Introduction
Epithelial-mesenchymal transition (EMT) refers to
the process by which differentiated epithelial cells transform into
mesenchymal cells by undergoing various biochemical changes
(1). In tumors, EMT involves a number
of processes, including invasion, metastasis, drug resistance and
development of anti-apoptotic features, which are the primary
causes of breast cancer-associated mortality.Successful inhibition
of these processes is expected to significantly improve breast
cancer prognosis (2). Fatty acid
synthase (FASN) is a key enzyme that catalyzes the synthesis of
long-chain saturated fatty acids (3).
Previous studies demonstrated that whenbreast cancer cells
underwent EMT, FASN expressionincreased (4), and EMT was reversed when FASN was
silenced with FASN short hairpin. (5). Osthole, an FASN inhibitor, was able to
eliminate phenotypes associated with EMT induced by hepatocyte
growth factor, including migration, invasion and metastasis in
breast cancer cells (6). Therefore,
FASN may serve an important role in EMT, but the mechanism is
unclear.
Vazquez-Martin et al (7) determined that overexpression of FASN and
the synthesis and aggregation of endogenous fatty acid induced the
transformation of epithelial cells to a tumor phenotype. This
change was dependent on ErbB1/ErbB2, which indicated an association
between ErbBs and FASN during EMT of tumor cells. The ErbB family,
including epidermal growth factor receptor (EGFR; ErbB1, HER1),
ErbB2 (Neu, HER2), ErbB3 (HER3) and ErbB4 (HER4) (8), has been shown to induce morphological
changes, survival, invasion and EMT in cells (9). ErbBs that are anchored in membrane lipid
rafts dimerize and become activated, thereby initiating relevant
downstream signaling pathways that induce EMT characteristics in
tumors (10). Destruction of lipid
rafts and reduced activity of the downstream signaling molecules
protein kinase B (Akt), extracellular signal-regulated kinase 1/2
(ERK1/2) and the FASN protein by docosahexaenoic acid, resulted in
inhibition of the ErbB2 signaling pathway and apoptosis of
transformed human mammary epithelial cells (11). FASN and its end product palmitate are
important components of lipid synthesis (12). Therefore, it was hypothesized that
FASN regulates the expression of ErbBs by affecting the structure
of membrane lipid rafts, and consequently affects downstream signal
pathways and the EMT phenotype. Due to the complex structure or
ErbBs, and functions in cancer, several studies have focused on
ErbB1 and ErbB2 in recent years and the development of targeted
inhibitors as a means of effective cancer treatment (13–15).
However, tumors have acquired resistance to those targeted
therapeutics (16). Whether ErbB3 or
ErbB4 have any significant function in cancer is debated (17–20), and
their association with the other ErbBs, FASN and
clinicopathological characteristics of breast cancer is currently
unclear. The present study investigated these associations using
MCF-7-MEK5 and its tumor tissues, which exhibited the EMT
phenotype. In the present study, the non-EMT isogenic MCF-7 cell
line and its tumor tissues served as controls. Cerulenin, a
specific inhibitor of FASN (21), was
used to assess the role of FASN. Western blot analysis was
performed to detect changes in the expression of FASN and ErbBs.
Immunohistochemistry was used to detect the expression of FASN and
ErbBs in 58 invasive ductal carcinoma samples and the association
of FASN expression withclinicopathological characteristics was
investigated.
Materials and methods
Ethical approval
The University of Sichuan Medical Institutional
Review Board approved the present study (approval no. K2015027).
Written informed consent was obtained from all study participants
prior to participation. The study was performed according to the
principles in the Declaration of Helsinki. All animal experiments
were performed in strict accordance with the recommendations in the
guide for the care and use of laboratory animals by the authority
of the People's Republic of China and approved by the Institutional
Animal Care and Use Committeeat Sichuan University (Sichuan,
China).
Patients
A total of 58 breast IDCs were collected from
patients subsequent to obtaining informed consent, at the
Department of Breast Surgery, West China Hospital of Sichuan
University (Sichuan, China) between May 2010 and Feb 2013. The mean
age of 58 patients was 50.4±11.3 years, and none of them had
undergone chemotherapy or radiotherapy prior to collection. All
specimens were fixed with 40 g/l formaldehyde at 4°C for 24 h,
embedded with paraffin and sectioned into serial slices (thickness,
4 µm).
Cell culture
Human breast cancer MCF-7 cells were purchased from
the Cell Bank of the Chinese Academy of Sciences (Shanghai, China),
and MCF-7-MEK5 (22) cells were
obtained from the Department of Immunology Laboratory, West China
College of Basic and Forensic Medicine of Sichuan University
(Sichuan, China). The cells were cultured in high glucose
Dulbecco's modified Eagle's medium complete medium (catalog no.
SH30022.01B), containing 100 ml/l fetal bovine serum (catalog no.
SV30087.01) and 10 ml/l penicillin and streptomycin (catalog no.
SV30010) (all from HyClone; GE Healthcare, Chicago, IL, USA) at
37°C under 50 ml/l CO2 saturated humidity.
Establishment of the breast cancer
orthotopic injection model
A total of 18 specific pathogen-free BALB/c nude
mice (5 weeks old; female; weighing 18–20 g) were provided by the
Experimental Animal Center at the West China School of Medicine,
Sichuan University (Sichuan, China). All mice were housed singly
with environmental conditions maintained at 21±1°C with a relative
humidity of 50±10% and 15 air charges/h under a 12-h light cycle
(12 h of light followed by 12 h of dark). Sawdust and wood shavings
were used as bedding. The mice were randomly divided into two
groups with 9 mice in each group. Each group was randomly divided
into an experimental subgroup (6 mice) and a control group (3
mice). Mice were allowed free access to food and water, and had a
12 h light/dark cycle. With an implanted 17 β-estradiol (E2) pellet
(0.36 mg/mouse; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany),
digested MCF-7 and MCF-7-MEK5 cells were diluted to
1×107/ml with Hank's balanced salt solution (14025092;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), and
orthotopically injected under the mammary fat pads of mice (0.1
ml/mouse). The mice were raised in fan-filtered units. When the
tumor grew to 100 mm3, the E2 pellets were removed and
160 mg/d/kg cerulenin (BML-G237-0025; Biomol GmbH, Hamburg,
Germany) was administered by intraperitoneal injection to mice in
the experimental group for 10 consecutive days. All mice were
carefully observed throughout the experimental period and should
signs consistent with severe suffering have been detected, those
mice would have been sacrificed immediately. At 5 days following
the last injection, the mice were sacrificed by cervical
dislocation and their tumor tissues were extracted.
Immunohistochemistry
Breast cancer tissues were fixed with 40 g/l
formaldehyde at 4°C for 24 h, embedded in paraffinand sectioned
into serial slices (thickness, 4 µm). Immunohistochemistry was
performed using the SPlink Detection kit (catalog no. SP9000;
ZSGB-BIO; OriGene Technologies, Inc., Beijing, China), according to
themanufacturer's instructions. For negative control, PBS solution
replaced primary antibodies. The following primary antibodies were
used: Mouse anti-human FASN (1:300; catalog no. sc-55580; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) and rabbit anti-human
ErbB-1 (1:200; catalog no. B8501; ImmunoWay Biotechnology Company,
Plano, TX, USA), ErbB-2 (1:200; catalog no. 29D8; Cell Signaling
Technology, Inc. Danvers, MA, USA), ErbB-3 (1:200; catalog no.
A0436; ABclonal Biotech Co., Ltd. Cambridge, MA, USA) and ErbB-4
(1:200; catalog no. A0749; ABclonal Biotech Co., Ltd.) monoclonal
antibodies. The paraffin sections were deparaffinizedin water. For
antigen retrieval, the sections were heated at 95–98°C for 10 min.
The sections were subsequently treated with 5 ml/l hydrogen
peroxide, and goat serum (SPlink Detection kit; ZSGB-BIO; Ori-Gene
Technologies, Inc.) was added to each section to block endogenous
peroxidase activity. The paraffin-embedded blocks were incubated at
37°C for 30 min. The sections were then incubated overnight at 4°C
with primary antibodies: Mouse anti-human FASN (1:300; cat. no.
sc-55580; Santa Cruz Biotechnology, Inc.) and rabbit anti-human
ErbB-1 (1:200; cat. no. B8501; ImmunoWay Biotechnology Company,
Plano, TX, USA), ErbB-2 (1:200; cat. no. 29D8; Cell Signaling
Technology, Inc.), ErbB-3 (1:200; cat. no. A0436; ABclonal Biotech
Co., Ltd. Cambridge, MA, USA) and ErbB-4 (1:200; cat. no. A0749;
ABclonal Biotech Co., Ltd.) monoclonal antibodies, followed by
incubation with the secondary and tertiary antibodies (SPlink
Detection kit; ZSGB-BIO; Ori-Gene Technologies, Inc.), at 37°C for
30 min. 3,3′-diaminobenzidine (Inspissation DAB kit; catalog no.
ZLI-9032; ZSGB-BIO; Ori-Gene Technologies, Inc.) was used to stain
the tissue sections at room temperaturefor between 3 and 5 min. The
sections were subsequently stained with 2 g/l hematoxylin at room
temperature for 4 min, differentiated with hydrochloric acid in
alcohol for 5 sec, flushed with tap water for 40 min, followed by
dehydration, clearing and mounting.
Analysis of immunohistochemical (IHC)
staining results
A total of five random fields of view were assessed
under ×400 magnification (Nikon ECLIPSE Ti-U, light microscope;
Nikon Corporation, Tokyo, Japan). The intensity of staining and the
percentage of positive cells were then recorded. Total points were
determined by adding the value for staining intensity and the
percentage of positive cells for each field of view. The sections
were divided according to staining intensity into negative, light
brown, brown and sepia, and scored as 0, 1, 2 and 3, respectively.
In terms of the percentage of positive cells, the sections were
categorized as negative, <10%, 11–50%, 51–80% and >80%, and
scored as 0, 1, 2, 3 and 4, respectively. Sections with total
points <4 were considered negative (−), 4 points as
weak-positive (+), 5 points as moderately-positive (++) and ≥6
points as strongly-positive (+++).
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was extracted from cells/tissues using
Tripure (cat. no. 1667165001; Roche Diagnostics, Basel,
Switzerland), and reverse transcription to cDNA was performed using
the RevertAid First Strand cDNA Synthesis kit (cat. no. K1621;
Fermentas; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Primer sequences (Table I) were designed using the online
software Primer3 (version 0.4.0; Whitehead Institute, Cambridge,
MA, USA). Primer sequence specificity was verified using the Basic
Local Alignment Search Tool (blast.ncbi.nlm.nih.gov/Blast.cgi), and the primers
were synthesized by Shanghai Bioengineering Co. (Shanghai, China).
RT-PCR was performed according to the manufacturer's protocol (cat.
no. k0221; Fermentas; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: 95°C for 10 min, followed
by 40 cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C for 30
sec. The primer annealing temperature was 60°C for all RT-PCR
procedures performed. Data were analyzed according to the
2−ΔΔCq method (23), and
all experiments were repeated three times.
 | Table I.Primers involved in reverse
transcription-polymerase chain reaction. |
Table I.
Primers involved in reverse
transcription-polymerase chain reaction.
| Gene name | Primer sequence
(5′-3′) | Product length
(bp) |
|---|
| GAPDH |
| 150 |
| F |
CTGCCCCCTCTGCTGATG |
|
| R |
TCCACGATACCAAAGTTGTCAT |
|
| FASN |
| 458 |
| F |
GCCTACTACATCGACTGCATCA |
|
| R |
TACTTGGCCTTGGGTGTGTACT |
|
| ErbB1 |
| 476 |
| F |
CCCTCAAGGAGATAAGTAATGG |
|
| R |
GTACTTCCAGACCAGGGTGTTGT |
|
| ErbB2 |
| 544 |
| F |
ACAGTCTACAAGGGCATCTGGA |
|
| R |
CCCACACAGTCACACCATAACT |
|
| ErbB3 |
| 251 |
| F |
AGGCTTTCAACATCCCACCTC |
|
| R |
GCCATTACAGCAGGAGTCATC |
|
| ErbB4 |
| 349 |
| F |
CTACGAGAGGTGTAGGGTGGT |
|
| R |
GAGCAGTCTTGGGTCATCATC |
|
Western blot analysis
Proteins extracted from MCF-7 and MCF-7-MEK5
cellsusing an radioimmunoprecipitation assaylysis buffer (cat. no.
P0013B; Beyotime Institute of Biotechnology, Shanghai, China) were
quantified and then denatured at 100°C for 5 min. SDS-PAGE (8% gel)
was performed, with 30 µg protein loaded into each well of the gel.
The proteins were then transferred to a 0.2 µm polyvinylidene
fluoride membrane (cat. no. 162-0177; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA), using a semi-dry electrophoresis apparatus for
90 min (FASN) or 40 min (ErbB1, ErbB2, ErbB3 and ErbB4 antibodies)
and then blocked with 50 ml/l low-fat milk solution at room
temperaturefor 1 h. The membrane was then rinsed with 1X TBS with
Tween-20 (TBST) and incubated with primary antibodies against ErbB1
(dilution, 1:200; cat. no. B8501; ImmunoWay Biotechnology), ErbB2
(dilution, 1:500; cat. no. 29D8; Cell Signaling Technology, Inc.),
ErbB3 (dilution, 1:500; cat. no. A0436; ABclonal Biotech Co., Ltd.,
Woburn, MA, USA) and ErbB4 (dilution, 1:500; cat. no. A0749;
ABclonal Biotech Co., Ltd.) or FASN (dilution, 1:1,000; cat. no.
sc-5558; Santa Cruz Biotechnology, Inc.) overnight at 4°C, or
primary antibody against β-actin (dilution, 1:1,000; cat. no.
ab8227; Abcam, Cambridge, MA, USA) at room temperature for 2 h.
Membranes were rinsed again with 1X TBST. Horseradish
peroxidase-labeled secondary antibodies (1:3,000; anti-mouse; cat.
no. 170-6516; 1:3,000; anti-rabbit; cat. no. 170-6515; Bio-Rad
Laboratories, Inc.) were added to the membrane and incubated at
room temperature for 1 h. Pierce™ ECL Western Blotting Substrate
kit (cat. no. 32209; Thermo Fisher Scientific, Inc.) was used to
detect target bands, which was performed according to the
manufacturer's protocol.
Statistical analysis
The data are presented as the mean ± standard
deviation. RT-PCR data were analyzed by using GraphPad Prism
version 5.0 for Windows (GraphPad Software, Inc. La Jolla, CA,
USA). A Student's t-test was used to detect the differences between
groups. Immunohistochemical staining data were analyzed using SPSS
13.0 statistical software (SPSS, Inc. Chicago, IL, USA). The
nonparametric rank sum test was used to detectinter-block
differences in pathological characteristics and a Spearmen
correlation analysis was applied to the correlation analysis of
positive expression between FASN and ErbB1-4. P<0.05 was
considered to indicate a statistically significant difference.
Results
mRNA and protein expression of ErbBs
in MCF-7 and MCF-7-MEK5 cell lines and tumor tissues
RT-PCR and western blot analyses were used to assess
the differential expression of FASN and ErbBs in MCF-7 and
MCF-7-MEK5 cell lines and tumor tissues. The FASN inhibitor,
cerulenin (with 20 µg/ml for 24 h), was added to cells in order to
determine the effect of FASN inhibition on the expression of ErbBs.
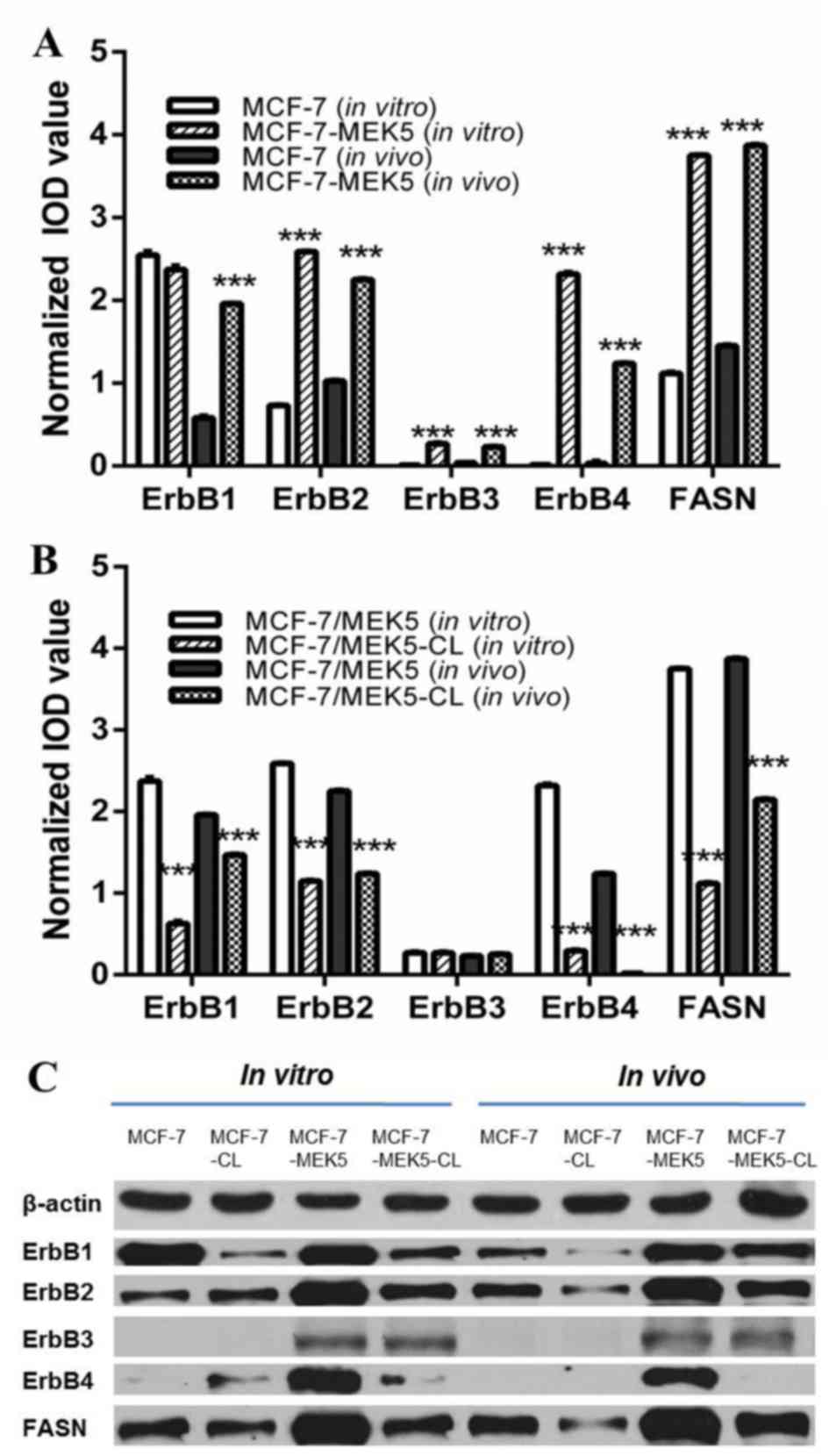
Compared with MCF-7 control cells, the levels of ErbB2-4 and FASN
mRNA expression were significantly upregulated in MCF-7-MEK5 cells.
Compared with MCF-7 cells, the expression of ErbB1, ErbB2, ErbB3,
ErbB4 and FASN was 0.92±0.04, 3.54±0.06, 28.1±1.20, 178±10.6 and
3.45±0.11-fold greater in MCF-7-MEK5 cells, respectively (Fig. 1A). FASN inhibition by cerulenin
treatment resulted in a significant decreased expression of ErbB1,
ErbB2 and ErbB4 in MCF-7-MEK5 cells. Compared with MCF-7 cells, the
expression of ErbB1, ErbB2, ErbB3 and ErbB4 decreased by 0.27±0.02,
0.44±0.00, 1.06±0.01 and 0.13±0.01-fold in MCF-7-MEK5 cells,
respectively. The expression of ErbBs and FASN mRNA was also
upregulated in MCF-7-MEK5 tumor tissues compared with MCF-7 cells.
Compared with MCF-7 tumor tissues, the expression of ErbB1, ErbB2,
ErbB3, ErbB4 and FASN was 3.37±0.19, 2.19±0.03, 5.57±0.24, 15.6±1.8
and 2.25±0.07-fold greater, in MCF-7-MEK5 cells, respectively.
Analysis of tumors from cerulenin-treated mice revealed that the
mRNA expression of ErbB1, ErbB2, ErbB3 and ErbB4 decreased by
0.75±0.01, 0.57±0.02, 1.09±0.01 and 0.013±0.001-fold, respectively,
compared with untreated control mice (Fig. 1B).
At the protein level, the expression of ErbB1 was
similar in MCF-7 and MCF-7-MEK5 cells. However, ErbB1 expression
was markedly upregulated in MCF-7-MEK5 tumor tissues when compared
with that of MCF-7 tumor tissues (Fig.
1C). ErbB1 expression was markedly downregulated in
cerulenin-treated MCF-7-MEK5 cells and tumor tissues compared with
MCF-7 cells. ErbB2 protein expression was higher in MCF-7-MEK5
cells and tumor tissues compared with the expression in MCF-7
cells. Expression of ErbB2 protein was markedly decreased in
MCF-7-MEK5 cells and tissues with cerulenin treatment.
No ErbB3 protein expression was observed in MCF-7
cells and tumor tissues, and there was very low expression in
MCF-7-MEK5 cells and tumor tissues. No significant change in ErbB3
was observed in cerulenin-treated MCF-7-MEK5 cells and tissues.
A very low ErbB4 expression was also observed in
MCF-7 cells, and it was not detected in the tumor tissues. However,
ErbB4 was highly expressed in MCF-7-MEK5 cells and tumor tissues,
and cerulenin treatment markedly decreased its expression in this
cell line.
Taken together, the present results suggestedan
association between the expression of ErbBs 1, 2 and 4, and FASN.
Furthermore, ErbBs were markedly increased in MCF-7-MEK5 cells and
tumor tissues that demonstrate EMT characteristics, and their
expression was notably decreased with FASN inhibition.
Expression of FASN and ErbBs in breast
IDCs and their association with clinicopathologicalparameters
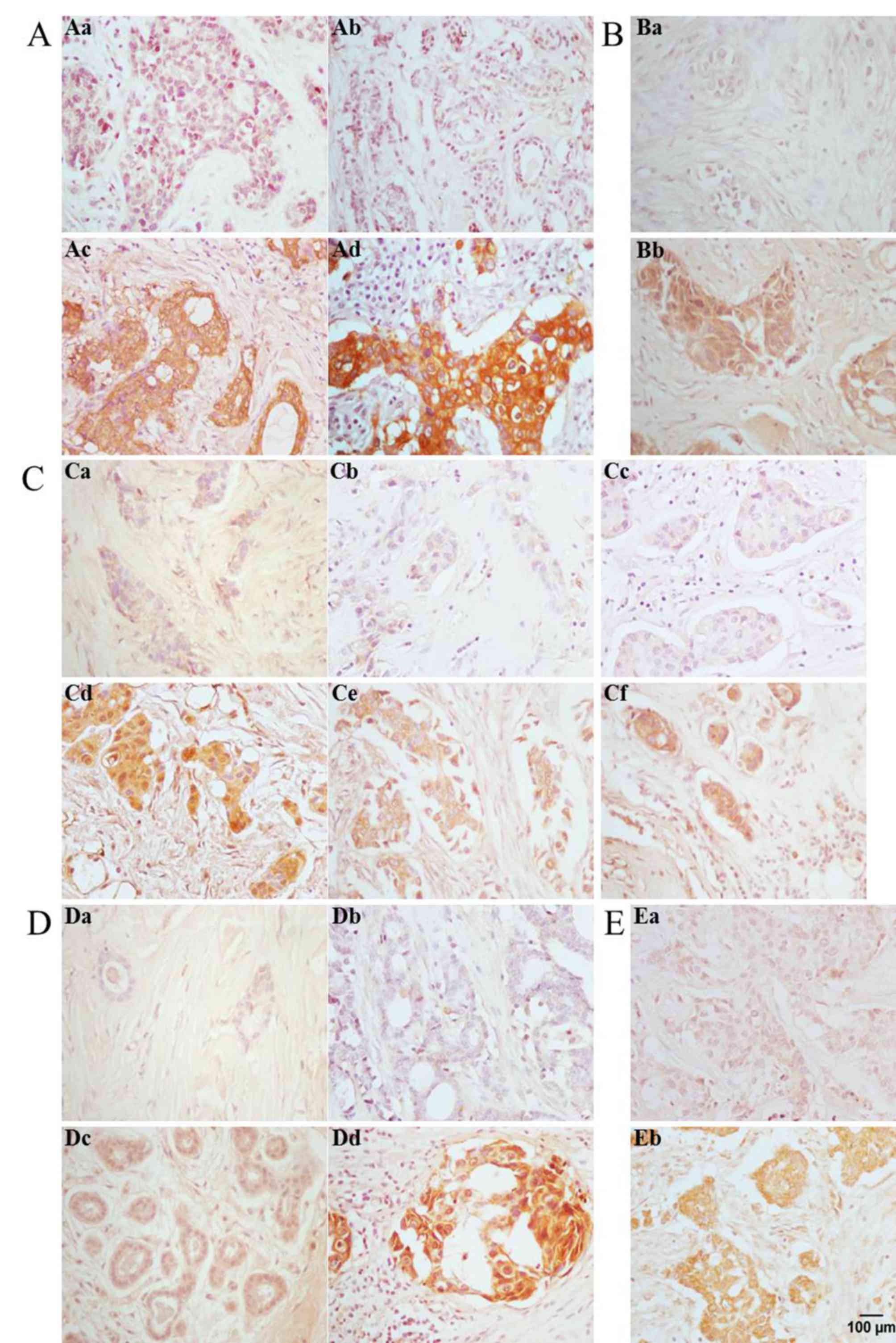
FASN was localized in the cell membrane and
cytoplasm of acinar and ductal epithelial cells of IDC, and its
expression was positively associated with lymphatic metastasis and
tumor size (Fig. 2A). The positive
expression rate of FASN, as assessed by IHC, was 77.5% in all 58
IDCs evaluated, and was higher in lymphatic metastases compared
with non-lymphatic metastases (P<0.05). In the case of tumor
size, the rate of FASN positive expression in larger tumors
(diameter >2 cm) was significantly higher compared with smaller
tumors (diameter ≤2 cm; P<0.05). Parameters, including age,
estrogen receptor (ER), progesterone receptor (PR) status and
clinical stage were also associated with FASN expression, as
differential expression was observed in these respective groups.
However, the differences were not statistically significant
(P>0.05; Table II).
 | Table II.Association between FASN expression
and clinicopathological parameters in IDC. |
Table II.
Association between FASN expression
and clinicopathological parameters in IDC.
|
| FASN
expression |
|---|
|
|
|
|---|
| Parameter | Negative (−) n,
% | Weak-positive (1+)
n, % | Medium-positive
(2+) n, % | Strong-positive
(3+) n, % | P-value | n |
|---|
| IDC |
|
|
|
|
| 58 |
| Age, years |
|
|
|
| 0.840 |
|
|
≤50 | 2 (5.0) | 8 (20.5) | 12 (30.8) | 17 (43.6) |
| 39 |
|
>51 | 2 (10.5) | 4 (21.0) | 7 (36.8) | 6 (31.6) |
| 19 |
| Lymph node
metastases |
|
|
|
| 0.025 |
|
|
Negative | 4 (16.0) | 5 (20.0) | 5 (20.0) | 11 (44) |
| 25 |
|
Positive | 0 | 7 (21.2) | 14 (42.4) | 12 (36.4) |
| 33 |
| IDC stage |
|
|
|
| 0.212 |
|
|
0-I | 2 (22.2) | 0 | 3 (33.3) | 4 (44.4) |
| 9 |
| II | 2 (6.3) | 9 (28.1) | 9 (28.1) | 12 (37.5) |
| 32 |
|
III–IV | 0 | 3 (17.6) | 7 (41.2) | 7 (41.2) |
| 17 |
| IDC size, cm |
|
|
|
| 0.048 |
|
| ≤2 | 3 (16.7) | 7 (38.9) | 7 (38.9) | 1 (5.5) |
| 18 |
|
>2 | 1 (2.5) | 5 (12.5) | 12 (30.0) | 22 (55) |
| 40 |
| ER |
|
|
|
| 0.220 |
|
|
Negative | 0 | 5 (27.8) | 5 (27.8) | 8 (44.4) |
| 18 |
|
Positive | 4 (10.0) | 7 (17.5) | 14 (35.0) | 15 (37.5) |
| 40 |
| PR |
|
|
|
| 0.538 |
|
|
Negative | 1 (5.3) | 5 (26.3) | 5 (26.3) | 8 (42.1) |
| 19 |
|
Positive | 3 (7.7) | 7 (17.9) | 14 (35.9) | 15 (38.5) |
| 39 |
ErbB1 expression was positively associated with
lymphatic metastasis in IDC, and was localized in the cell membrane
and cytoplasm of acinar and ductal epithelial cells of IDC
(Fig. 2B). Immunohistochemistry
analysis determined that the rate of positive ErbB1 expression was
72.4% in all 58 IDCs evaluated, and was higher in lymphatic
metastases compared with non-lymphatic metastases (78.8 vs. 64%;
P<0.01). Similarly, while an association between ErbB1
expression and age, ER and PR status and clinical stage was
observed, the differences were not statistically significant
(P>0.05; Table III).
 | Table III.Associations between ErbB1 expression
and clinicopathological parameters in IDC. |
Table III.
Associations between ErbB1 expression
and clinicopathological parameters in IDC.
|
| ErbB1
expression |
|---|
|
|
|
|---|
| Parameter | Negative (−) n,
% | Weak-positive (1+)
n, % | Medium-positive
(2+) n, % | Strong-positive
(3+) n, % | P-value | n |
|---|
| IDC |
|
|
|
|
| 58 |
| Age,
years |
|
|
|
| 0.547 |
|
|
≤50 | 10 (25.6) | 15 (38.5) | 12 (30.8) | 2 (5.1) |
| 39 |
|
>51 | 6 (31.6) | 5 (26.3) | 8 (42.1) | 0 |
| 19 |
| Lymph node
metastases |
|
|
|
| 0.005 |
|
|
Negative | 9 (36) | 13 (52) | 3 (12) | 0 |
| 25 |
|
Positive | 7 (21.2) | 7 (21.2) | 17 (51.5) | 2(6) |
| 33 |
| IDC stage |
|
|
|
| 0.160 |
|
|
0-I | 6 (40) | 5 (33.3) | 4 (26.6) | 0 |
| 15 |
| II | 10 (33.3) | 8 (26.7) | 10 (33.3) | 2 (6.7) |
| 30 |
|
III–IV | 0 | 7 (53.8) | 6 (46.2) | 0 |
| 13 |
| IDC size |
|
|
|
| 0.199 |
|
| ≤2
cm | 8 (44.4) | 4 (22.2) | 6 (33.3) | 0 |
| 18 |
| >2
cm | 8 (20) | 16 (40) | 14 (35) | 2 (5) |
| 40 |
| ER |
|
|
|
| 0.411 |
|
|
Negative | 3 (16.7) | 7 (38.9) | 8 (44.4) | 0 |
| 18 |
|
Positive | 13 (32.5) | 13 (32.5) | 12 (30) | 2 (5) |
| 40 |
| PR |
|
|
|
| 0.166 |
|
|
Negative | 3 (15.8) | 6 (31.6) | 10 (52.6) | 0 |
| 19 |
|
Positive | 13 (33.3) | 14 (35.9) | 10 (25.6) | 2 (5.1) |
| 39 |
ErbB2 expression was positively associated with
lymphatic metastasis, clinical stage and tumor size in IDC, and was
localized in the cell membrane and cytoplasm of acinar and ductal
epithelial cells of IDC (Fig. 2C).
The rate of positive ErbB2 expression, as determined by IHC, was
65.5% in all 58 IDCs evaluated, and was higher in lymphatic
metastases compared with non-lymphatic metastases (78.8 vs. 48%;
P<0.01). The rate of positive ErbB2 expression in advanced
clinical stages (stages II, III and IV; 65.1%) was significantly
increased compared with early stages (stages 0-I, 40%; P<0.05).
The rate of ErbB2 positive expression in larger tumors (diameter
>2 cm; 100%) was considerably higher compared with smaller
tumors (diameter ≤2 cm; 83.3%; P<0.05). Differential expression
of ErbB2 in relation to patient age and ER and PR status was
observed. However, the differences were not statistically
significant (P>0.05; Table
IV).
 | Table IV.Association between ErbB2 expression
and clinicopathological parameters in IDC. |
Table IV.
Association between ErbB2 expression
and clinicopathological parameters in IDC.
|
| ErbB2
expression |
|---|
|
|
|
|---|
| Parameter | Negative (−) n,
% | Weak-positive (1+)
n, % | Medium-positive
(2+) n, % | Strong-positive
(3+) n, % | P-value | n |
|---|
| IDC |
|
|
|
|
| 58 |
| Age,
years |
|
|
|
| 0.860 |
|
|
≤50 | 12 (30.8) | 2 (5.1) | 8 (20.5) | 17 (43.6) |
| 39 |
|
>51 | 8 (42.1) | 1 (5.3) | 3 (15.8) | 7 (36.8) |
| 19 |
| Lymph node
metastases |
|
|
|
| 0.002 |
|
|
Negative | 13 (52) | 2 (8) | 7 (28) | 3 (12) |
| 25 |
|
Positive | 7 (21.2) | 1 (3.0) | 4 (12.1) | 21 (63.6) |
| 33 |
| IDC stage |
|
|
|
| 0.028 |
|
|
0-I | 9 (60.0) | 1 (6.6) | 3 (20) | 2 (13.3) |
| 15 |
| II | 10 (33.3) | 0 | 8 (26.7) | 12 (40) |
| 30 |
|
III–IV | 5 (38.5) | 0 | 0 | 8 (61.5) |
| 13 |
| IDC size |
|
|
|
| 0.028 |
|
| ≤2
cm | 4 (22.2) | 3 (16.7) | 5 (27.8) | 6 (33.3) |
| 18 |
| >2
cm | 16 (40) | 0 | 6 (15) | 18 (45) |
| 40 |
| ER |
|
|
|
| 0.262 |
|
|
Negative | 9 (50) | 0 | 2 (11.1) | 7 (38.9) |
| 18 |
|
Positive | 11 (27.5) | 3 (7.5) | 9 (22.5) | 17 (42.5) |
| 40 |
| PR |
|
|
|
| 0.282 |
|
|
Negative | 9 (47.4) | 0 | 2 (10.5) | 8 (42.1) |
| 19 |
|
Positive | 11 (28.2) | 3 (7.7) | 9 (23.1) | 16 (41.0) |
| 39 |
ErbB3 expression was positively associated with
lymphatic metastasis and ER status in IDC, and was localized in the
cell membrane and cytoplasm of acinar and ductal epithelial cells
of IDC (Fig. 2D). The rate of
positive ErbB3 expression was 79.3% in all 58 IDCs evaluated, but
was higher in lymphatic metastases compared with non-lymphatic
metastases (81.8 vs. 76%; P=0.01), and increased in ER-positive
compared with ER-negative IDCs (75 vs. 39.9%; P<0.05).
Differential expression of ErbB3 in relation to patient age, tumor
size, PR status and clinical stage was observed. However, the
differences were not statistically significant (P>0.05; Table V).
 | Table V.Association between ErbB3 expression
and clinicopathological parameters in IDC. |
Table V.
Association between ErbB3 expression
and clinicopathological parameters in IDC.
|
| ErbB3
expression |
|---|
|
|
|
|---|
| Parameter | Negative (−) n,
% | Weak-positive (1+)
n, % | Medium-positive
(2+) n, % | Strong-positive
(3+) n, % | P-value | n |
|---|
| IDC |
|
|
|
|
| 58 |
| Age,
years |
|
|
|
| 0.802 |
|
|
≤50 | 8 (20.5) | 10 (25.6) | 14 (35.9) | 7 (17.9) |
| 39 |
|
>51 | 4 (21.1) | 3 (15.8) | 9 (47.4) | 3 (15.8) |
| 19 |
| Lymph node
metastases |
|
|
|
| 0.010 |
|
|
Negative | 6 (24) | 10 (40) | 8 (32) | 1(4) |
| 25 |
|
Positive | 6 (18.2) | 3 (9.1) | 15 (45.5) | 9 (27.3) |
| 33 |
| IDC stage |
|
|
|
| 0.062 |
|
|
0-I | 5 (33.3) | 4 (26.7) | 3 (20) | 3 (20) |
| 15 |
| II | 6 (20) | 8 (26.7) | 11 (36.7) | 5 (16.7) |
| 30 |
|
III–IV | 1 (7.7) | 1 (7.7) | 9 (69.2) | 2 (15.4) |
| 13 |
| IDC size |
|
|
|
| 0.229 |
|
| ≤2
cm | 6 (33.3) | 4 (22.2) | 4 (22.2) | 4 (22.2) |
| 18 |
| >2
cm | 6 (15) | 9 (22.5) | 19 (47.5) | 6 (15) |
| 40 |
| ER |
|
|
|
| 0.031 |
|
|
Negative | 11 (61.1) | 5 (27.8) | 2 (11.1) | 0 |
| 18 |
|
Positive | 10 (25) | 8 (20) | 12 (30) | 10 (25) |
| 40 |
| PR |
|
|
|
| 0.102 |
|
|
Negative | 2 (10.5) | 5 (26.3) | 11 (57.9) | 1 (5.3) |
| 19 |
|
Positive | 10 (25.6) | 8 (20.5) | 12 (30.8) | 9 (23.1) |
| 39 |
ErbB4 expression was positively associated with
lymphatic metastasis in IDC, and was localized in the cell membrane
and cytoplasm of acinar and ductal epithelial cells of IDC
(Fig. 2E). Immunohistochemistry
analysis determined that the rate of positive ErbB4 expression was
60.3% in all 58 IDCs evaluated and was higher () in lymphatic
metastases compared with non-lymphatic metastases (69.7 vs. 48.0%;
P<0.01). Differential expression of ErbB4 in relation to patient
age, tumor size, ER and PR status and clinical stage was observed.
However, the differences were not statistically significant
(P>0.05; Table VI).
 | Table VI.Association between ErbB4 expression
and clinicopathological parameters in IDC. |
Table VI.
Association between ErbB4 expression
and clinicopathological parameters in IDC.
|
| ErbB4
expression |
|---|
|
|
|
|---|
| Parameter | Negative (−) n,
% | Weak-positive (1+)
n, % | Medium-positive
(2+) n, % | Strong-positive
(3+) n, % | P-value | n |
|---|
| IDC |
|
|
|
|
| 58 |
| Age,
years |
|
|
|
| 0.610 |
|
|
≤50 | 15 (38.5) | 13 (33.3) | 10 (25.6) | 1 (2.6) |
| 39 |
|
>51 | 8 (42.1) | 4 (21.1) | 7 (36.8) | 0 |
| 19 |
| Lymph node
metastases |
|
|
|
| 0.001 |
|
|
Negative | 13 (52) | 11 (44) | 1 (4) | 0 |
| 25 |
|
Positive | 10 (30.0) | 6 (18.2) | 16 (48.5) | 1 (3) |
| 33 |
| IDC stage |
|
|
|
| 0.266 |
|
|
0-I | 8 (53.3) | 4 (26.7) | 3 (20) | 0 |
| 15 |
| II | 12 (40) | 8 (26.7) | 9 (30) | 1 (3.3) |
| 30 |
|
III–IV | 3 (23) | 5 (38.5) | 5 (38.5) | 0 |
| 13 |
| IDC size |
|
|
|
| 0.628 |
|
| ≤2
cm | 8 (44.4) | 6 (33.3) | 4 (22.2) | 0 |
| 18 |
| >2
cm | 15 (37.5) | 11 (27.5) | 13 (32.5) | 1 (2.5) |
| 40 |
| ER |
|
|
|
| 0.457 |
|
|
Negative | 5 (27.8) | 6 (33.3) | 7 (38.9) | 0 |
| 18 |
|
Positive | 18 (45) | 11 (27.5) | 10 (25) | 1 (2.5) |
| 40 |
| PR |
|
|
|
| 0.118 |
|
|
Negative | 4 (21.1) | 8 (42.1) | 7 (36.8) | 0 |
| 19 |
|
Positive | 19 (48.7) | 9 (23.1) | 10 (25.6) | 1 (2.6) |
| 39 |
Relevance of effect of FASN expression
on ErbB1, ErbB2 and ErbB4 expression
Spearman's correlation analysis demonstrated that
the correlation between FASN expression and ErbB1, ErbB2 and ErbB4
was statistically significant (FASN and ErbB1, P<0.01; FASN and
ErbB2, P<0.05; FASN and ErbB4, P<0.05). In addition, the
association between the expression of all ErbBs (ErbB1, ErbB2,
ErbB3 and ErbB4) was also statistically significant (P<0.01;
Table VII).
 | Table VII.P-values of correlation analysis of
positive expression between FASN and ErbB1-4 in invasive ductal
carcinoma. |
Table VII.
P-values of correlation analysis of
positive expression between FASN and ErbB1-4 in invasive ductal
carcinoma.
| FASN | ErbB1 | ErbB2 | ErbB3 | ErbB4 |
|---|
| FASN | 0.002 | 0.016 | 0.055 | 0.016 |
| ErbB1 |
| <0.001 | <0.001 | <0.001 |
| ErbB2 |
|
| <0.001 | <0.001 |
| ErbB3 |
|
|
| <0.001 |
| ErbB4 |
|
|
|
|
Discussion
The present paper had three major findings from
studies using breast cancer cells MCF7 and MCF-7-MEK5 cells (with
EMT phenotype) in vivo and in vitro. First, it was
observed that compared with MCF7 cells, mesenchymal and invasive
MCF-7-MEK5 cells highly expressed FASN and ErbB1-4. In addition,
ErbB1-4 expression was directly associated with FASN expression,
which indicated that FASN and ErbBs may act as oncogenes, essential
in promoting EMT of breast cancer cells. Secondly, it was revealed
that that the expression of FASN and ErbB1-4 in IDC was associated
with lymphatic metastasis. Thirdly, it was demonstrated that ErbB1,
ErbB2 and ErbB4 expression wasassociated with FASN expression
during EMT in MCF-7-MEK5 breast cancer cells and in breast cancer
tissues. These results confirmed the hypothesis that FASN affects
the EMT phenotypes of breast cancer cells and such malignant
processes as invasion and metastasis through regulating the
expression of ErbB1, 2 and 4, while ErbB3 was not subjected to the
regulation of FASN.
Previous studies have demonstrated that
overexpression or mutation of ErbBs is associated with various
types of malignant tumors (24),
including breast (25), head and neck
(26), pancreatic (27), spongioblastoma (28), lung (29) and gastriccancer (30) and carcinoma of the rectum (31). ErbB1 and ErbB2 in breast cancer are
generally considered oncogenes and are essential for cancer cell
EMT, proliferation, migration, survival and metastasis (32). ErbB1 and ErbB2 inhibitors are able to
prevent EMT of cancer cells and reduce their migration and invasion
(33). However, there remains great
debate in the field concerning the functionality of ErbB3 and ErbB4
in tumors (17–20). Previous studies have suggested that
ErbB3 functions as an oncogene (34,35). ErbB3
and its ligand, heregulin-β1, were able to promote the migration
and invasion of cancer cells via activation of the phosphoinositide
3-kinase (PI3K) pathway, and induced EMT of breast cancer cells
(36). Small interfering RNA-mediated
inhibition of ErbB3 eliminated EMT, migration and invasion of colon
cancer cells, and induced apoptosis (37). Other studies have argued that ErbB3
functions as a tumor suppressor (18,19,38–41).
Decreased expression of ErbB3 was associated with a malignant
change in phenotypeinvolving EMT, invasion and metastasis of tumor
cells and chemoresistance (38–41). The
sensitivity of breast and pancreatic cancer cells to the
anti-carcinogen elisideps in was increased with ErbB3
overexpression. In addition, drug tolerance was observed with
decreased ErbB3 expression (40).
ErbB3 expression was relatively low in KB and Hep-2 cells (oral and
laryngeal epidermoid cancer cells, respectively) that exhibit EMT
characteristics. These cells were resistant to gefitinib. However,
treatment with the antineoplastic drug vorinostat was able to
reverse EMT induced by upregulatedErbB3 and E-cadherin (41).
The role of ErbB4 in cancer is also controversial
(18), and a number of studies have
shown that ErbB4 exhibit oncogenic activity (42,43). ErbB4
overexpression or mutation induced tyrosine phosphorylation and
activation of downstream signaling pathways (44), and its overexpression was shown to
serve a role in breast cancer oncogenesis (45). Cluster of differentiation 146-mediated
modification of ErbB3 and ErbB4 expression on the surface of breast
cancer cells resulted in activated signaling, increased EMT and
increased drug resistance of cells (46). Other studies demonstrated that ErbB4
acts as a tumor suppressor (47,48), is
deficient in invasive malignant tumors, including prostate cancer,
pancreatic cancer and laryngocarcinoma, and is associated with an
improved prognosis in neck, ovarian and breast cancers (49). In breast cancer, high ErbB4 expression
promoted the sensitivity of tumor cells to hormone therapy
(50). A previous study has
identified that microRNA-193a-3p/5p-mediated downregulation of
ErbB4 resulted in suppression of EMT, migration and invasion of
human non-small-cell lung cancer, by activation of the ErbB4/PIK3
regulatory subunit 3/mammaliantargetofrapamycin/S6 kinase 2
signaling pathway (51). The present
study demonstrated that ErbB1-4 expression is directly associated
with FASN expression, which indicates that FASN and ErbBs may act
as oncogenes, essential in promoting EMT of breast cancer.
IHC results indicated that the rate of ErbB1
expression in IDC was 72.4 and 65.5% in ErbB2, which were slightly
higher than the values reported in previous studies of in
situ (14–65%) (52,53) and invasive breast cancer (10–34%)
(54,55). The rates of FASN, ErbB3 and ErbB4
expression were 75.5, 79.3 and 60.3%, respectively, which were in
agreement with previous findings (56–58). The
present results also revealed that the levels of ErbB1, ErbB2,
ErbB3 and ErbB4 were associated with each other and were consistent
with the results of Abd El-Rehim et al (54) in breast cancer and that of Silva et
al (59) in head and neck
squamous cell carcinomas.
In addition, the present study demonstrated that the
expression of FASN, ErbB1, ErbB2, ErbB3 and ErbB4 in IDC was
associated with lymphatic metastasis, which confirmed the results
of cytological detection analysis in the present study. Previous
studies suggested that high levels of FASN (60), ErbB1 (52,54,55), ErbB2
(54,55,59), ErbB3
(54,55) and ErbB4 (54,55,61)
associated with lymphatic metastasis and other clinicopathological
characteristics. However, other studies demonstrated that the
levels of ErbB1 (62), ErbB3 and
ErbB4 (63,64) in breast cancer were independent of
lymphatic metastasis. In the present study, a positive association
was observed between ErbB2 expression and the clinical stages of
IDC, tumor size and ER positivity, which was in agreement with a
number of the previously published studies (54,55,58,64).
However, other studies have demonstrated that tumor size and
clinical stages were independent of ErbB2 expression, and ErbB3 was
independent of ER positivity.
Therefore, the studies regarding the association
between ErbBs and clinicopathological characteristics of breast
cancer appear to be inconsistent. There are several possible
reasons for the differences in the findings, which are associated
with variations between the studies, including: i) the use of
different scoring systems of positive expression rate; ii) use of
different antibodies; iii) variation in the genetic backgrounds of
the study populations; iv) variations in the breast cancer subtypes
collected for analysis, including in situ breast cancer,
invasive breast cancer and invasive ductal carcinoma; v) different
number of samples collected for analysis (between 51 and 6,046
cases) (52–66); and vi) differences in detection
methods.
As oncogenes associated with metabolism, membrane
microdomains composed of phospholipids generated by FASN, serve as
anchoring sites for ErbBs and other receptor tyrosine kinases, and
perform key roles in connecting ErbBs to downstream molecules
(67). In the present study,
toinvestigate the function and relevance of FASN and ErbBs in tumor
cell EMT, MCF-7-MEK5 cells were treated with the FASN inhibitor
cerulenin. The findings indicated that ErbB1, ErbB2 and ErbB4
levels were significantly downregulated with FASN inhibition.
However, there was no effect on the levels of ErbB3. Additionally,
a positive association was observed between ErbB1, ErbB2, ErbB4 and
FASN expression in 58 IDC samples. Samples expressing ErbB1, ErbB2
and ErbB4 also overexpressed FASN, which suggested that ErbB1,
ErbB2 and ErbB4 were associated with FASN during EMT and breast
cancer malignancy.
Binding of ErbBs with their respective ligands
initiates the formation homo- and heterodimers, and
autophosphorylation of tyrosine residues on their intracellular
domain, which activates downstream PI3K/AKT or
Ras/Raf/mitogen-activated protein kinase kinase/ERK1/2 signaling
pathways (68). It has been
demonstrated that ErbB-mediated activation of these pathways lead
to cellular changes in EMT, migration and invasion (69). Additionally, ErbB1-ErbB2 heterodimers
promoted migration and invasion of MCF-10A cells (70). Overexpression of ErbB1 and ErbB2 was
associated with poor prognostic features and decreased 5-year
disease-free survival rates (55).
The simultaneous overexpression of the intracellular sequence motif
of ErbB1 (CYT2 mICD) and ErbB4 in a breast cancer cell line
significantly increased cellular invasion (71). There is no soluble ligand for ErbB2,
although it has a neuregulin domain similar to ErbB3. Therefore,
ErbB2 frequently heterodimerizes with other ErbBs to promote EMT,
migration, invasion and survival (72–74).
Targeted inhibition of ErbB2 expression or its tyrosine kinase
activity may effectively treat ErbB4-dependent breast cancer, even
tumor cells which do not express ErbB2. Furthermore, silencing of
ErbB2 or ErbB4 led to a significant decrease in anchorage
independence and cell motility of breast cancer cells (42). Therefore, ErbB heterodimers are able
to significantly increase EMT, migration and invasion of various
types of tumor cells.
Previous studies have focused more on the
association between FASN and ErbB1 and ErbB2 dimers in tumors
(7,67,75–77), and
have demonstrated that cross-talk between FASN and ErbB1-ErbB2
expression may be associated with metastasis progression in
malignant ovarian cancers (67,75,77).
Inhibition of FASN affected the synthesis of phospholipids, which
indirectly destroyed the polymerization of ErbB1 and ErbB2 on the
cell membrane (10–11). There are few studies investigating the
effect of FASN on the expression of ErbB3, ErbB4 and their dimers.
However, studies have shown that tumors that co-express ErbB1,
ErbB2 and ErbB4 present an unfavorable outcome compared with other
groups (54), and ErbB3 evade
inhibition by HER-family tyrosine kinase inhibitors in vitro
and in tumors in vivo (74,78).
Together, the present results significantly expanded
the understanding of FASN and ErbB1-4 function in EMT breast cancer
cells, particularly when they are over expressed, and revealed an
association between FASN and ErbB1, 2 and 4 expression in
vitro and in vivo. Therefore, based on the findings of
the present and previously reported studies, it was hypothesized
that the long chain fatty acid synthesized by FASN may affect the
expression and function of ErbB1, ErbB2 and ErbB4, as well as their
polymerized dimers, via formation of microstructures on the cell
membrane, therefore regulating the process of tumor cell EMT,
invasion and migration. ErbB3 may regulate EMT, invasion and
metastasis of the tumor cells via a FASN-independent pathway. These
results additionally indicated that it may be advantageous to
inhibit FASN and ErbB1, 2 and 4 for treatment of certain subsets of
patients with breast cancer who positively FASN, ErbB1, 2 or 4.
Acknowledgements
The present study was funded by a grant from Natural
Science Foundation of China (grant no. J1103604). The sponsors had
no role in study design, data collection and analysis, decision to
publish or preparation of the manuscript. The authors thank the
Immunology Laboratory of Sichuan University (Sichuan, China) for
supplying breast cancer cells, and thank West China Hospital of
Sichuan University (Sichuan, China) for supplying breast cancer
specimens and clinicopathological characteristics materials.
Glossary
Abbreviations
Abbreviations:
|
IDC
|
invasive ductal carcinoma
|
|
FASN
|
fatty acid synthase
|
|
EMT
|
epithelial-mesenchymal transition
|
|
ErbB
|
human epidermal growth factor
receptor
|
|
EGFR
|
epidermal growth factor receptor
|
|
HER-2/neu
|
human epidermal growth factor receptor
2
|
|
ER
|
estrogen receptor
|
|
PR
|
progesterone receptor
|
|
VEGF
|
vascular endothelial growth factor
|
|
VEGFR2
|
vascular endothelial growth factor
receptor 2
|
|
MEK5
|
mitogen-activated protein kinase
(MAPK) kinase 5
|
|
ERK1/2
|
extracellular signal-regulated kinase
1/2
|
|
AKT
|
protein kinase B
|
References
|
1
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weigelt B, Peterse JL and van't Veer LJ:
Breast cancer metastasis: Markers and models. Nat Rev Cancer.
5:591–602. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Swinnen JV, Van Veldhoven PP, Timmermans
L, De Schrijver E, Brusselmans K, Vanderhoydonc F, van de Sande T,
Heemers H, Heyns W and Verhoeven G: Fatty acid synthase drives the
synthesis of phospholipids partitioning into detergent-resistant
membrane microdomains. Biochem Biophys Res Commun. 302:898–903.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li J, Dong L, Wei D, Wang X, Zhang S and
Li H: Fatty acid synthase mediates the epithelial-mesenchymal
transition of breast cancer cells. Int J Biol Sci. 10:171–180.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Flavin R, Peluso S, Nguyen PL and Loda M:
Fatty acid synthase as a potential therapeutic target in cancer.
Future Oncol. 6:551–562. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hung CM, Kuo DH, Chou CH, Su YC, Ho CT and
Way TD: Osthole suppresses hepatocyte growth factor (HGF)-induced
epithelial-mesenchymal transition via repression of the
c-Met/Akt/mTOR pathway in human breast cancer cells. J Agric Food
Chem. 59:9683–9690. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vazquez-Martin A, Colomer R, Brunet J,
Lupu R and Menendez JA: Overexpression of fatty acid synthase gene
activates HER1/HER2 tyrosine kinase receptors in human breast
epithelial cells. Cell Prolif. 41:59–85. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Burgess AW: EGFR family: Structure
physiology signalling and therapeutic targets. Growth Factors.
26:263–274. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lindsey S and Langhans SA: Epidermal
growth factor signaling in transformed cells. Int Rev Cell Mol
Biol. 314:1–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Menendez JA, Vellon L and Lupu R:
Targeting fatty acid synthase-driven lipid rafts: A novel strategy
to overcome trastuzumab resistance in breast cancer cells. Med
Hypotheses. 64:997–1001. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ravacci GR, Brentani MM, Tortelli T Jr,
Torrinhas RS, Saldanha T, Torres EA and Waitzberg DL: Lipid raft
disruption by docosahexaenoic acid induces apoptosis in transformed
human mammary luminal epithelial cells harboring HER-2
overexpression. J Nutr Biochem. 24:505–515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Menendez JA and Lupu R: Fatty acid
synthase and the lipogenic phenotype in cancer pathogenesis. Nat
Rev Cancer. 7:763–777. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moulder SL, Yakes FM, Muthuswamy SK,
Bianco R, Simpson JF and Arteaga CL: Epidermal growth factor
receptor (HER1) tyrosine kinase inhibitor ZD1839 (Iressa) inhibits
HER2/neu (erbB2)-overexpressing breast cancer cells in vitro and in
vivo. Cancer Res. 61:8887–8895. 2001.PubMed/NCBI
|
|
14
|
Arora A and Scholar EM: Role of tyrosine
kinase inhibitors in cancer therapy. J Pharmacol Exp Ther.
315:971–979. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xia W, Gerard CM, Liu L, Baudson NM, Ory
TL and Spector NL: Combining lapatinib (GW572016), a small molecule
inhibitor of ErbB1 and ErbB2 tyrosine kinases, with therapeutic
anti-ErbB2 antibodies enhances apoptosis of ErbB2-overexpressing
breast cancer cells. Oncogene. 24:6213–6221. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Roskoski R Jr: ErbB/HER protein-tyrosine
kinases: Structures and small molecule inhibitors. Pharmacol Res.
87:42–59. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gullick WJ: The c-erbB3/HER3 receptor in
human cancer. Cancer Surv. 27:339–349. 1996.PubMed/NCBI
|
|
18
|
Gullick WJ: c-erbB-4/HER4: Friend or foe?
J Pathol. 200:279–281. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Knowlden JM, Gee JM, Seery LT, Farrow L,
Gullick WJ, Ellis IO, Blamey RW, Robertson JF and Nicholson RI:
c-erbB3 and c-erbB4 expression is a feature of the endocrine
responsive phenotype in clinical breast cancer. Oncogene.
17:1949–1957. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fry WH, Kotelawala L, Sweeney C and
Carraway KL III: Mechanisms of ErbB receptor negative regulation
and relevance in cancer. Exp Cell Res. 315:697–706. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Omura S: The antibiotic cerulenin, a novel
tool for biochemistry as an inhibitor of fatty acid synthesis.
Bacteriol Rev. 40:681–697. 1976.PubMed/NCBI
|
|
22
|
Zhou C, Nitschke AM, Xiong W, Zhang Q,
Tang Y, Bloch M, Elliott S, Zhu Y, Bazzone L, Yu D, et al:
Proteomic analysis of tumor necrosis factor-alpha resistant human
breast cancer cells reveals a MEK5/Erk5-mediated
epithelial-mesenchymal transition phenotype. Breast Cancer Res.
10:R1052008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Roskoski R Jr: The ErbB/HER family of
protein-tyrosine kinases and cancer. Pharmacol Res. 79:34–74. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Salatino M, Schillaci R, Proietti CJ,
Carnevale R, Frahm I, Molinolo AA, Iribarren A, Charreau EH and
Elizalde PV: Inhibition of in vivo breast cancer growth by
antisense oligodeoxynucleotides to type I insulin-like growth
factor receptor mRNA involves inactivation of ErbBs, PI-3K/Akt and
p42/p44 MAPK signaling pathways but not modulation of progesterone
receptor activity. Oncogene. 23:5161–5174. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rogers SJ, Harrington KJ, Rhys-Evans P,
O-Charoenrat P and Eccles SA: Biological significance of c-erbB
family oncogenes in head and neck cancer. Cancer Metastasis Rev.
24:47–69. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Banerjee S, Wang Z, Xu H, Zhang
L, Mohammad R, Aboukameel A, Adsay NV, Che M, Abbruzzese JL, et al:
Antitumor activity of epidermal growth factor receptor-related
protein is mediated by inactivation of ErbB receptors and nuclear
factor-kappaB in pancreatic cancer. Cancer Res. 66:1025–1032. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Clark PA, Iida M, Treisman DM, Kalluri H,
Ezhilan S, Zorniak M, Wheeler DL and Kuo JS: Activation of multiple
ERBB family receptors mediates glioblastoma cancer stem-like cell
resistance to EGFR-targeted inhibition. Neoplasia. 14:420–428.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Engelman JA, Zejnullahu K, Gale CM,
Lifshits E, Gonzales AJ, Shimamura T, Zhao F, Vincent PW, Naumov
GN, Bradner JE, et al: PF00299804, an irreversible pan-ERBB
inhibitor, is effective in lung cancer models with EGFR and ERBB2
mutations that are resistant to gefitinib. Cancer Res.
67:11924–11932. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Allgayer H, Babic R, Gruetzner KU,
Tarabichi A, Schildberg FW and Heiss MM: c-erbB-2 is of independent
prognostic relevance in gastric cancer and is associated with the
expression of tumor-associated protease systems. J Clin Oncol.
18:2201–9. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee JW, Soung YH, Seo SH, Kim SY, Park CH,
Wang YP, Park K, Nam SW, Park WS, Kim SH, et al: Somatic mutations
of ERBB2 kinase domain in gastric, colorectal, and breast
carcinomas. Clin Cancer Res. 12:57–61. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ueno NT and Zhang D: Targeting EGFR in
triple negative breast cancer. J Cancer. 2:324–328. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang ZG, Wei JM, Qin CF, Hao K, Tian XD,
Xie K, Xie XH and Yang YM: Suppression of the epidermal growth
factor receptor inhibits epithelial-mesenchymal transition in human
pancreatic cancer PANC-1 cells. Dig Dis Sci. 57:1181–1189. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Holbro T, Beerli RR, Maurer F, Koziczak M,
Barbas CF III and Hynes NE: The ErbB2/ErbB3 heterodimer functions
as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor
cell proliferation. Proc Natl Acad Sci USA. 100:8933–8938. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stern DF: ERBB3/HER3 and ERBB2/HER2 duet
in mammary development and breast cancer. J Mammary Gland Biol
Neoplasia. 13:215–223. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Smirnova T, Zhou ZN, Flinn RJ, Wyckoff J,
Boimel PJ, Pozzuto M, Coniglio SJ, Backer JM, Bresnick AR,
Condeelis JS, et al: Phosphoinositide 3-kinase signaling is
critical for ErbB3-driven breast cancer cell motility and
metastasis. Oncogene. 31:706–715. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Beji A, Horst D, Engel J, Kirchner T and
Ullrich A: Toward the prognostic significance and therapeutic
potential of HER3 receptor tyrosine kinase in human colon cancer.
Clin Cancer Res. 18:956–968. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ebbing EA, Steins A, Fessler E, Stathi P,
Lesterhuis WJ, Krishnadath KK, Vermeulen L, Medema JP, Bijlsma MF
and van Laarhoven HWM: Esophageal adenocarcinoma cells and
xenograft tumors exposed to Erb-b2 receptor tyrosine kinase 2 and 3
inhibitors activate transforming growth factor beta signaling,
which induces epithelial to mesenchymal transition.
Gastroenterology. 153:63–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
McEvoy LM, O'Toole SA, Spillane CD, Martin
CM, Gallagher MF, Stordal B, Blackshields G, Sheils O and O'Leary
JJ: Identifying novel hypoxia-associated markers of chemoresistance
in ovarian cancer. BMC Cancer. 15:5472015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Teixidó C, Marés R, Aracil M, Cajal Ramón
y S and Hernández-Losa J: Epithelial-mesenchymal transition markers
and HER3 expression are predictors of elisidepsin treatment
response in breast and pancreatic cancer cell lines. PLoS One.
8:e536452013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bruzzese F, Leone A, Rocco M, Carbone C,
Piro G, Caraglia M, Di Gennaro E and Budillon A: HDAC inhibitor
vorinostat enhances the antitumor effect of gefitinib in squamous
cell carcinoma of head and neck by modulating ErbB receptor
expression and reverting EMT. J Cell Physiol. 226:2378–2390. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mill CP, Zordan MD, Rothenberg SM,
Settleman J, Leary JF and Riese DJ II: ErbB2 is necessary for ErbB4
ligands to stimulate oncogenic activities in models of human breast
cancer. Genes Cancer. 2:792–804. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bae JA, Kho DH, Sun EG, Ko YS, Yoon S, Lee
KH, Ahn KY, Lee JH, Joo YE, Chung IJ, et al: Elevated coexpression
of KITENIN and the ErbB4 CYT-2 isoform promotes the transition from
colon adenoma to carcinoma following APC loss. Clin Cancer Res.
22:1284–1294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Prickett TD, Agrawal NS, Wei X, Yates KE,
Lin JC, Wunderlich JR, Cronin JC, Cruz P, Rosenberg SA and Samuels
Y: Analysis of the tyrosine kinome in melanoma reveals recurrent
mutations in ERBB4. Nat Genet. 41:1127–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wali VB, Gilmore-Hebert M, Mamillapalli R,
Haskins JW, Kurppa KJ, Elenius K, Booth CJ and Stern DF:
Overexpression of ERBB4 JM-a CYT-1 and CYT-2 isoforms in transgenic
mice reveal isoform-specific roles in mammary gland development and
carcinogenesis. Breast Cancer Res. 16:5012014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Imbert AM, Garulli C, Choquet E, Koubi M,
Aurrand-Lions M and Chabannon C: CD146 expression in human breast
cancer cell lines induces phenotypic and functional changes
observed in epithelial to mesenchymal transition. PLoS One.
7:e437522012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu Y, Song L, Ni H, Sun L, Jiao W, Chen
L, Zhou Q, Shen T, Cui H, Gao T and Li J: ERBB4 acts as a
suppressor in the development of hepatocellular carcinoma.
Carcinogenesis. 38:465–473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gallo RM, Bryant IN, Mill CP, Kaverman S
and Riese DJ II: Multiple functional motifs are required for the
tumor suppressor activity of a constitutively-active ErbB4 mutant.
J Cancer Res Ther Oncol. 1:102013.PubMed/NCBI
|
|
49
|
Uberall I, Kolár Z, Trojanec R, Berkovcová
J and Hajdúch M: The status and role of ErbB receptors in human
cancer. Exp Mol Pathol. 84:79–89. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ghayad SE, Vendrell JA, Ben Larbi S,
Dumontet C, Bieche I and Cohen PA: Endocrine resistance associated
with activated ErbB system in breast cancer cells is reversed by
inhibiting MAPK or PI3K/Akt signaling pathways. Int J Cancer.
126:545–562. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu T, Li J, Yan M, Liu L, Lin H, Zhao F,
Sun L, Zhang Y, Cui Y, Zhang F, et al: MicroRNA-193a-3p and −5p
suppress the metastasis of human non-small-cell lung cancer by
downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway.
Oncogene. 34:413–423. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Suo Z, Emilsen E, Tveit KM and Nesland JM:
Type 1 protein tyrosine kinases in benign and malignant breast
lesions. Histopathology. 33:514–521. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Walker RA and Dearing SJ: Expression of
epidermal growth factor receptor mRNA and protein in primary breast
carcinomas. Breast Cancer Res Treat. 53:167–176. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
El-Rehim Abd DM, Pinder SE, Paish CE, Bell
JA, Rampaul RS, Blamey RW, Robertson JF, Nicholson RI and Ellis IO:
Expression and co-expression of the members of the epidermal growth
factor receptor (EGFR) family in invasive breast carcinoma. Br J
Cancer. 91:1532–1542. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Badovinac-Crnjevic T, Jakic-Razumovic J,
Podolski P, Pleština S, Sarčević B, Munjas R and Vrbanec D:
Significance of epidermal growth factor receptor expression in
breast cancer. Med Oncol. 28 Suppl 1:S121–S128. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Porta R, Blancafort A, Casòliva G, Casas
M, Dorca J, Buxo M, Viñas G, Oliveras G and Puig T: Fatty acid
synthase expression is strongly related to menopause in early-stage
breast cancer patients. Menopause. 21:188–191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Suo Z, Berner HS, Risberg B, Karlsson MG
and Nesland JM: Estrogen receptor-alpha and C-ERBB-4 expression in
breast carcinomas. Virchows Arch. 439:62–69. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Witton CJ, Reeves JR, Going JJ, Cooke TG
and Bartlett JM: Expression of the HER1-4 family of receptor
tyrosine kinases in breast cancer. J Pathol. 200:290–297. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Silva SD, Cunha IW, Younes RN, Soares FA,
Kowalski LP and Graner E: ErbB receptors and fatty acid synthase
expression in aggressive head and neck squamous cell carcinomas.
Oral Dis. 16:774–780. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Long QQ, Yi YX, Qiu J, Xu CJ and Huang PL:
Fatty acid synthase (FASN) levels in serum of colorectal cancer
patients: Correlation with clinical outcomes. Tumour Biol.
35:3855–3859. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lodge AJ, Anderson JJ, Gullick WJ, Haugk
B, Leonard RC and Angus B: Type 1 growth factor receptor expression
in node positive breast cancer: Adverse prognostic significance of
c-erbB-4. J Clin Pathol. 56:300–304. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lewis S, Locker A, Todd JH, Bell JA,
Nicholson R, Elston CW, Blamey RW and Ellis IO: Expression of
epidermal growth factor receptor in breast carcinoma. J Clin
Pathol. 43:385–389. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chiu CG, Masoudi H, Leung S, Voduc DK,
Gilks B, Huntsman DG and Wiseman SM: HER-3 overexpression is
prognostic of reduced breast cancer survival: A study of 4046
patients. Ann Surg. 251:1107–1116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Fujiwara S, Ibusuki M, Yamamoto S,
Yamamoto Y and Iwase H: Association of ErbB1-4 expression in
invasive breast cancer with clinicopathological characteristics and
prognosis. Breast Cancer. 21:472–481. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lim ST, Yu JH, Park HK, Moon BI, Ko BK and
Suh YJ: A comparison of the clinical outcomes of patients with
invasive lobular carcinoma and invasive ductal carcinoma of the
breast according to molecular subtype in a Korean population. World
J Surg Oncol. 12:562014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zengel B, Yararbas U, Duran A, Uslu A,
Elıyatkın N, Demırkıran MA, Cengiz F, Şimşek C, Postacı H, Vardar E
and Durusoy R: Comparison of the clinicopathological features of
invasive ductal, invasive lobular, and mixed (invasive ductal +
invasive lobular) carcinoma of the breast. Breast Cancer.
22:374–381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Grunt TW, Wagner R, Grusch M, Berger W,
Singer CF, Marian B, Zielinski CC and Lupu R: Interaction between
fatty acid synthase- and ErbB-systems in ovarian cancer cells.
Biochem Biophys Res Commun. 385:454–459. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Suda K, Tomizawa K and Mitsudomi T:
Biological and clinical significance of KRAS mutations in lung
cancer: An oncogenic driver that contrasts with EGFR mutation.
Cancer Metastasis Rev. 29:49–60. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Stivarou T and Patsavoudi E: Extracellular
molecules involved in cancer cell invasion. Cancers (Basel).
7:238–265. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhan L, Xiang B and Muthuswamy SK:
Controlled activation of ErbB1/ErbB2 heterodimers promote invasion
of three-dimensional organized epithelia in an ErbB1-dependent
manner: Implications for progression of ErbB2-overexpressing
tumors. Cancer Res. 66:5201–5208. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kiuchi T, Ortiz-Zapater E, Monypenny J,
Matthews DR, Nguyen LK, Barbeau J, Coban O, Lawler K, Burford B,
Rolfe DJ, et al: The ErbB4 CYT2 variant protects EGFR from
ligand-induced degradation to enhance cancer cell motility. Sci
Signal. 7:ra782014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Baselga J and Swain SM: Novel anticancer
targets: Revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer.
9:463–475. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Spears M, Taylor KJ, Munro AF, Cunningham
CA, Mallon EA, Twelves CJ, Cameron DA, Thomas J and Bartlett JM: In
situ detection of HER2:HER2 and HER2:HER3 protein-protein
interactions demonstrates prognostic significance in early breast
cancer. Breast Cancer Res Treat. 132:463–470. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sergina NV, Rausch M, Wang D, Blair J,
Hann B, Shokat KM and Moasser MM: Escape from HER-family tyrosine
kinase inhibitor therapy by the kinase-inactive HER3. Nature.
445:437–441. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Tomek K, Wagner R, Varga F, Singer CF,
Karlic H and Grunt TW: Blockade of fatty acid synthase induces
ubiquitination and degradation of phosphoinositide-3-kinase
signaling proteins in ovarian cancer. Mol Cancer Res. 9:1767–1779.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Vazquez-Martin A, Fernandez-Real JM,
Oliveras-Ferraros C, Navarrete JM, Martin-Castillo B, Del Barco S,
Brunet J and Menendez JA: Fatty acid synthase activity regulates
HER2 extracellular domain shedding into the circulation of
HER2-positive metastatic breast cancer patients. Int J Oncol.
35:1369–1376. 2009.PubMed/NCBI
|
|
77
|
Cai Y, Wang J, Zhang L, Wu D, Yu D, Tian
X, Liu J, Jiang X, Shen Y, Zhang L, et al: Expressions of fatty
acid synthase and HER2 are correlated with poor prognosis of
ovarian cancer. Med Oncol. 32:3912015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Mirschberger C, Schiller CB, Schräml M,
Dimoudis N, Friess T, Gerdes CA, Reiff U, Lifke V, Hoelzlwimmer G,
Kolm I, et al: RG7116, a therapeutic antibody that binds the
inactive HER3 receptor and is optimized for immune effector
activation. Cancer Res. 73:5183–5194. 2013. View Article : Google Scholar : PubMed/NCBI
|