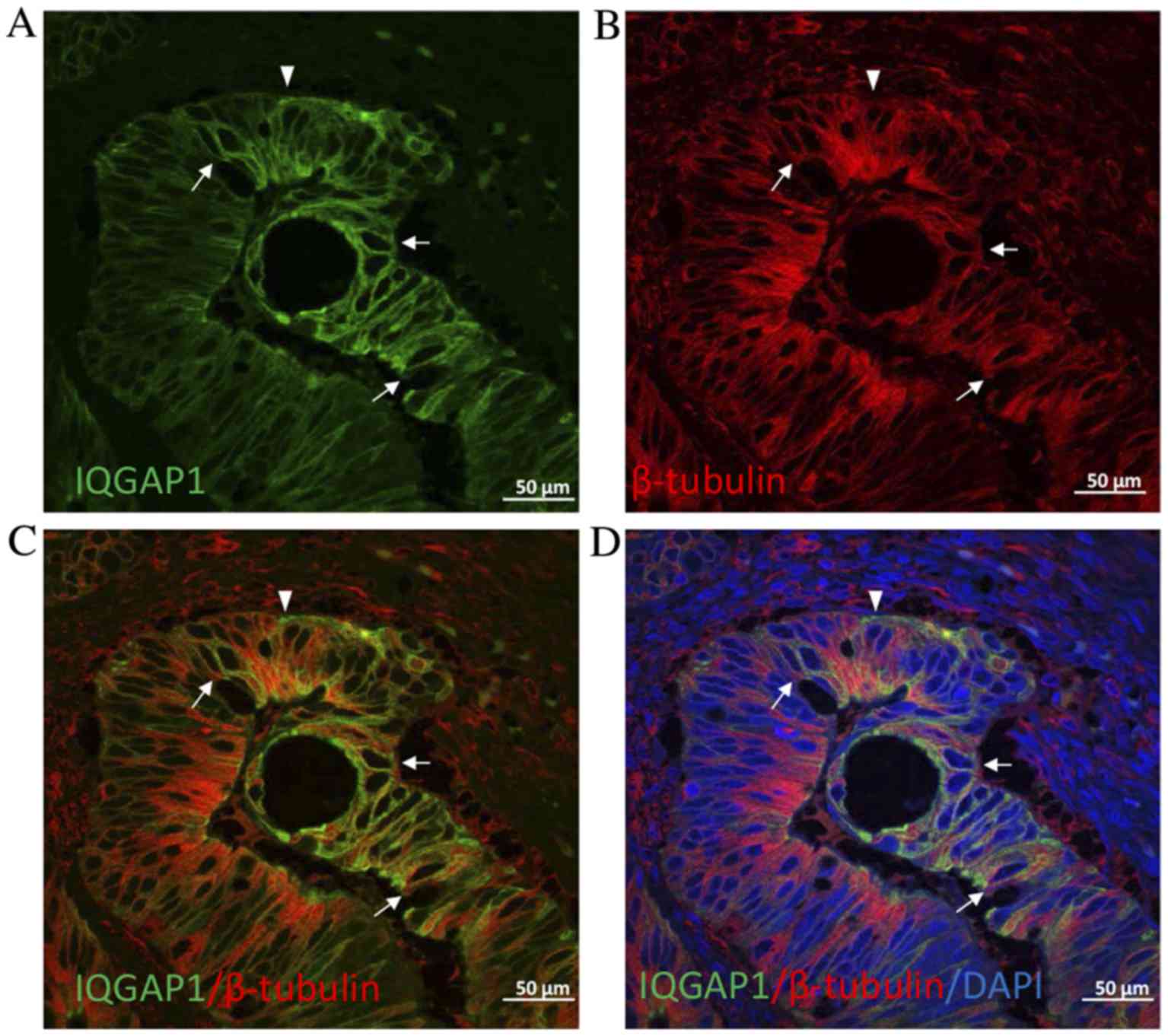
Given the key role of IQGAP1 in the regulation of
the cytoskeleton, double immunofluorescence experiments were
performed on CRC tissue sections to localize IQGAP1 and β-tubulin.
As presented in Fig. 4, IQGAP1
co-localized with microtubules at the cytoplasmic face of the
nuclear envelope in certain cells (arrows). However, in other
cells, the pole of the cell was stained for IQGAP1 with no label
for β-tubulin. Nucleolar staining was often observed throughout the
fields of observation (Figs.
1–4).
Discussion
The present study aimed to study the expression of
IQGAP1 in CRC and liver metastases following the administration of
adjuvant CT using immunohistochemistry. In contrast to normal
cells, in which IQGAP1 was homogeneously distributed, cancer cells
presented variable expression patterns, ranging from no expression
to whole cell, high level of expression (Fig. 1A-C). As shown in Fig. 1C, maximum expression was observed at
the growing front of the tumor gland indicating the expanding
direction, which corresponded to the apical region of the expanding
cell in which nuclei were in the opposite pole. Furthermore, double
labeling by IQGAP1 and β-tubulin demonstrated that in certain tumor
cells the association of each protein disappeared (Fig. 4), indicating clear points where
epithelial to mesenchymal cell transition (EMT) occurs. This
supports the role of IQGAP scaffold proteins in the regulation of
membrane dynamics by coupling the cortical actin meshwork to
microtubules via plus-end binding proteins and other proteins
involved in intracellular signaling that, ultimately, results in
modulation of more complex functions, including cytokinesis, cell
migration, cell growth or survival. Specifically, IQGAP1 is
observed in actin-dependent membrane structures (membrane ruffles
implicated in cell locomotion and lamellipodia) interacting with
the plus-end binding proteins cytoplasmic linker protein-170 and
adenomatous polyposis coli (APC), tethering microtubules to the
actin network (13,14). Furthermore, in the nuclear envelope of
certain tumor cells, co-expression of IQGAP1 protein and β-tubulin
was observed (Fig. 4). This
co-localization of IQGAP1 with the microtubule network at the
cytoplasmic face of the nuclear envelope has previously been
described by Johnson and Henderson (15) in MCF-7 (breast cancer epithelial
cells), HT29 (colon cancer epithelial cells) and NIH3T3 (non-tumor
embryonic fibroblasts) cell lines, and was also correlated with a
possible role for IQGAP1 in cell polarization and migration events,
in addition to cell cycle-associated nuclear envelope
assembly/disassembly. Johnson and Henderson (15) suggested that interactions between
IQGAP1 and microtubules may tether these cytoskeletal networks to
perinuclear actin via the plus-end protein APC and/or interaction
with other nuclear envelope proteins to regulate the microtubule
organizing center and nuclear positioning for cell polarization
during cell migration, a key process in tumorigenesis and
carcinogenesis.
In the current study, increased expression and
altered localization of IQGAP1 from the cytoplasm to the plasma
membrane was observed in several tumor cells compared with healthy
tissue (Figs. 1D, F and H, 2E and 4) may
serve to decrease AJ stability, favoring dissociation of the tumor
cells (12). IQGAP1 at the plasma
membrane regulates the stability of the AJ complex, which is
necessary for proper apical-basal polarity of epithelial cells that
disappears during the EMT process (16).
In the present study, the co-localization of CD34
and IQGAP1 in several vessels (Fig.
3A-E) is indicative of a role for IQGAP1 in tumor
vasculogenesis and/or in vascular invasion. Nakhaei-Nejad et
al (17) studied the involvement
of endothelial IQGAP1 in leukocyte transendothelial migration, and
demonstrated, by RNAi silencing of IQGAP1 in human umbilical vein
endothelial cells, that IQGAP1 and interendothelial
junction-associated microtubules were involved in remodeling
interendothelial junctions to facilitate lymphocyte diapedesis
under physiological shear stress. Furthermore, Yamaoka-Tojo et
al (18) reported that IQGAP1 is
a novel vascular endothelial growth factor receptor 2 (VEGFR2)
binding protein in quiescent endothelial cells and is important for
the establishment of VE-cadherin-based cell-cell contacts, and
suggested that IQGAP1 may function as a scaffold linking VEGFR2 to
the β-catenin/VE-cadherin compound at the AJ (18).
PCNA is a DNA clamp that increases the processivity
of DNA polymerase δ in eukaryotic cells, which is used as a marker
of proliferating cells (19–21). The immunohistochemical experiments of
the present study revealed that certain cells co-expressed IQGAP1
and PCNA in the cytoplasm, whereas others expressed IQGAP1 in the
cytoplasm and PCNA in the nucleus, which is indicative of S phase
(22). In addition, CD34+
microvessels of CRC and metastatic sections co-express PCNA and
IQGAP, while the majority of cancer cells do not contain
intranuclear PCNA, though they may contain cytoplasmic, pointing to
a quiescent phase while waiting for energy to be supplied in order
for the carcinogenic process to progress (23).
As shown in Fig. 3F-M,
during the synthesis of PCNA, PCNA and IQGAP1 are simultaneously
expressed in the cytoplasm of cells within the liver. PCNA then
enters into the nucleus and S phase of mitosis begins. A role for
IQGAP1 in regulating early S phase replication events has been
recently proposed. Johnson et al (23) identified nuclear localization of
IQGAP1 in several mammalian cell lines. This nuclear localization
was low in asynchronous cells, but was significantly increased in
cells arrested in G1/S phase (23).
The authors suggested that the protein enters the nucleus at G1/S
phase and exits in late S phase. Furthermore, nuclear IQGAP1 was
identified to function as part of a complex with replication
protein A 32 kDa subunit and PCNA, suggesting a functional role for
IQGAP1 in the reinitiation of S phase following DNA replication
arrest (23).
As observed in the current study, nucleoli staining
is often observed throughout the field of microscopic observation,
which is consistent with Bielak-Zmijewska et al (24). The authors observed the presence of
IQGAP1 in mouse oocyte nuclei, forming a ring around the nucleolus
only in transcriptionally active oocytes, but not in
transcriptionally silent ones or in growing oocytes treated with
the transcription inhibitor α-amanitin, indicating an association
between IQGAP1 expression in the nucleolus and RNA synthesis
(24).
In conclusion, extremely few studies have analyzed
IQGAP1 expression in colorectal tumors, and none have yet addressed
IQGAP1 expression in CRC and its associated liver metastases. To
the best of our knowledge, the present study is the first to
provide a detailed analysis of IQGAP1 expression in CRC tissue
sections and metastasized liver tissue samples, which were resected
following oxaliplatin-based CT. Despite the homogeneous IQGAP1
staining pattern observed in healthy colon tissue sections, the CRC
tissues exhibited heterogeneous expression (in terms of
localization and intensity), which was more marked in the
metastasized liver sections resected following CT treatment.
However, more notable findings are the described effects over the
cellular and subcellular distribution and its implications in the
cancer biology. Therefore, IQGAP1 may be a novel cancer antigen, a
surrogate biomarker of responses to CT, a candidate for the
development of new clinical diagnostics and a new target for
anti-cancer therapeutics.
Acknowledgements
The present study was supported by grants from the
Health Institute of Charles III, Spain (no. FIS PI11/00114) and
Microscopy was funded by a joint grant from the Insular Council of
Tenerife, Spain; the Ministry of Science and Technology, Spain; and
the European Regional Development Fund 2003/2004, Belgium (no.
IMBRAIN-FP7-REGPOT-2012-31637).
Glossary
Abbreviations
Abbreviations:
|
AJ
|
adherens junction
|
|
APC
|
adenomatous polyposis coli
|
|
CRC
|
colorectal adenocarcinoma
|
|
CT
|
chemotherapy
|
|
EMT
|
epithelial mesenchymal transition
|
|
FOLFOX
|
folinic acid, leucovorin,
5-fluorouracil and oxaliplatin
|
|
IQGAP1
|
IQ-motif containing GTPase activating
protein 1
|
|
MAPK
|
mitogen-activated protein kinase
|
|
PCNA
|
proliferating cell nuclear antigen
|
|
PWCs
|
peripheral white cells
|
References
|
1
|
André T, Boni C, Mounedji-Boudiaf L,
Navarro M, Tabernero J, Hickish T, Topham C, Zaninelli M, Clingan
P, Bridgewater J, et al: Oxaliplatin, fluorouracil, and leucovorin
as adjuvant treatment for colon cancer. N Engl J Med.
350:2343–2351. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu R, Zhou B, Fung PC and Li X: Recent
advances in the treatment of colon cancer. Histol Histopathol.
21:867–872. 2006.PubMed/NCBI
|
|
3
|
Morales M, Ávila J, González-Fernández R,
Boronat L, Soriano ML and Martín-Vasallo P: Differential
transcriptome profile of peripheral white cells to identify
biomarkers involved in oxaliplatin induced neuropathy. J Pers Med.
4:282–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rotoli D, Morales M, Del Carmen Maeso M,
Del Pino García M, Morales A, Avila J and Martín-Vasallo P:
Expression and localization of the immunophilin FKBP51 in
colorectal carcinomas and primary metastases, and alterations
following oxaliplatin-based chemotherapy. Oncol Lett. 12:1315–1322.
2016.PubMed/NCBI
|
|
5
|
Johnson M, Sharma M and Henderson BR:
IQGAP1 regulation and roles in cancer. Cell Signal. 21:1471–1478.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
White CD, Brown MD and Sacks DB: IQGAPs in
cancer: A family of scaffold proteins underlying tumorigenesis.
FEBS Lett. 583:1817–1824. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Erickson JW, Cerione RA and Hart MJ:
Identification of an actin cytoskeletal complex that includes IQGAP
and the Cdc42 GTPase. J Biol Chem. 272:24443–24447. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roy M, Li Z and Sacks DB: IQGAP1 is a
scaffold for mitogen-activated protein kinase signaling. Mol Cell
Biol. 25:7940–7952. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McDonald KL, O'Sullivan MG, Parkinson JF,
Shaw JM, Payne CA, Brewer JM, Young L, Reader DJ, Wheeler HT, Cook
RJ, et al: IQGAP1 and IGFBP2: Valuable biomarkers for determining
prognosis in glioma patients. J Neuropathol Exp Neurol. 66:405–417.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nabeshima K, Shimao Y, Inoue T and Koono
M: Immunohistochemical analysis of IQGAP1 expression in human
colorectal carcinomas: Its overexpression in carcinomas and
association with invasion fronts. Cancer Lett. 176:101–109. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Holck S, Nielsen HJ, Hammer E, Christensen
IJ and Larsson LI: IQGAP1 in rectal adenocarcinomas: Localization
and protein expression before and after radiochemotherapy. Cancer
Lett. 356:556–560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takemoto H, Doki Y, Shiozaki H, Imamura H,
Utsunomiya T, Miyata H, Yano M, Inoue M, Fujiwara Y and Monden M:
Localization of IQGAP1 is inversely correlated with intercellular
adhesion mediated by e-cadherin in gastric cancers. Int J Cancer.
91:783–788. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fukata M, Watanabe T, Noritake J, Nakagawa
M, Yamaga M, Kuroda S, Matsuura Y, Iwamatsu A, Perez F and Kaibuchi
K: Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170.
Cell. 109:873–885. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Watanabe T, Wang S, Noritake J, Sato K,
Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T and Kaibuchi
K: Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin
filaments during cell polarization and migration. Dev Cell.
7:871–883. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Johnson MA and Henderson BR: The
scaffolding protein IQGAP1 co-localizes with actin at the
cytoplasmic face of the nuclear envelope: Implications for
cytoskeletal regulation. Bioarchitecture. 2:138–142. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Noritake J, Watanabe T, Sato K, Wang S and
Kaibuchi K: IQGAP1: A key regulator of adhesion and migration. J
Cell Sci. 118:2085–2092. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakhaei-Nejad M, Zhang QX and Murray AG:
Endothelial IQGAP1 regulates efficient lymphocyte transendothelial
migration. Eur J Immunol. 40:204–213. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamaoka-Tojo M, Tojo T, Kim HW, Hilenski
L, Patrushev NA, Zhang L, Fukai T and Ushio-Fukai M: IQGAP1
mediates VE-cadherin-based cell-cell contacts and VEGF signaling at
adherence junctions linked to angiogenesis. Arterioscler Thromb
Vasc Biol. 26:1991–1997. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guzińska-Ustymowicz K, Pryczynicz A,
Kemona A and Czyzewska J: Correlation between proliferation
markers: PCNA, Ki-67, MCM-2 and antiapoptotic protein Bcl-2 in
colorectal cancer. Anticancer Res. 29:3049–3052. 2009.PubMed/NCBI
|
|
20
|
Bleau AM, Agliano A, Larzabal L, de
Aberasturi AL and Calvo A: Metastatic dormancy: A complex network
between cancer stem cells and their microenvironment. Histol
Histopathol. 29:1499–1510. 2014.PubMed/NCBI
|
|
21
|
Moldovan GL, Pfander B and Jentsch S:
PCNA, the maestro of the replication fork. Cell. 129:665–679. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Connolly KM and Bogdanffy MS: Evaluation
of proliferating cell nuclear antigen (PCNA) as an endogenous
marker of cell proliferation in rat liver: A dual-stain comparison
with 5-bromo-2′-deoxyuridine. J Histochem Cytochem. 41:1–6. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Johnson M, Sharma M, Brocardo MG and
Henderson BR: IQGAP1 translocates to the nucleus in early S-phase
and contributes to cell cycle progression after DNA replication
arrest. Int J Biochem Cell Biol. 43:65–73. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bielak-Zmijewska A, Kolano A, Szczepanska
K, Maleszewski M and Borsuk E: Cdc42 protein acts upstream of
IQGAP1 and regulates cytokinesis in mouse oocytes and embryos. Dev
Biol. 322:21–32. 2008. View Article : Google Scholar : PubMed/NCBI
|