Introduction
Renal cell carcinoma (RCC) is the third most common
urological cancer (1). A number of
patients with RCC develop metastatic disease and the 5-year
survival rate in these patients just 2% (2). Therefore, further screening and
investigations into novel treatment methods are highly
warranted.
MicroRNAs (miRNAs/miRs) are non-coding small RNAs of
~19–25 nt in length which are cleaved from 70–100 nt-long hairpin
precursor (pre)-miRNAs by the enzyme ribonuclease 3 (Drosha)
(3,4).
miRNAs have essential functions in the development and
establishment of cell identity, and aberrant metabolism or
expression of miRNAs has been associated with human disease,
including cancer (4). There are an
increasing number of reports implicating aberrant expression of
certain miRNAs, including miR-21, 17–92,-15,-16,-141 and let-7, in
tumor growth, carcinogenesis and response to chemotherapy in
various malignancies (3–10). miR-21, which is overexpressed in
various cancer types, is one of the most widely studied miRNAs in
cancer (11–13). The overexpression of miR-21 has been
implicated in various processes associated with carcinogenesis,
including the inhibition of apoptosis (6), promotion of cell proliferation (9) and stimulation of tumor growth (10).
Programmed cell death 4 (PDCD4) has been
demonstrated to be an inhibitor of neoplastic transformation. The
PDCD4 gene was identified in the mouse epidermal clonal genetic
variant JB6 cell system as a 64 kDa protein that is preferentially
expressed in tumor promoter-resistant cells, but suppressed in
tumor promoter-sensitive cells undergoing neoplastic transformation
(14). PDCD4 levels were continuously
reduced in the colon and colorectal adenocarcinoma (15,16). PDCD4
inhibits activator protein (AP)-1 transactivation (17), stalls translation machinery (18), decreases benign and malignant tumor
progression (19), and regulates
lymphoma initiation and autoimmune inflammation (20). Subsequent investigations demonstrated
that a loss of PDCD4 expression was associated with tumor
progression in carcinomas of the lung, colon, prostate and breast
(21).
Previous bioinformatics analyses have demonstrated
that PDCD4 contains a miR-21 binding site and acts as a tumor
suppressor through the regulation of various processes associated
with cancer progression, including cell proliferation, invasion,
metastasis and neoplastic transformation (21–23).
Notably, Asangani et al (11)
studied 10 colorectal cell lines and observed an inverse
correlation between miR-21 and PDCD4 protein expression. Lu et
al (24) demonstrated that
translation of the tumor suppressor gene, PDCD4, is negatively
regulated by miR-21 in HEK-293T, MCF-7 and JB6 cell lines, and
provided evidence that the miR-21 gene functions as an oncogene to
promote cell transformation. In human hepatocellular carcinoma cell
lines, overexpression of miR-21 did not cause degradation of PDCD4
mRNA, but significantly inhibited its protein expression (25). It has been demonstrated that knockdown
of miR-21 upregulates PDCD4 expression leading to increased
apoptotic cell death in glioblastoma cells (26), in addition to suppressing invasion and
metastasis in colorectal cancer cells (22) and esophageal squamous cell carcinoma
(27). Further studies have confirmed
the regulation of PDCD4 by miR-21 in colon, breast and bladder
carcinoma (22), cholangiocarcinoma
(23), esophageal carcinoma (27) and glioblastoma (26). Consistent with these results, Li et
al (28) demonstrated that miR-21
is significantly overexpressed in RCC tissue and cell lines, and
that PDCD4 is negatively regulated by miR-21.
To the best of our knowledge, no previous studies
have elucidated the roles of and associations between miR-21 and
PDCD4 in an animal RCC model. Therefore, the aim of the present
study was to determine the roles of and interactions between PDCD4
and miR-21 in a nude mouse renal cancer model, and the effects of
silencing PDCD4 on RCC tumor cell growth and invasion.
Materials and methods
Ethics statement
The present study was performed in strict accordance
with the recommendations of the Guide for the Care and Use of
Laboratory Animals of the National Institutes of Health (29). The protocol used in the present study
was approved by the Committee on the Ethics of Animal Experiments
at Henan University of Science and Technology (Henan, China;
approval no. 20140126). All surgical procedures were performed
under 1% sodium pentobarbital anesthesia 70 mg/kg, and all efforts
were made to minimize suffering. As a humane endpoint, mice were
euthanized if they met any of the following conditions: i) When
they exhibited loss of >20% of body weight; ii) when the tumor
mass >10% of body weight; iii) when an increased respiratory
rate and/or effort was observed; iv) if a loss of skin elasticity
was observed; v) if the mice exhibited the inability to access food
or water. Mice were injected with an excess of sodium pentobarbital
anesthetic (150 mg/kg, 1%) as the method of euthanasia.
Rearing of nude mice and cell
culture
BALB/c nude mice (n=24; male) were obtained from the
Laboratory Animal Center of the Academy of Military Medical
Sciences (Shanghai, China). These BALB/c nude mice were 5–6 weeks,
weight 20–23 g, and they were fed for 1 week prior to the
experiment under the food condition of SPF at 20–26°C, relative
humidity 40–70% and 12 h light-dark cycle. All food was treated
with high temperature steam disinfection (45 min, 120°C). All water
was acidified by hydrochloric acid and adjusted to a pH between 2.5
and 2.8. Renal cell adenocarcinoma 786-O cells were obtained from
the Xiehe Cell Bank of the Chinese Academy of Medical Sciences
(Beijing, China) and were cultured under the conditions recommended
by the cell bank. Briefly, the 786-O cells were cultured as a
monolayer in Keratinocyte Serum-Free medium (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 0.05
mg/ml bovine pituitary extract (Invitrogen; Thermo Fisher
Scientific, Inc.), 5 ng/ml human recombinant epidermal growth
factor (Invitrogen; Thermo Fisher Scientific, Inc.) and 10% fetal
bovine serum (FBS; Atlanta Biologicals Inc., Lawrenceville, GA,
USA), 50 mg/ml penicillin and 50 mg/ml streptomycin (both
Invitrogen; Thermo Fisher Scientific, Inc.). Cells were maintained
in an incubator with a humidified atmosphere and 5% CO2
at 37°C. Subconfluent 786-O cells (60–70% confluence) were treated
with genistein (25 mM; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) and 10% dimethyl sulfoxide (15 ml).
RNA interference assay
The 786-O cells (1×106 cells, 60–80%
confluence) were incubated in a 6-well tissue culture dish without
antibiotics for 24 h prior to transfection. The 786-O cells were
transfected with PDCD4 siRNA and the negative control siRNA, and
the effects of silencing PDCD4 on tumor cell growth, proliferation
and invasion were investigated. The small interfering RNA (siRNA)
transfection reagent complexes (#AM16708A; Invitrogen; Thermo
Fisher Scientific, Inc.) were mixed with Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions and subsequently added to the cells.
The sequences of the siRNAs used in the present study were as
follows: NC siRNA (sense, 5′-GCUGCUUTGGACAAGGCUATC-3′; antisense,
5′-UAGCCUAGUCCAAAGCAGCAT-3′) PDCD4 siRNA (sense,
5′-GCUGCUUUGGACAAGGCUATT-3′; antisense,
5′-UAGCCUUGUCCAAAGCAGCTT-3′). After 6 h of incubation at 37°C, the
medium was replaced and the cells were cultured in RPMI-1640
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%
heat-inactivated FBS for various time periods. At the same time,
cells were transfected with control siRNA as control group.
Nude mouse renal cancer model
The male nude mice were randomly assigned into the
following three groups to investigate the effect of miR-21:
Negative control (NC; n=8), miR-21 mimic (n=8) and miR-21 inhibitor
(n=8). A 0.1 ml 786-O cell suspension (1×106 cells) was
subcutaneously transplanted into the armpits of the mice, which
were subsequently injected daily with NC siRNA (#AM17110;
Invitrogen; Thermo Fisher Scientific, Inc.), pre-miR-21 (mimic) or
anti-miR-21 (inhibitor); (#A25576; Invitrogen; Thermo Fisher
Scientific, Inc.). The sequences of the primers as follows:
pre-miR-21 primer sequence, sense, 5′-CATCCTUCUTGAAGUGACUC-3′ and
antisense, 5′-CGCUCUAUGACGUAUGGAGGU-3′; anti-miR-21 primer
sequence, sense, 5′-GATCCAUCUTCGAAGUGACTT-3′ and antisense,
5′-UGCUCUTUGACGUAUGGAGTT-3′; NC siRNA primer sequence, sense,
5′-UUCACCGUACGUCUCACCUGT-3′ and antisense,
5′-ACUGGAACCUCUCGCGGAATT-3′. MTT assays (Roche Diagnostics GmbH,
Mannheim, Germany) were performed to detect cell viability. Cells
were seeded into 96-well plates (6.0×103 cells/well) and left at
normal culture conditions. Cells were incubated with MTT (5 mg/ml
per well) for 4 h. Dimethyl sulfoxide was used to dissolve the
formazan crystals. PBS was used as a control. Absorbance at a
wavelength of 490 nm was measured using the Infinite M200 PRO
multimode microplate reader (Tecan Benelux BVBA, Mechelen,
Belgium). Cell viability was detected prior to the injection of
cells to ensure that the cells were in logarithmic phase. A total
of 16 days after transplantation, the tumor formation rate was 100%
and the mice were sacrificed, with the tumors collected and
weighed. The expression of miR-21 and PDCD4 mRNA in the cancer
tissues was analyzed using reverse transcription-quantitative
polymerase chain reaction (RT-qPCR).
The expression of PDCD4 protein in cancer tissues
was examined using immunohistochemistry and western blotting.
BALB/c mice were randomly assigned into two groups as follows: NC
(n=8) and PDCD4 siRNA (n=8). The 786-O cells were subcutaneously
transplanted into the armpits of the mice, and this was followed by
daily injections of NC siRNA or PDCD4 siRNA (#AM16708A; Invitrogen;
Thermo Fisher Scientific, Inc.). NC siRNA sense,
5′-GCUGCUUTGGACAAGGCUATC-3′ and antisense,
5′-UAGCCUAGUCCAAAGCAGCAT-3′; PDCD4 siRNA sense,
5′-GCUGCUUUGGACAAGGCUATT-3′ and antisense,
5′-UAGCCUUGUCCAAAGCAGCTT-3′. The tumors were removed from the mice
and weighed 16 days after the transplantation. Sodium pentobarbital
(150 mg/kg, 1%) was injected into each mouse as the mode of
euthanisia.
Immunohistochemistry
For immunohistochemical analysis, mouse cancer
tissues were fixed with 10% buffered formalin and embedded in
paraffin. The sections (2-µm-thick) were cut 1 day prior to use.
All sections were deparaffinized and dehydrated with graded ethyl
alcohol (99, 95, 85 and 75%). The sections were then washed for 10
min in phosphate-buffered saline (PBS; pH 7.2; 37°C). The
endogenous peroxidase activity was quenched by incubation in
methanol containing 3% H2O2 for 10 min at
room temperature, then heated for 30 min at 95°C to repair antigens
and finally washed with PBS. To maximize immunohistochemistry
signals, the following two strategies were used: Antigen retrieval
in citrate buffer and signal amplification with biotinylated
tyramide. The tissue sections were incubated overnight at 4°C with
PDCD4 antibody (dilution, 1:100; #ab105998; Abcam, Cambridge, UK).
Detection was subsequently performed using biotinylated goat
anti-rabbit antibody (dilution, 1:5,000; #SAB4504290;
Sigma-Aldrich; Merck KGaA) and the tissue sections were incubated
at 37°C for 20 min. Diaminobenzidine was used as a chromogen, and
the slide was counterstained with Mayer's hematoxylin. The results
were independently observed under a fluorescent microscope (Olympus
BX41; Olympus Corporation, Tokyo, Japan) at ×400 magnification by
two pathologists. For the NC, the primary antibody was replaced by
10% non-immune goat serum.
Western blot analysis
Western blot analyses were performed to determine
the expression of PDCD4. Total protein was extracted from tissues
using a Total Protein Extraction kit (Nanjing KeyGen Biotech Co.,
Ltd., Nanjing, China) according to the manufacturer's
recommendations. The concentration of protein was measured using a
BCA Assay kit (Nanjing KeyGen Biotech Co., Ltd.). The protein
samples were separated on a 10% polyacrylamide gel using SDS-PAGE
and transferred onto a hybond polyvinylidene difluoride membrane
(GE Healthcare Bio-Sciences, Pittsburgh, PA, USA). The membranes
were subsequently blocked in 5% fat-free milk at room temperature
for 2 h. Following incubation with rabbit or goat primary
antibodies directed against PDCD4 (1:10,000; cat no. ab80590;
Abcam) or GAPDH (1:200; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) at 4°C overnight, the membranes were probed with goat
anti-rabbit (#SAB2502080; Sigma-Aldrich; Merck KGaA) or mouse
anti-goat secondary antibodies (#G8795; Sigma-Aldrich; Merck KGaA)
at a dilution of 1:5,000 at room temperature for 2 h. The signals
were detected using a Super Enhanced Chemiluminescence Plus kit
(Nanjing KeyGen Biotech Co., Ltd.) and quantified using UVP
software (BioDoc-It® Imager System; UVP, LLC, Upland,
CA, USA). The integrated optical density (IOD) ratio
IODPDCD4/IODGAPDH was used to indicate the
relative expression of PDCD4 protein at a wavelength of 280 nm.
RT-qPCR
Total RNA was extracted from tumors using an RNA
Isolation kit (CWbiotech Co., Ltd., Beijing, China) following the
manufacturer's protocol. Stem-loop RT-qPCR for mature miR-21 was
performed as previously described (28). RT-qPCR for PDCD4 was performed using
Power SYBR® Green PCR Master mix (Agilent Technologies,
Inc., Santa Clara, CA, USA) in a final volume of 20 µl, comprising
of 100 ng cDNA, 10 µl master mix, 1 µl ROX and 0.4 pmol/µl of each
primer. qPCR cycling conditions were as follows: 95°C for 2 min,
and then 95°C for 15 sec and 55°C for 30 sec, for 40 cycles,
followed by 60°C for 1 min. The melting curve was 65–95°C. Human U6
mRNA was used for normalization for the stem-loop RT-qPCR and GAPDH
was used for normalization for the PDCD4 RT-qPCR. Fluorescent
signals were normalized to these internal reference genes, and the
threshold cycle (Cq) was set within the exponential phase of the
PCR. The relative gene expression was calculated by comparing cycle
times for each target PCR. The target PCR Cq values were normalized
by subtracting the U6 or GADPH Cq value, which provided the ΔCq
value. The relative expression level between treatments was then
calculated using the following equation: Relative gene
expression=2(ΔCqsample−ΔCqcontrol) (30).
Primers
For miR-21 RT-qPCR, the primer sequence was 5-TAG
CTT ATC AGA CTG ATG TTGA-3, and reverse 5-AAC GCT TCA CGA ATT TGC
GT-3. The other primer sequences for RT-qPCR were as follows: U6
forward, 5-CTC GCT TCG GCA GCA CA-3 and reverse, 5-AAC GCT TCA CGA
ATT TGC GT-3; PDCD4 forward, 5-AGG CCG AGG TGG GCG GAT CAC TTG A-3
and reverse, 5-GCC ACC ATG CCT GGC TAC T-3; and GAPDH forward,
5-CCT CTG ACT TCA ACA GCG ACA C-3 and reverse, 5-TGG TCC AGG GGT
CTT ACT CC-3.
EdU incorporation cell proliferation
assay
Transfected 786-O cells were plated in 24-well
plates at density of 4×104 cells/well, allowed to adhere
for 5 h, washed with PBS and then incubated in serum-free RPMI-1640
containing 10 µmol/l EdU (Guangzhou RiboBio Co., Ltd., Guangzhou,
China) for 2 h at 37°C. The cells were subsequently washed with
PBS, and then fixed and permeabilized in PBS containing 2%
formaldehyde, 0.5% Triton X-100, and 300 mmol/l sucrose for 15 min
at 37°C. Following washing with PBS, the cells were blocked using
10% FBS in PBS at ambient temperature for 30 min, and incorporated
EdU was detected by incubation with a fluorescent azide coupling
solution (#C10310-3; Apollo; Guangzhou RiboBio Co. Ltd) for 30 min
at ambient temperature. The cells were washed three times with PBS
containing 0.05% Tween-20, incubated with the DNA staining dye
Hoechst 33342 for 30 min and washed in PBS. Images were captured
using a fluorescent microscope, and the nuclear fluorescent
intensity was calculated from ≥50 non-S phase cells randomly
selected in five different fields of view.
Soft agar colony formation assay
The bottom layer of the 15-cm plate (0.6% low-melt
agarose) was prepared with RPMI-1640 medium containing 10% FBS, 100
U/ml penicillin and 100 mg/ml streptomycin, in addition to
5×102 of the transfected 786-O cells. The top layer of
the plate (0.3% low-melt agarose) was prepared with RPMI 1640
medium containing 10% FBS, 100 U/ml penicillin and 100 mg/ml
streptomycin, in addition to 5×102 of the transfected
786-O cells. Plates were incubated at 37.8°C with 5% CO2
in a humidified incubator. Images of the plates were captured on
day 14, and the number of colonies was quantified using Quantity
One software (version 4.0.3; Bio-Rad Laboratories, Inc., Hercules,
CA, USA). The assays were performed 3 times. The data are presented
as mean average.
Cell invasion assays
Cell invasion assays were performed using a
Transwell chamber (BD Biosciences, Franklin Lakes, NJ, USA).
Transfected NC siRNA cells were used as a control group. The assays
were performed 3 times. In the invasion assay, 2×104
transfected cells in serum-free medium was seeded into the top
chamber, which was pre-coated with Matrigel (BD Biosciences).
Following incubation for 24 h at 37°C, the membranes were fixed
using methyl alcohol (100%) and then stained with 0.1% crystal
violet. The number of cells that passed through the membranes were
counted under a light microscope.
Statistical analysis
SPSS software (version 13.0; SPSS, Inc., Chicago,
IL, USA) was employed for the analysis of all data. Data are
expressed as the mean ± standard deviation. A Student's t-test were
used to determine the significance of the differences between
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
Silencing of miR-21 inhibits tumor
growth in the mouse renal cancer model
To investigate whether the miR-21 inhibitor
inhibited tumor growth, the weight of tumors were measured
following miR-21 inhibitor treatment. Fig. 1A presents images of the tumors from
the nude mice models in the three groups (NC, miR-21 mimic and
miR-21 inhibitor). Compared with the NC, the weight of the tumors
in the miR-21 inhibitor group was significantly decreased
(P<0.05; Fig. 1B). The weight of
the tumors in the miR-21 mimic group was significantly increased
compared with the NC (P<0.05; Fig.
1B).
Downregulation of miR-21 expression
increases PDCD4 expression in the mouse renal cancer model
To evaluate the association between PDCD4 and miR-21
in the nude mouse RCC models, whether the transplantation of 786-O
cells with miR-21 mimic or miR-21 inhibitor affected the expression
of PDCD4 was determined. Downregulation of endogenous miR-21 by the
miR-21 inhibitor resulted in a significant increase in the
expression of PDCD4 protein compared with the NC group (P<0.05;
Fig. 1C and 1D). By contrast, there
was a significant decrease in the levels of PDCD4 protein in the
miR-21 mimic group compared with the NC group (P<0.05; Fig. 1C and D). The expression of was
significantly upregulated in the miR-21 mimic group compared with
the NC group, whilst it was significantly reduced in the miR-21
inhibitor group (P<0.05; Fig. 1E).
However, there was no significant difference in the expression of
PDCD4 mRNA between the three groups (P>0.05; Fig. 1F).
Immunohistochemical analysis of PDCD4 was performed
(Fig. 2), indicating that PDCD4 was
localized to the cytoplasm. Reduced or complete loss of PDCD4
expression was detected in the miR-21 mimic group (Fig. 2A). The immunocytochemical staining
depth was moderate in the NC group (Fig.
2B). By contrast, strong immunopositivity for PDCD4 was
observed in the miR-21 inhibitor group (Fig. 2C).
Silencing of PDCD4 induces tumor cell
proliferation, colony formation, migration and invasion
The effect of silencing PDCD4 on cell proliferation
was investigated by transfecting PDCD4 siRNA and control siRNA into
786-O cells. To examine whether there was a change in the number of
proliferating 786-O cells following transfection, the cells were
labeled with EdU to measure active DNA synthesis and Hoechst 33342
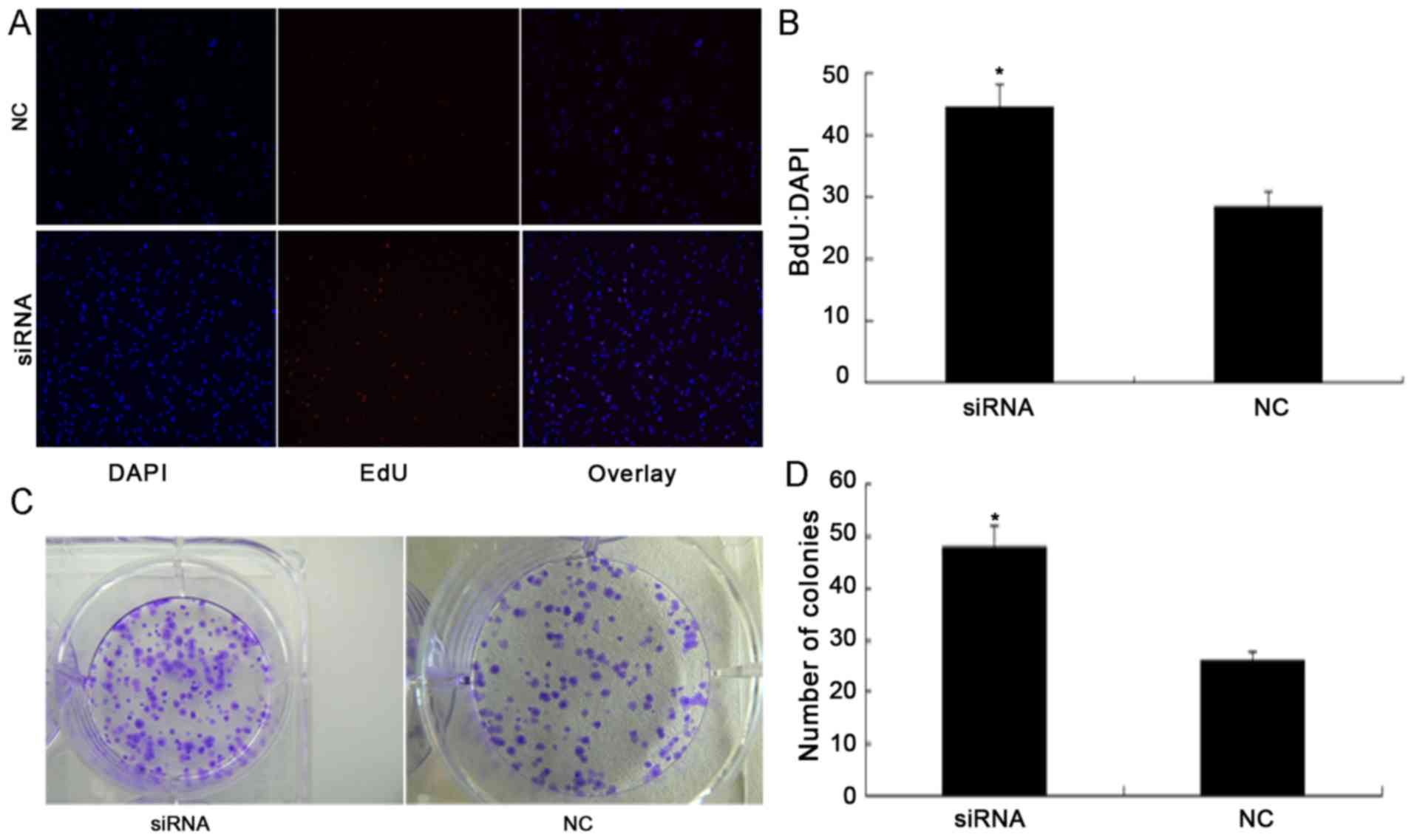
to illustrate the nuclei of all cells (Fig. 3A). It was identified that silencing of
PDCD4 significantly promoted cell proliferation. According to the
results of fluorescent microscopic analysis, the mean percentage of
newly formed cells that incorporated EdU was 28.6% in the NC siRNA
group and 44.7% in the PDCD4 siRNA-transfected cells (P<0.05;
Fig. 3B).
A colony formation assay was performed to determine
whether PDCD4 knockdown promoted the colony forming ability of
786-O cells (Fig. 3C). The colony
formation assay demonstrated that the total number of colonies in
the PDCD4 siRNA-transfected group was significantly increased
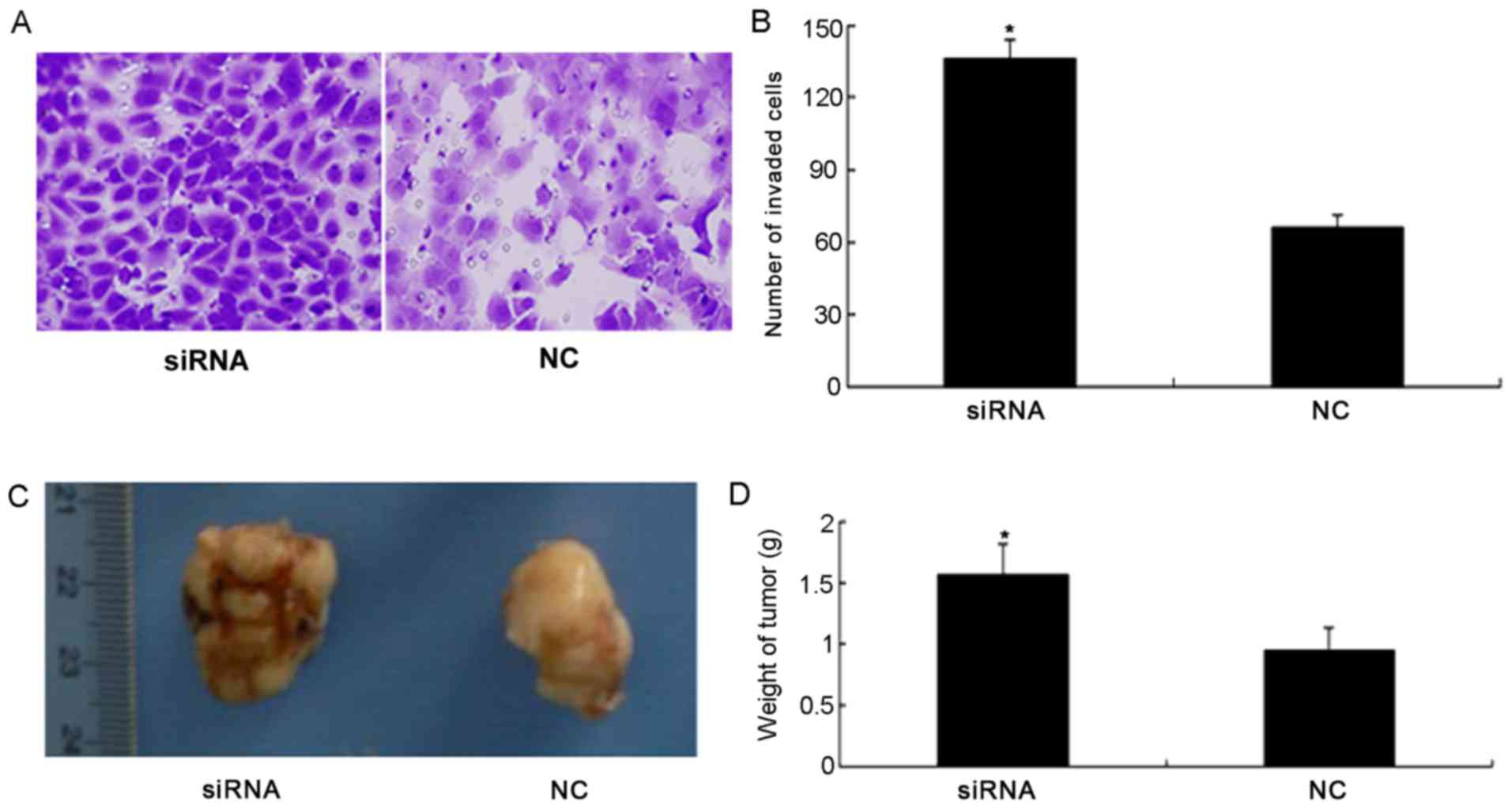
compared with the NC group (P<0.05; Fig. 3D). Furthermore, the results of the
invasion assay demonstrated that silencing the expression of PDCD4
significantly increased invasion of 786-O cells compared with the
NC group (P<0.05; Fig. 4A and
B).
Silencing of PDCD4 induces tumor
growth in the mouse renal cancer model
To investigate whether PDCD4 silencing induces tumor
growth, tumor weight was measured following PDCD4 siRNA and NC
treatment. Fig. 4C illustrates the
tumors from the nude mouse models. Compared with the NC group,
tumor weight in the PDCD4 siRNA group was significantly increased
(P<0.05; Fig. 4D).
Discussion
To the best of our knowledge, the present study is
the first to demonstrate that PDCD4 is negatively regulated by
miR-21, in addition to being able to suppress tumor growth and
metastasis in a nude mouse renal cancer model. In a previous study,
it was revealed that there is a conserved target site for miR-21
within the PDCD4 3′ untranslated region at nucleotides 228–249
(11). It has also been reported that
miR-21 is able to regulate the Ras/mitogen-activated protein kinase
kinase/extracellular signal-regulated kinase signaling pathway and
therefore affect tumor formation (31). Additionally, a meta-analysis has
indicated that miR-21 is able to act as an important biomarker for
the prognosis of various types of cancer (32). Xu et al (33) demonstrated that the downregulation of
miR-21 increased the sensitivity of lung cancer cells to cisplatin
in vitro and in vivo. Wang et al (34) reported that miR-21 expression was
significantly increased in hepatocellular carcinoma tissues
compared with normal adjacent liver tissues. Furthermore, Li et
al (28) demonstrated that the
tumor suppressor PDCD4 was negatively regulated at a
post-transcriptional level by miR-21, and that miR-21 induced cell
proliferation and invasion/metastasis in RCC. PDCD4 expression is
also significantly positively associated with RCC metastasis, and
tumor stage and grade (35).
To further investigate the association between PDCD4
and miR-21 in vivo, BALB/c male nude mice and 786-O cells
were used in the present study to establish a nude mouse renal
cancer model. It was identified that tumor weight in the miR-21
inhibitor group was significantly decreased compared with the NC
group. Compared with the NC group, tumor weight in the miR-21 mimic
group was significantly increased. Subsequently, the expression of
PDCD4 in the NC group, miR-21 inhibitor group and miR-21 mimic
group was analyzed by western blotting. There was loss or reduced
expression of PDCD4 protein in the miR-21 mimic group; however,
PDCD4 protein was highly expressed in the miR-21 inhibitor group
compared with the NC group. Similar results were revealed by
immunohistochemistry. In addition, there was no significant
difference between PDCD4 mRNA levels between the three groups
(miR-21 mimic, miR-21 inhibitor and negative control). The results
of the present study, which were similar to the previously
published study of human renal cancer tissue and cell lines
(28), demonstrated that miR-21
downregulates PDCD4 at a post-transcriptional level, and promotes
cell colony formation and proliferation in the nude mouse renal
cancer model.
PDCD4 has been known to be a tumor suppressor gene
and potential target for anticancer therapies for several years
(11,14,16–21).
Reduced PDCD4 expression has been reported in >5 types of human
tumors, including those of the lung, brain, breast, colon and
pancreas (11). In colon cancer cell
lines, Wang et al (36) has
demonstrated that downregulation of PDCD4 leads to an increase in
colon carcinoma cell invasion. Furthermore, the knockdown of PDCD4
in the study by Wang et al (36) was associated with a significant
reduction in E-cadherin expression and accumulation of active
β-catenin in the nucleus of these cells. In the same series of
experiments, PDCD4 knockdown resulted in an increase in activator
protein (AP)-1-dependent transcription. These results indicate that
reduced PDCD4 expression promotes cancer cell invasion, and that
this is associated with the activation of β-catenin, E-cadherin and
AP-1-dependent transcription. Another study demonstrated that
decreased PDCD4 expression was significantly associated with the
clinical stage of adenoid cystic carcinoma (37). Li et al (35) observed that decreased PDCD4 expression
was significantly positively associated with metastasis, and tumor
stage and grade in RCC.
In the present study, the effect of PDCD4 on miR-21
expression was investigated. PDCD4 siRNA was transfected into 786-O
cells, and PDCD4 mRNA and protein levels were found to be
significantly decreased compared with the NC group. Silencing PDCD4
also significantly promoted 786-O cell proliferation, migration and
invasion, similar to the effects observed after miR-21
overexpression.
In conclusion, the results of the present study and
previous studies indicate that PDCD4 and miR-21 serve important
roles in RCC. The results also suggest that the promotion of PDCD4
expression or inhibition of miR-21 expression may constitute
effective novel therapeutic strategies for the treatment of renal
cancer.
Acknowledgements
The present study was supported by the Henan
Provincial Science and Technology Plan Foundation (grant no.
B2014225013).
References
|
1
|
van Spronsen DJ, Mulders PF and De Mulder
PH: Novel treatments for metastatic renal cell carcinoma. Crit Rev
Oncol Hematol. 55:177–191. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hammond SM: MicroRNAs as oncogenes. Curr
Opin Genet Dev. 16:4–9. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meng F, Henson R, Lang M, Wehbe H,
Maheshwari S, Mendell JT, Jiang J, Schmittgen TD and Patel T:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Roldo C, Missiaglia E, Hagan JP, Falconi
M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A and
Croce CM: MicroRNA expression abnormalities in pancreatic endocrine
and acinar tumors are associated with distinctive pathologic
features and clinical behavior. J Clin Oncol. 24:4677–4684. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in breast
cancer cells. J Biol Chem. 283:1026–1033. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cmarik JL, Min H, Hegamyer G, Zhan S,
Kulesz-Martin M, Yoshinaga H, Matsuhashi S and Colburn NH:
Differentially expressed protein Pdcd4 inhibits tumor
promoter-induced neoplastic transformation. Proc Natl Acad Sci USA.
96:14037–14042. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee S, Bang S, Song K and Lee I:
Differential expression in normal-adenoma-carcinoma sequence
suggests complex molecular carcinogenesis in colon. Oncol Rep.
16:747–754. 2006.PubMed/NCBI
|
|
16
|
Mudduluru G, Medved F, Grobholz R, Jost C,
Gruber A, Leupold JH, Post S, Jansen A, Colburn NH and Allgayer H:
Loss of programmed cell death 4 expression marks adenoma-carcinoma
transition, correlates inversely with phosphorylated protein kinase
B, and is an independent prognostic factor in resected colorectal
cancer. Cancer. 110:1697–1707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang HS, Jansen AP, Nair R, Shibahara K,
Verma AK, Cmarik JL and Colburn NH: A novel transformation
suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB
or ODC transactivation. Oncogene. 20:669–676. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang HS, Jansen AP, Komar AA, Zheng X,
Merrick WC, Costes S, Lockett SJ, Sonenberg N and Colburn NH: The
transformation suppressor Pdcd4 is a novel eukaryotic translation
initiation factor 4A binding protein that inhibits translation. Mol
Cell Biol. 23:26–37. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jansen AP, Camalier CE and Colburn NH:
Epidermal expression of the translation inhibitor programmed cell
death 4 suppresses tumorigenesis. Cancer Res. 65:6034–6041. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hilliard A, Hilliard B, Zheng SJ, Sun H,
Miwa T, Song W, Göke R and Chen YH: Translational regulation of
autoimmune inflammation and lymphoma genesis by programmed cell
death 4. J Immunol. 177:8095–8102. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
LaRonde-LeBlanc N, Santhanam AN, Baker AR,
Wlodawer A and Colburn NH: Structural basis for inhibition of
translation by the tumor suppressor Pdcd4. Mol Cell Biol.
27:147–156. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baffa R, Fassan M, Volinia S, O'Hara B,
Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM and
Rosenberg A: MicroRNA expression profiling of human metastatic
cancers identifies cancer gene targets. J Pathol. 219:214–221.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Selaru FM, Olaru AV, Kan T, David S, Cheng
Y, Mori Y, Yang J, Paun B, Jin Z, Agarwal R, et al: MicroRNA-21 is
overexpressed in human cholangiocarcinoma and regulates programmed
cell death 4 and tissue inhibitor of metalloproteinase 3.
Hepatology. 49:1595–1601. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu Z, Liu M, Stribinskis V, Klinge CM,
Ramos KS, Colburn NH and Li Y: MicroRNA-21 promotes cell
transformation by targeting the programmed cell death 4 gene.
Oncogene. 27:4373–4379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou L, Yang ZX, Song WJ, Li QJ, Yang F,
Wang DS, Zhang N and Dou KF: MicroRNA-21 regulates the migration
and invasion of a stem-like population in hepatocellular carcinoma.
Int J Oncol. 43:661–669. 2013.PubMed/NCBI
|
|
26
|
Chen Y, Liu W, Chao T, Zhang Y, Yan X,
Gong Y, Qiang B, Yuan J, Sun M and Peng X: MicroRNA-21
down-regulates the expression of tumor suppressor PDCD4 in human
glioblastoma cell T98G. Cancer Lett. 272:197–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hiyoshi Y, Kamohara H, Karashima R, Sato
N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M
and Baba H: MicroRNA-21 regulates the proliferation and invasion in
esophageal squamous cell carcinoma. Clin Cancer Res. 15:1915–1922.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li X, Xin S, He Z, Che X, Wang J, Xiao X,
Chen J and Song X: MicroRNA-21 (miR-21) post-transcriptionally
downregulates tumor suppressor PDCD4 and promotes cell
transformation, proliferation, and metastasis in renal cell
carcinoma. Cell Physiol Biochem. 33:1631–1642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Care and Use of Laboratory Animals of the
National Institutes of Health. 8th. Washington (DC): National
Academies Press (US); 2011
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hatley ME, Patrick DM, Garcia MR,
Richardson JA, Bassel-Duby R, van Rooij E and Olson EN: Modulation
of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell.
18:282–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou X, Wang X, Huang Z, Wang J, Zhu W,
Shu Y and Liu P: Prognostic value of miR-21 in various cancers: An
updating meta-analysis. PLoS One. 9:e1024132014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu L, Huang Y, Chen D, He J, Zhu W, Zhang
Y and Liu X: Downregulation of miR-21 increases cisplatin
sensitivity of non-small-cell lung cancer. Cancer Genet.
207:214–220. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang WY, Zhang HF, Wang L, Ma YP, Gao F,
Zhang SJ and Wang LC: miR-21 expression predicts prognosis in
hepatocellular carcinoma. Clin Res Hepatol Gastroenterol.
38:715–719. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Xin S, Yang D, Li X, He Z, Che X,
Wang J, Chen F, Wang X and Song X: Down-regulation of PDCD4
expression is an independent predictor of poor prognosis in human
renal cell carcinoma patients. J Cancer Res Clin Oncol.
138:529–535. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Q, Sun Z and Yang HS: Downregulation
of tumor suppressor Pdcd4 promotes invasion and activates both
beta-catenin/Tcf and AP-1-dependent transcription in colon
carcinoma cells. Oncogene. 27:1527–1535. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qi C, Shao Y, Li N, Zhang C, Zhao M and
Gao F: Prognostic significance of PDCD4 expression in human
salivary adenoid cystic carcinoma. Med Oncol. 30:4912013.
View Article : Google Scholar : PubMed/NCBI
|