Introduction
Glioma is considered as the largest group of primary
brain malignant tumor in adults, which shows an aggressive nature
and is very likely to spread to the surrounding brain tissues
(1,2).
Although considerable progress had been made in surgical and
anticancer therapy, the prognosis of glioma is still unfavourable.
Glioblastoma multiforme (GBM), the glioma histology type reported
to be the most malignant, results in a life expectancy of 10–12
months after diagnosis and it ranks the third fatal malignant tumor
following lung and pancreatic cancer (3,4). As the
mechanism and prognostic factors of glioma is still unclear, there
is no effective and specific treatments for glioma so far (5). Therefore, developing new diagnostic
approaches will be helpful to the early diagnosis and treatment for
gilomas (6).
HDGF is an acidic heparin-binding growth factor
which was first purified from the medium of human hepatoma cell
line Huh-7 (7). In recent years, A
various biological roles of HDGF have been found, including the
effect on promoting mitosis and vascular development (8). The results were similar to those found
in other studies that HDGF played an important roles in promoting
cancer cell proliferation, vascular formation, invasion, and
metastasis in several malignant tumor, such as oral squamous cell
carcinoma, esophageal cancer, colon carcinoma as well as lung and
stomach cancer. (9–13) Moreover, pathological analysis
indicated that the over expression of HDGF is significantly related
to poor outcome of multiple cancer types, such as pancreatic cancer
(14), hepatocellular carcinoma
(15) and gastric cancer (16). However, the role of HDGF in the
prognosis of human gliomas is still unclear.
To address this problem, immunohistochemistry
staining, western blotting analysis and RT-PCR were used to
evaluate the expression of HDGF protein and mRNA respectively in
130 patients with primary gliomas. The correlation between HDGF
expression and these clinicopathological characteristics were
statistically evaluated. Then, multivariate liner regression was
also used to evaluate their effect on patients survival time.
Materials and methods
Patients and tissue samples
The study had obtained the approval from the Ethics
Committee of Tangdu Hospital, Fourth Military Medical University,
Xi'an, China. According to the ethical standards, informed consents
were sighed by all subjects and the samples were handled
anonymous.
Fresh glioma samples were obtained from totally 130
patients who was diagnosed with glioma at the Department of
Neurosurgery, Tangdu Hospital from June 2009 to June 2013.
Radiotherapy or chemotherapy was not performed for the subjects
before surgery. Intraoperative histological examination was
performed to make a definite diagnosis of glioma. Patients received
adjuvant treatment after surgery according to a uniform guideline
depending on the stage of disease. Histopathologically
classification of the glioma samples were performed depending on
the WHO classification (17). 26, 32,
40 and 32 patients were classified as WHO grade I, II, III and IV
respectively.
Specimens got from each patient were divided into
two parts. One part was made into paraffin sections by fixing
tissues in formalin and then imbedding them in paraffin. Another
was stored at −80°C immediately after surgery for posterior Western
Blot and qRT-PCR. Fifteen patients with intractable epilepsy were
involved in the study and the nonneoplastic brain tissues obtained
from them were taken as control.
The database of electronic medical record system was
used to collect clinical information and we set the date of surgery
as the starting point for survival analyses. Patients died of other
reasons not related to glioma served as censored data. Follow-up
was terminated until June 18, 2016.
Quantitative Real-Time PCR
TRIzol reagent (Invitrogen, Carlsbad, US) was used
to isolate total RNA of glioma specimen following the operating
instruction. qRT-PCR was conducted in CFX96™ PCR System (Bio-rad).
PCR was done with the following primers: HDGF forward primer
5′-TGCTCCTACCCACGCAGATT-3′, reverse primer
5′-GGCCAACCCAGAGTTGGAA-3′; β-actin: Sense,
5′-CTACAATGAGCTGCGTGTGGC-3′; antisense,
5′-CAGGTCCAGACGCAGGATGGC-3′. β-actin was used to normalize the
targets as a standard.
Western blotting analyses
Samples were lysed by lysis buffer for 30 min and
then centrifuged (12,000 rpm) for 20 min. Protein quantitation was
performed by the procedure of BCA Protein Assay kit (Beyotime Inst,
Biotech, China). Samples were separated with 12% SDS-PAGE and
transferred to nitrocellulose (NC) membrane by electrophoresis.
Then, 5% skim milk was used to block the membranes for 1 h, and
incubated with the suitable primary rabbit anti-human HDGF antibody
(Abcam, USA) overnight at 4°C. After washed by TBST, the HRP
adjointed secondary antibodies (Jackson, USA) was used to incubate
membranes for 1 h. Then, membranes were washed and the blots were
visualized using enhanced chemiluminescence reagents (Millipore,
USA). Bands were digitally scanned and analyzed using Image J
software and the intensity signal was recorded for further
statistical analysis.
Immunohistochemistry analyses
The slices were deparaffinized by a group of xylene
and then dexylene by a group of ethanol with graded concentrations.
Then they were incubated in 0.01 M citrate buffer (pH=6.0) for
antigen retrieval by heating the tissues slices in pressure cooker
for 5 min. Once the slices cooled to room temperature, the activity
of endogenous enzyme was blocked by soaking the slices in a
humidified chamber contained with 3% hydrogen peroxide for 10 min.
After a brief wash in distilled water, they were incubated with 10%
donkey serum (Abcam) and then the primary antibody were prepared to
appropriate concentration using PBS. Antibodies adopted in our
study include: Primary rabbit anti-human HDGF antibody (Santa Cruz,
USA) and anti-human Ki-67 antibody (Santa Cruz, USA). Slices were
incubated in a humidified chamber at 4°C overnight. Following that,
slices were incubated with goat anti-rabbit immunoglobulin G
antibody (Santa Cruz, USA) conjugated by horseradish peroxidase for
30 min. Diaminobenzidine (DAB) staining and hematoxylic
counterstaining were performed to show the location of HDGF in the
glioma specimen. Two experienced neuropathologists, blinded to
clinical information, rated the percentage of positive nuclei
staining of the stained slices. The level of HDGF expression was
defined as follows: Negative staining was classified as Level 0.
More than 60% of positive staining was considered as level 2 and
the rest of slices were graded as level 1.
Statistical analyses
SPSS 13.0 software was applied to perform all
statistical analyses. The relationship between HDGF levels and
clinicopathologic data was analyzed by the χ2 test. Data
of western blotting analyses and qRT-PCR were dealt by using
one-way classification of ANOVA followed by Bonferroni's test. The
Kaplan-Meier method was used to generate survival curves and
further analysis was performed using the log-rank test.
Multivariable linear regression was adopted to analyze the effects
of HDGF, age, gender, WHO grade and KPS on prognosis. A P-value of
less than 0.05 was regarded as having statistical difference.
Results
Increased expression of HDGF mRNA in
glioma tissues
The expression of HDGF mRNA was obviously increased
in the glioma than in intractable epileptic brain (*P<0.05).
Further statistical analysis was conducted to assess the
relationship between HDGF mRNA expression and various clinical
pathological features (Table I).
Interestingly, HDGF mRNA expression was augmented as the WHO grades
increased (*P<0.05) and was higher in subjects whose Ki-67 index
≥20% (*P<0.05) and KPS <80 (*P<0.05).
 | Table I.Association of HDGF mRNA expression
with various. |
Table I.
Association of HDGF mRNA expression
with various.
| Clinicopathological
features | No. of cases | HDGF mean (SD) | P-value |
|---|
| Tissue type |
|
|
|
|
Control | 15 | 0.051 (0.079) | <0.05 |
|
Glioma | 130 | 2.437 (0.190) |
|
| WHO grade |
|
|
|
| I | 26 | 0.793 (0.009) | <0.05 |
| II | 32 | 1.635 (0.217) |
|
| III | 35 | 3.178 (0.316) |
|
| IV | 37 | 3.893 (0.427) |
|
| Ki-67 index |
|
|
|
|
<20% | 64 | 1.736 (0.109) | <0.05 |
| ≥20% | 66 | 3.987 (0.520) |
|
| KPS |
|
|
|
| ≥80 | 66 | 1.523 (0.215) | <0.05 |
|
<80 | 64 | 3.197 (0.296) |
|
Increased expression of HDGF protein
in glioma tissues
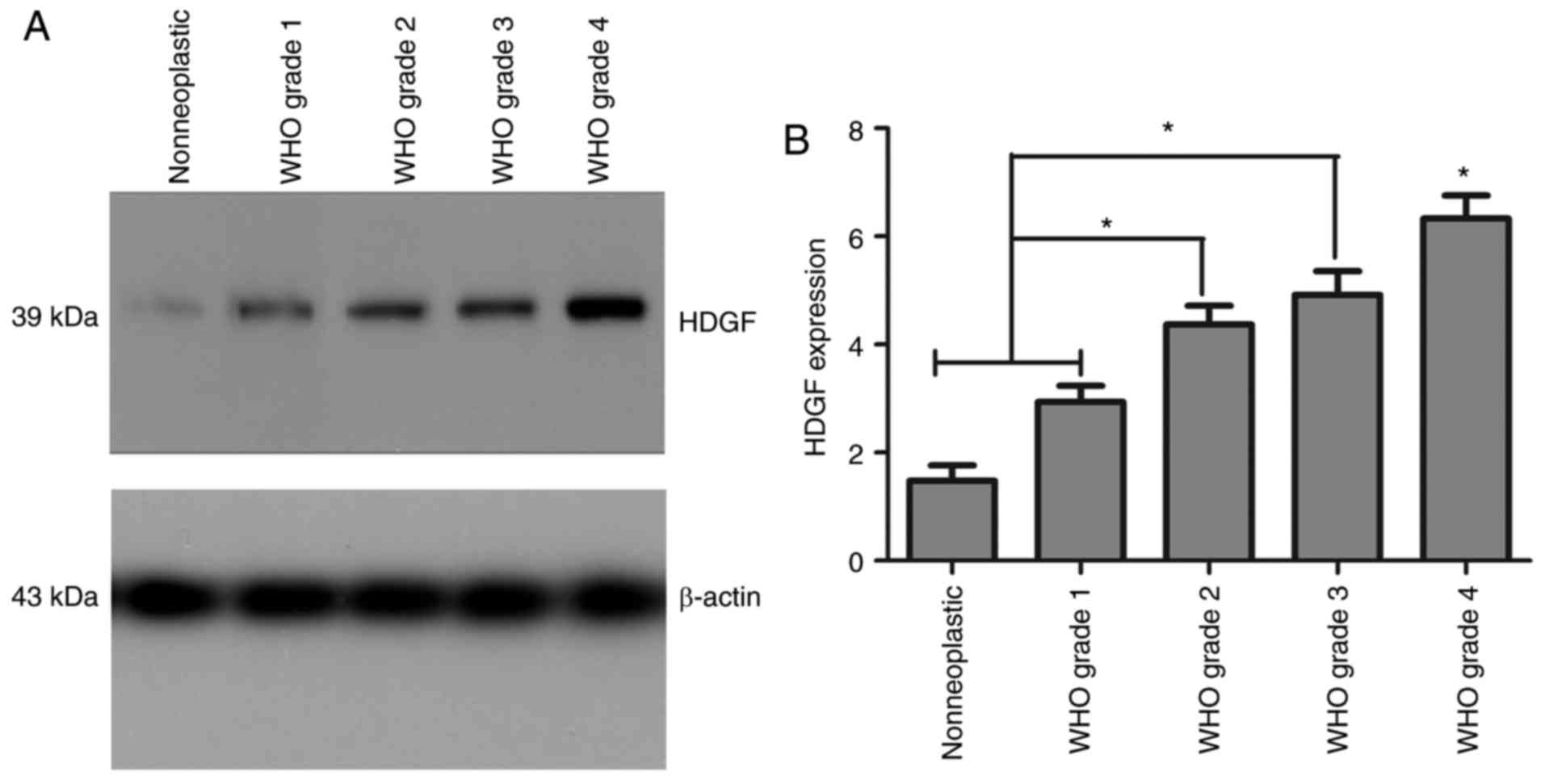
Western blotting indicated that the expressions of
HDGF protein were obviously higher in both the high (WHO III–IV)
and low (WHO I–II) grade glioma groups compared with normal brain
tissue group (*P<0.01). Moreover, in the high-grade glioma
group, the expression of HDGF protein expression was obviously
higher compared with the low-grade glioma group (*P<0.01). But
no statistical difference was observed between grade II and grade
III group (P>0.05). (Fig. 1).
Positive rate of HDGF in glioma
samples
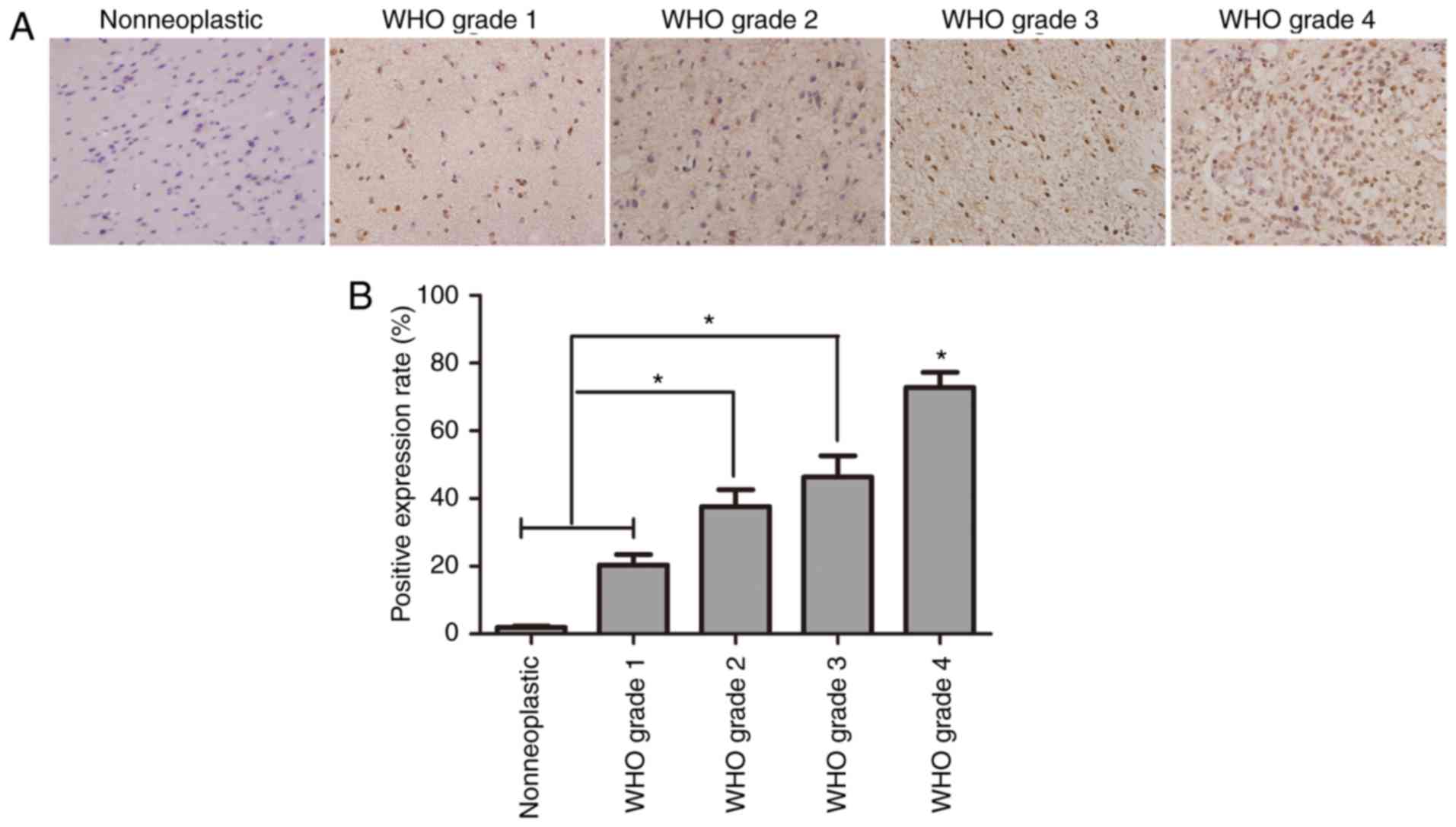
The results of immunohistochemistry indicated a
positive result of HDGF in glioma cells (Fig. 2A). The positive rate of HDGF in the
control group and grade I–IV glioma groups was 1.96, 20.40, 37.64,
46.35 and 72.76%, respectively. These outcomes illustrated that the
positive rate of HDGF was evidently higher in the WHO II–IV group
than in WHO I and control groups (P<0.001). However, no
statistical difference was observed between WHO II and III groups
(P>0.05) (Fig. 2B).
Relationship between the HDGF
expression and clinical pathologic parameters
The association of HDGF immunostaining with the
clinical pathological parameters of glioma patients was summarized
in Table II. As is shown in the
table, the expression of HDGF was not markedly influenced by gender
or age (P>0.05). In comparison, it was closely related to the
WHO grade of gliomas and the KPS. The quantity of HDGF expression
was significantly higher in glioma tissues with Ki-67 index ≥20%,
KPS <80 and grades II ~IV than in those with Ki-67 index
<20%, KPS ≥80 and grades I (Table
II; *P<0.05).
 | Table II.Association of HDGF protein expression
with various clinicopathological features. |
Table II.
Association of HDGF protein expression
with various clinicopathological features.
|
|
| HDGF expression
(n) |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | No. of cases | Level 1 and 0 | Level 2 | P-value |
|---|
| WHO grade |
|
|
|
|
| I | 26 | 14 | 12 | <0.05 |
| II | 32 | 10 | 22 |
|
|
III | 35 | 6 | 29 |
|
| IV | 37 | 4 | 33 |
|
| Age |
|
|
|
|
|
<55 | 69 | 17 | 52 | NS |
|
≥55 | 61 | 15 | 46 |
|
| Gender |
|
|
|
|
|
Male | 67 | 20 | 47 | NS |
|
Female | 63 | 16 | 47 |
|
| Ki-67 index |
|
|
|
|
|
<20% | 64 | 49 | 15 | <0.05 |
|
≥20% | 66 | 18 | 48 |
|
| KPS |
|
|
|
|
|
≥80 | 66 | 32 | 34 | <0.05 |
|
<80 | 64 | 10 | 54 |
|
Increase in HDGF protein expression
indicates bad prognosis of patients with gliomas
The complete follow-up data obtained from 130
patients with gliomas and the results of HDGF expression level was
used for survival analysis. 102 glioma patients (78.5%) died during
follow-up (80 from the HDGF high expression group (level 2) and 22
from the HDGF low expression group (level 0 and 1)). Among the 102
dead patients, 6 died because of accidents or other diseases not
directly related to gliomas (4 from HDGF high expression group
(level 2) and 2 from the HDGF low expression group (level 0 and
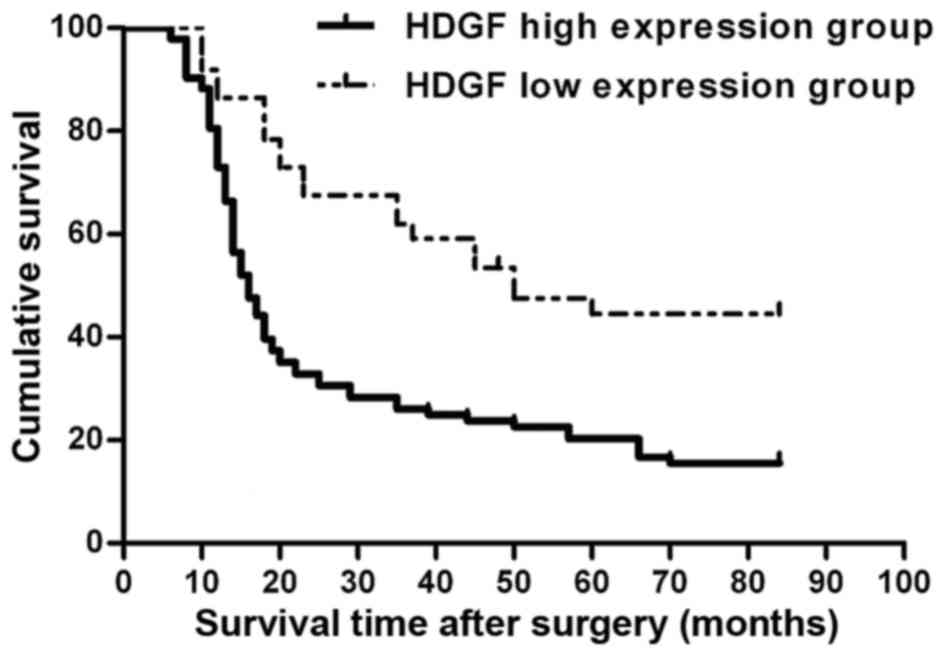
1)). In the univariate survival analysis, the cumulative survival
curve was plotted by using the Kaplan-Meier method and the
difference in survival was determined by the log-rank method. The
findings revealed that subjects with high level of HDGF had an
obviously shorter survival time than patients with low HDGF
expression level (P<0.001; Fig.
3). The average survival period of subjects with high and low
HDGF expression were 16.6±2.0 and 49.8±1.5 months (log rank test:
*P<0.01) respectively. Further more, the effect of age, gender,
WHO grade, KPS and HDGF on prognosis was evaluated by multivariable
linear regression. The results in Table
III indicated that the WHO grade (HR=1.781, 95%CI: 1.145–2.770,
P=0.01), KPS (HR=1.952, 95%CI: 1.251–3.048, P=0.006), Ki-67
(HR=2.671, 95%CI: 1.827–4.727, P<0.001) and HDGF expression
(HR=4.028, 95%CI: 2.542–6.380, P<0.001) were significantly
correlated with the prognosis of glioma patients, but no effect was
found on age and gender.
 | Table III.Multivariate Cox regression
analysis. |
Table III.
Multivariate Cox regression
analysis.
| Parameter | Hazard ratio | 95% confidence
interval | P-value |
|---|
| Age | 0.923 | 0.614–1.691 | 0.61 |
| Gender | 0.986 | 0.648–1.785 | 0.55 |
| WHO grade | 1.781 | 1.145–2.770 | 0.01 |
| KPS score | 1.952 | 1.251–3.048 | 0.006 |
| Ki-67 index | 2.671 | 1.827–4.727 | <0.001 |
| HDGF | 4.028 | 2.542–6.380 | <0.001 |
Discussion
Despite huge progress in developing the diagnostic
methods and strategies for therapy, such as radiation treatment and
chemotherapy, glioma is still one of the most lethal cancer in
human (18,19). The average survival period of patients
with glioma is less than 2 years and the 5-year survival rate is no
more than 3%, which ranks the lowest among all cancers (20). Thus, it is urgent to develop novel
diagnostic methods and effective treatment strategies. In recent 2
decades, extensive studies have identified HDGF as an important
regulator that are critical to various biological processes, such
as regeneration, growth, remodeling, mitosis promotion, vascular
formation, transcriptional regulation, differentiation and
apoptosis (21–26). The crucial role of HDGF overexpression
on tumor progression and prognosis has been revealed in multiple
cancer types, such as gastric cancer (16), hepatocellular carcinoma (15), pancreatic cancer (14), as well as lung and esophageal cancer
(14,27). However, its role in human gliomas is
still unknown.
In order to deal with the problem, 130 samples of
human gliomas were collected to examine the HDGF expression and
analyze the association between its expression and
clinicopathological characteristics. Our data indicated that HDGF
expression, at both protein and mRNA levels, was found to be more
obviously up-regulated in glioma tissues than in intractable
epileptic brain tissue without tumor. Moreover, high expression of
HDGF was closely related to several clinicopathological parameters,
including WHO grades II~IV, Ki-67 index ≥20% or KPS <80
(*P<0.05). These outcomes may indicate an important role of HDGF
in genesis or development of glioma.
Prior studies have mainly focused on the function of
HDGF in other malignant tumors and accumulating evidence has
revealed the effect of HDGF as a vital biomarker on cancer
diagnosis and prognosis. Lots of studies have demonstrated that the
over-expression of HDGF might play an important role in metastasis
and eventually lead to poor results in various metastatic tumors.
HDGF expression is significantly higher in breast cancer tissues
and has a positive correlation with bad result severity, histology
grades and tumor sizes. Thus, it is a strong predictor of the
median survival time for breast cancer patients (28). Similar results were observed in
several other types of cancer, including gastric cancer (14), lung cancer (26), pancreatic cancer (15) and esophageal carcinoma (14). For human glioma, current studies were
mostly focused on the mechanism of carcinogenesis induced by HDGF.
Hsu et al concluded that HDGF is a mitogenic growth factor
in glioma progression (29). Zhang
et al revealed that the knockdown of HDGF significantly
inhibited tumorigenesis as well as colony formation, migration and
invasion of U87 glioma cells (23).
Song et al's observed in their early studies that knocking
out of HDGF obviously inhibited the formation, development and
spread of glioma cell as well as restored the expression of
E-cadherin and inhabited the biomarkers of mesenchymal cell such as
β-catenin and N-cadherin and vimentin. They also found that HDGF
probably participated in the activation of PI3K/Akt and TGF-β
signaling pathways (30). In accord
with these studies, our research also confirmed the carcinogenic
role of HDGF as its expression, at both protein and mRNA levels,
was up-regulated to a greater degree in glioma than in brain tissue
without tumor. Moreover, the effect of HDGF expression on survival
period of glioma patients was statistically analyzed. As a result,
negative correlation was found between them. In addition, the
results of multivariable linear regression suggested that WHO
grade, KPS, Ki-67 and HDGF expression were closely related to
glioma patients' prognosis. We have several innovations compared
with these prior studies. These researches mostly based on glioma
cell lines and animal as well as collected clinical features like
age and gender. While we adopted glioma tissues of human brain in
our study and more clinical data like Karnofsky performance Status
(KPS) and Ki-67 index was collected in our research except for age
and gender. So our research are more clinically relevant and
tightly associated to human glioma.
Considering all of the results, animal experiments
should be conducted by utilizing molecular biotechniques to
evaluate the role of HDGF gene regulation on the development and
invasion of glioma. Which may provide much more theoretical
foundations for investigating prognostic and therapeutic potential
of HDGF for glioma patients.
Glossary
Abbreviations
Abbreviations:
|
WHO
|
world health organization
|
|
KPS
|
Karnofsky performance score
|
|
qRT-PCR
|
Quantitative Real-Time PCR
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Claes A, Idema AJ and Wesseling P: Diffuse
glioma growth: A guerilla war. Acta Neuropathol. 114:443–458. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 15
Suppl 2:ii1–ii56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wick W, Platten M and Weller M: New
(alternative) temozolomide regimens for the treatment of glioma.
Neuro Oncol. 11:69–79. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bao S, Wu Q, Li Z, Sathornsumetee S, Wang
H, McLendon RE, Hjelmeland AB and Rich JN: Targeting cancer stem
cells through L1CAM suppresses glioma growth. Cancer Res.
68:6043–6048. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Roversi G, Pfundt R, Moroni RF, Magnani I,
van Reijmersdal S, Pollo B, Straatman H, Larizza L and Schoenmakers
EF: Identification of novel genomic markers related to progression
to glioblastoma through genomic profiling of 25 primaryglioma cell
lines. Oncogene. 25:1571–1583. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nakamura H, Kambe H, Egawa T, Kimura Y,
Ito H, Hayashi E, Yamamoto H, Sato J and Kishimoto S: Partial
purification and characterization of human hepatoma-derived growth
factor. Clin Chim Acta. 183:273–284. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shih TC, Tien YJ, Wen CJ, Yeh TS, Yu MC,
Huang CH, Lee YS, Yen TC and Hsieh SY: MicroRNA-214 downregulation
contributes to tumor angiogenesis by inducing secretion of the
hepatoma-derived growth factor in human hepatoma. J Hepatol.
57:584–591. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin YW, Li CF, Chen HY, Yen CY, Lin LC,
Huang CC, Huang HY, Wu PC, Chen CH, Chen SC and Tai MH: The
expression and prognostic significance of hepatoma-derived growth
factor in oral cancer. Oral Oncol. 48:629–635. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mao J, Xu Z, Fang Y, Wang H, Xu J, Ye J,
Zheng S and Zhu Y: Hepatoma-derived growth factor involved in the
carcinogenesis of gastric epithelial cells through promotion of
cell proliferation by Erk1/2 activation. Cancer Sci. 99:2120–2127.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liao F, Dong W and Fan L: Apoptosis of
human colorectal carcinoma cells is induced by blocking
hepatoma-derived growth factor. Med Oncol. 27:1219–1226. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Meng J, Xie W, Cao L, Hu C and Zhe Z:
shRNA targeting HDGF suppressed cell growth and invasion of
squamous cell lung cancer. Acta Biochim Biophys Sin (Shanghai).
42:52–57. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamamoto S, Tomita Y, Hoshida Y, Morii E,
Yasuda T, Doki Y, Aozasa K, Uyama H, Nakamura H and Monden M:
Expression level of hepatoma-derived growth factor correlates with
tumor recurrence of esophageal carcinoma. Ann Surg Oncol.
14:2141–2149. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Uyama H, Tomita Y, Nakamura H, Nakamori S,
Zhang B, Hoshida Y, Enomoto H, Okuda Y, Sakon M, Aozasa K, et al:
Hepatoma-derived growth factor is a novel prognostic factor for
patients with pancreatic cancer. Clin Cancer Res. 12:6043–6048.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu TH, Huang CC, Liu LF, Lin PR, Liu SY,
Chang HW, Changchien CS, Lee CM, Chuang JH and Tai MH: Expression
of hepatoma-derived growth factor in hepatocellular carcinoma.
Cancer. 98:1444–1456. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamamoto S, Tomita Y, Hoshida Y, Takiguchi
S, Fujiwara Y, Yasuda T, Doki Y, Yoshida K, Aozasa K, Nakamura H
and Monden M: Expression of hepatoma-derived growth factor is
correlated with lymph node metastasis and prognosis of gastric
carcinoma. Clin Cancer Res. 12:117–122. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Soffietti R, Bertero L, Pinessi L and Rudà
R: Pharmacologic therapies for malignant glioma: A guide for
clinicians. CNS Drugs. 28:1127–1137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu CX, Lin GS, Lin ZX, Zhang JD, Chen L,
Liu SY, Tang WL, Qiu XX and Zhou CF: Peritumoral edema on magnetic
resonance imaging predicts a poor clinical outcome in malignant
glioma. Oncol Lett. 10:2769–2776. 2015.PubMed/NCBI
|
|
20
|
Chen J, Li Y, Yu TS, McKay RM, Burns DK,
Kernie SG and Parada LF: A restricted cell population propagates
glioblastoma growth after chemotherapy. Nature. 488:522–526. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Narron JV, Stoops TD, Barringhaus K,
Matsumura M and Everett AD: Hepatoma-derived growth factor is
expressed after vascular injury in the rat and stimulates smooth
muscle cell migration. Pediatr Res. 59:778–783. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Okuda Y, Nakamura H, Yoshida K, Enomoto H,
Uyama H, Hirotani T, Funamoto M, Ito H, Everett AD, Hada T and
Kawase I: Hepatoma-derived growth factor induces tumorigenesis in
vivo through both direct angiogenic activity and induction of
vascular endothelial growth factor. Cancer Sci. 94:1034–1041. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang A, Long W, Guo Z and Cao BB:
Downregulation of hepatoma-derived growth factor suppresses the
malignant phenotype of U87 human glioma cells. Oncol Rep. 28:62–68.
2012.PubMed/NCBI
|
|
24
|
Enomoto H, Yoshida K, Kishima Y, Kinoshita
T, Yamamoto M, Everett AD, Miyajima A and Nakamura H:
Hepatoma-derived growth factor is highly expressed in developing
liver and promotes fetal hepatocyte proliferation. Hepatology.
36:1519–1527. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oliver JA and Al-Awqati Q: An endothelial
growth factor involved in rat renal development. J Clin Invest.
102:1208–1219. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cilley RE, Zgleszewski SE and Chinoy MR:
Fetal lung development: Airway pressure enhances the expression of
developmental genes. J Pediatr Surg. 35:113–118. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ke Y, Zhao W, Xiong J and Cao R:
Downregulation of miR-16 promotes growth and motility by targeting
HDGF in non-small cell lung cancer cells. J Biomed Res.
587:3153–3157. 2013.
|
|
28
|
Chen X, Yun J, Fei F, Yi J, Tian R, Li S
and Gan X: Prognostic value of nuclear hepatoma-derived growth
factor (HDGF) localization in patients with breast cancer. Pathol
Res Pract. 208:437–443. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hsu SS, Chen CH, Liu GS, Tai MH, Wang JS,
Wu JC, Kung ML, Chan EC and Liu LF: Tumorigenesis and prognostic
role of hepatoma-derived growth factor in human gliomas. J
Neurooncol. 107:101–109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Song Y, Hu Z, Long H, Peng Y, Zhang X, Que
T, Zheng S, Li Z, Wang G, Yi L, et al: A complex mechanism for
HDGF-mediated cell growth, migration, invasion, and TMZ
chemosensitivity in glioma. J Neurooncol. 119:285–295. 2014.
View Article : Google Scholar : PubMed/NCBI
|