Introduction
Colorectal cancer (CRC), as the third commonest
malignant tumor globally, has thrown great threaten to public
health (1,2). Despite many efforts have been made to
improve the efficiency of diagnosis and treatment of CRC, the
prognosis still remains unsatisfied. CRC is a multi-step process
involving the dyregulation of multiple genes; therefore,
investigations on the precise mechanisms underlying the initiation
and progression of CRC are essential.
lncRNAs, a class of RNAs longer than 200 nucleotides
with limited or no protein-coding ability, have attracting more and
more attention of researchers. lncRNAs have been reported to be
involved in a variety of biological processes, including
tumorigenesis through regulating oncogenes or tumor suppressors at
several levels (3–12). For example, Xia et al (13) demonstrated that long noncoding RNA
papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3)
inhibited proliferation and invasion of glioma cells by suppressing
the Wnt/β-catenin signaling pathway. Su et al (14) illustrated that long noncoding RNA
BLACAT1 indicated a poor prognosis of colorectal cancer and
affected cell proliferation by epigenetically silencing p15. Pei
et al (15) reported that
downregulation of lncRNA CASC2 promoted cell proliferation and
metastasis of bladder cancer by activating the Wnt/β-catenin
signaling pathway. And also many lncRNAs, for instance, ROR,
BLACAT1, ANCR, TUG1, are identified to be dysregulated and play
critical role in the initial and progression of CRC (14,16–18).
FAM83H-AS1 has been identified to play critical roles in lung
cancer and breast cancer (19,20).
However, its biological function in CRC has not been thoroughly
studied. Notch signaling has been reported to play important roles
in regulating cell proliferation, differentiation and death
(21–23).
In the present study, we revealed that FAM83H-AS1
was upregulated both in CRC tissues and cells for the first time.
Further experiments demonstrated that silenced FAM83H-AS1 could
suppress cell proliferation and such function was at least
partially mediated by regulating Notch signal pathway.
Materials and methods
Patients and specimens
Human primary CRC tissues and their paired adjacent
tissues were obtained from 40 patients at the Sichuan Cancer
Hospital and Institute, Sichuan Cancer Center, School of Medicine,
University of Electronic Science and Technology of China. None of
these patients had received local or systemic treatment before the
operation. All of the tissues were stored at −80°C. An experienced
pathologist assessed the differentiation grade, pathological stage,
grade and nodal status. The clinical pathological parameters
involved in the analysis included age, sex, tumor differentiation,
distant metastasis, TNM stage and tumor size (defined based on the
diameter of tumor). The tumor differentiation is defined as
following: Well, tumor cell differentiation close to normal cell;
moderately, tumor cell is poorly differentiated but still retain
some vestiges of originally tissues; and poor, tumor cell is very
poorly differentiated with no vestiges of originally tissues. The
TNM classification is defined as fowling: T is defined as primary
tumor (Tx: The primary tumor is not determined;
T0: No original tumor evidence; Tis: carcinoma in
situ; T1: Tumor invading tunica submucosa;
T2: Tumor invading intestinal wall muscularis;
T3: Tumor invading serosa or primary tumor located at
the colon or rectum without serosa or tumor located at colon or
rectum adjacent tissues; T4: Primary tumor invading
through the peritoneal or invading other ogans). N is defined as
regional lymph nodes (Nx: Regional lymph node is not
determined; N0: No regional lymph node metastasis;
N1: 1–3 regional lymph nodes metastasis; N2:
4 or more regional lymph nodes metastasis). M is defined as distant
metastasis (M0: No metastasis; M1:
metastasis). TNM stage includes I–IV. All the written informed
consents were obtained from patients. The study protocol was
approved by the Ethics Committee of the Sichuan Cancer Hospital and
Institute, Sichuan Cancer Center, School of Medicine, University of
Electronic Science and Technology of China.
Cell culture
All human colonic cancer cell lines including SW480,
LoVo, HCT116, HT29 and the human colonic epithelial cells HCoEpiC
were obtained from the American Type Culture Collection (ATCC;
Manassas, VA, USA). Cells were cultured in RPMI-1640 supplemented
with 10% fetal bovine serum at 37°C in a incubator containing 5% of
CO2.
Cell transfection
CRC cells were placed with a desired cell number and
24 h later, target shRNA and non-targeting controls (shRNA;
Dharmacon, Inc., Lafayette, CO, USA) were transfected with a final
concentration of 10 nM. Lipofectamine RNAiMAX reagent and OptiMEM
medium were used for the cell interference assays on the basis of
the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA).
SMART pool of FAM83H-AS1 shRNA (Dharmacon, Inc.) was used in this
study to form silenced FAM83H-AS1 (sh-FAM83H-AS1). The interference
efficiency was examined by reverse transcription-quantitative
polymerase chain reaction (RT-qPCR). Cell lines stably interfering
FAM83H-AS1 were transfected with the plasmid sh-FAM83H-AS1, and
screened with G418 (10 mg/ml) for two months.
RT-qPCR
Total RNA was severally extracted from tumor tissues
and cell lines by using a Trizol kit (Invitrogen). cDNA was
subsequently synthesized from total RNA by using an Omniscript RT
kit (Qiagen, Valencia, CA, USA) under the manufacturer's
instructions. RT-PCR reaction was conducted on the Mastercycler ep
realplex (Eppendorf 2S; Eppendorf, Hamburg, Germany). A
25-µl-reaction mixture contained 1 µl of cDNA from samples, 12.5 µl
of 2X Fast EvaGreen™ qPCR Master Mix, 1 µl primers (10 mM), and
10.5 µl of RNase/DNase-free water. The Ct value was defined as the
cycle number at which the fluorescence intensity reached a certain
threshold where amplification of each target gene was within the
linear region of the reaction amplification curves. Glyceraldheyde
3-phosphate dehydrogenase (GAPDH) was taken as internal controls.
Relative mRNA expression of FAM83H-AS1 was calculated by using the
2−ΔΔCt method. The primers for FAM83H-AS1 were: forward,
5′-TAGGAAACGAGCGAGCCC-3′ and reverse, 5′-GCTTTGGGTCTCCCCTTCTT-3′.
The primers for GAPDH were: forward, 5′-GGGAGCCAAAAGGGTCAT-3′ and
reverse, 5′-GAGTCCTTCCACGATACCAA-3′.
Cell viability
Cell viability was assessed via
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-trtrazolium bromide (MTT)
assay. Cells (5×103 cells/well) transfected with
indicated vector were seeded in a 96-well flat-bottomed plate for
24 h and cultured in the normal medium. At 0, 24, 48, 72 and 96 h
after transfection, the MTT solution (5 mg/ml, 20 µl) was added to
each well. Following incubation for 4 h, the media was removed and
100 µl DMSO was added to each well. The relative number of
surviving cells was assessed by measuring the optical density (OD)
of cell lysates at 560 nm. All assays were performed in
triplicate.
5-ethynyl-2′-deoxyuridine (EdU)
proliferation assay
The EdU proliferation assay was conducted using
Cell-Light EdU Apollo 567 In Vitro Imaging kit (RiboBio,
Guangzhou, China) under the introduction of the manufracture's
instructions. Briefly, approximately 5×103 cells/wells
with indicated treatments were seeded into 96-well plates. After 24
h, 100 µl medium with 50 µM EdU was added into each well and
incubated for 2 h under 37°C. Afterwards, cells were fixed with 4%
paraformaldehyde, then stained with Hoechst 33342 and Apollo
reaction cocktail. Images were taken by applying fluorescence
microscopy (Nikon Corporation, Tokyo, Japan) and then merged by
Photoshop 6.0 software. EdU-positive cells and total cells were
counted within each field.
Colony formation assay
Cells (500 cells/well) transfected with indicated
vector were plated in 6-well plates and incubated at 37°C. Two
weeks later, the cells were fixed and stained with 0.1% of crystal
violet. The number of visible colonies was counted manually.
Western bolt analysis and
antibodies
Total protein lysates were separated in 10% of
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE), and were electrophoretically transferred to
polyvinylidene difluoride membranes (Roche Diagnostics GmbH,
Mannheim, Germany). Protein loading was estimated by using mouse
anti-GAPDH monoclonal antibody. The membranes were blotted with 10%
of non-fat milk in TBST for 2 h at room temperature, washed and
then probed with the rabbit anti-Notch1 (1:2,000 dilution), Hes1
(1:2,000 dilution), and GAPDH (1:3,000 dilution), overnight (8 h)
at 4°C, followed by treatment with secondary antibody conjugated to
horseradish peroxidase for 2 h at room temperature. The proteins
were detected by using an enhanced chemiluminescence system and
exposed to X-ray film. All antibodies were purchased from Abcam
(Cambridge, MA, USA).
Statistical analysis
Data were shown as the means ± standard error of at
least three independent experiments. The SPSS 17.0 software (SPSS,
Inc., Chicago, IL, USA) was used for statistical analysis. Two
group comparisons were performed with a Student's t-test. Multiple
group comparisons were analyzed with one-way ANOVA. Statistically
significant positive correlation between the expression level of
FAM83H-AS1 and that of Notch1 or Hes1 in CRC tissues was analyzed
by Spearman's correlation analysis. The Pearson χ2 test
was used to evaluate the relationship between the expression of
FAM83H-AS1 and clinical features. Kaplan-Meier method was used to
compare the overall survival curves between high-FAM83H-AS1 and
low-FAM83H-AS1 expression groups via the log-rank test. P<0.05
was considered to indicate a statistically significant
difference.
Results
FAM83H-AS1 and Notch1 were upregulated
in CRC tissues and cell lines
To explore the biological function of FAM83H-AS1 and
its relationship with Notch signaling in CRC, we first measured the
levels of FAM83H-AS1, Notch1 and Hes1 in the CRC tissues and
corresponding normal tissues by RT-qPCR. As shown in Fig. 1A, compared with corresponding normal
tissues, the levels of FAM83H-AS1, Notch1 and Hes1 were
significantly increased in CRC tissues. And the FAM83H-AS1 showed
significantly positive correlation with Notch1 and Hes1, analyzed
by Spearman's correlation analysis (Fig.
1B). Then, the levels of FAM83H-AS1, Notch1 and Hes1 were also
determined in four human colonic cancer cell lines including SW480,
LoVo, HCT116, HT29 and a human colonic epithelial cell HCoEpiC. As
presented in Fig. 1C, the expression
levels of the three molecules were obviously increased in all the
four CRC cell lines in comparison to the human colonic epithelial
cell. The consistent expression levels of FAM83H-AS1, Notch1 and
Hes1 both in CRC tissues and cell lines indicated that FAM83H-AS1
might be involved in the progression of CRC and implied the
association between FAM83H-AS1 and Notch signal pathway.
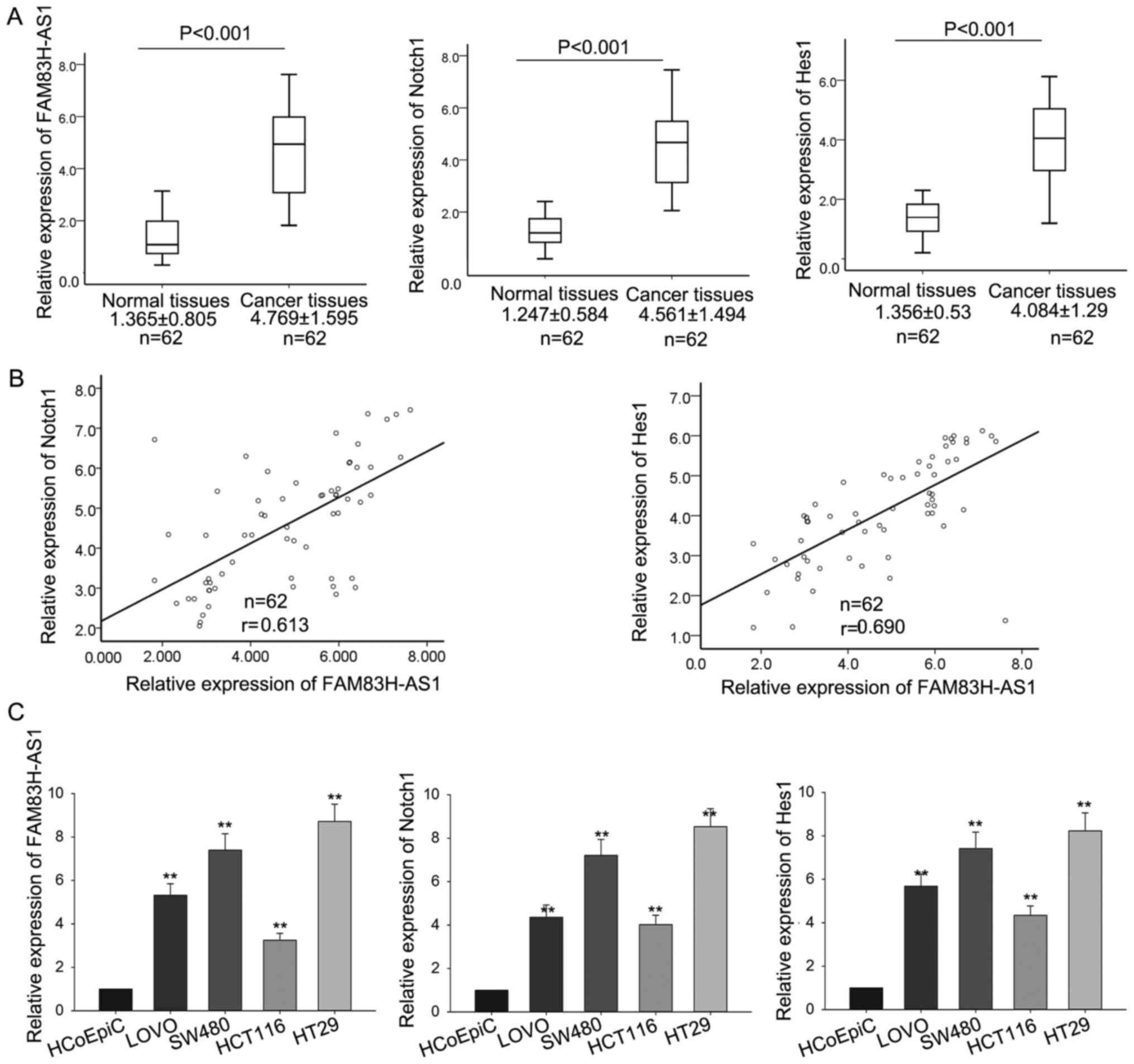 | Figure 1.FAM83H-AS1 and Notch1 were upregulated
in CRC tissues and cell lines. (A) The levels of FAM83H-AS1, Notch1
and Hes1 in CRC tissues and the corresponding normal tissues were
measured by RT-qPCR. (B) The correlation of FAM83H-AS1 with Notch1
and Hes1 was analyzed by Spearman's correlation analysis. (C) The
levels of FAM83H-AS1, Notch1 and Hes1 were also determined by
RT-qPCR in four human colonic cancer cell lines including SW480,
LοVο, HCT116, HT29, and the human colonic epithelial cell HCoEpiC
(control). Data are presented as the mean ± standard deviation of
at least three independent experiments. **P<0.01 vs. control
group. FAM83H-AS1, FAM83H antisense RNA 1 (head to head); CRC,
colorectal carcinoma; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; Hes1, Hes family basic-helix-loop-helix
transcription factor 1. |
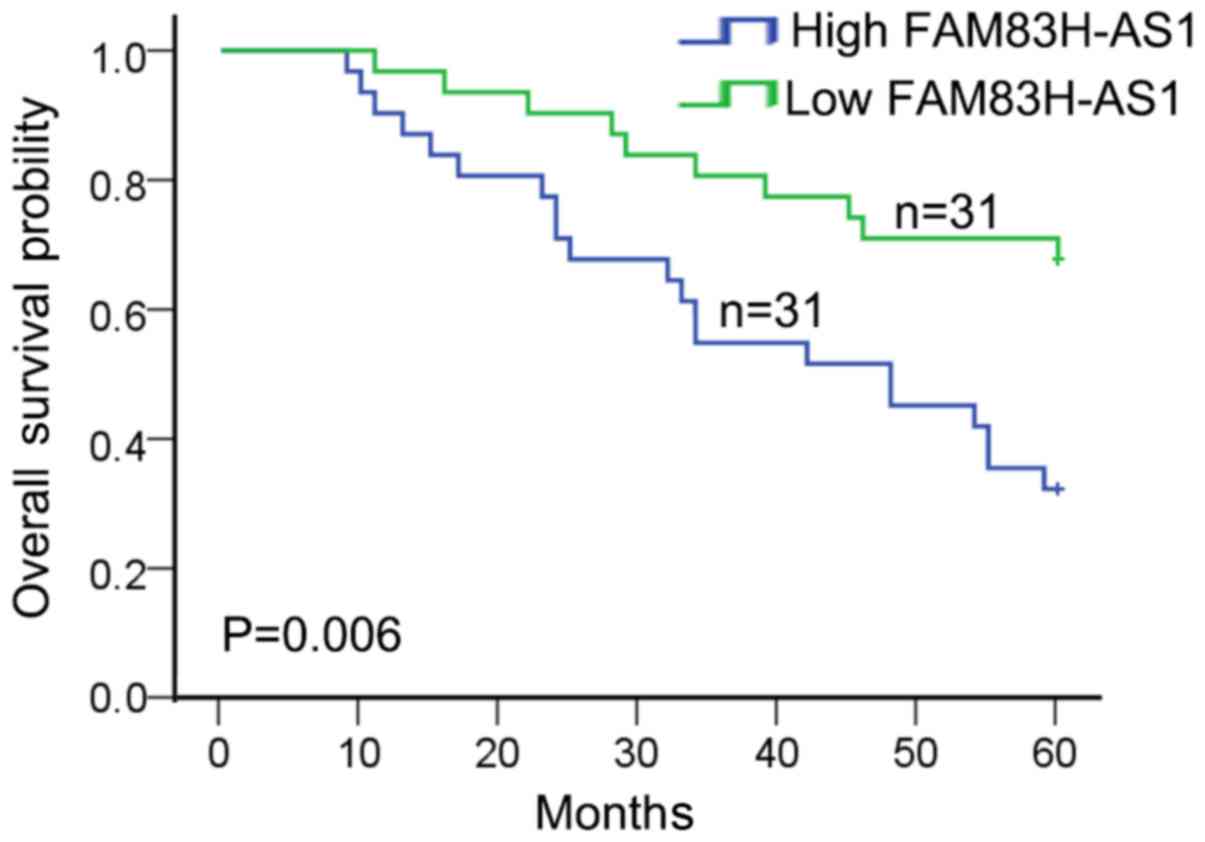
High level of FAM83H-AS1 was
associated with clinical features and poor prognosis
To detect the clinical significance of FAM83H-AS1
expression in CRC, 62 patients were divided into highly-expressed
FAM83H-AS1 group (n=31) and low-expressed FAM83H-AS1 group (n=31)
according to the cut-off value, which was defined as the median of
the cohort. As revealed in Table I,
high expression of FAM83H-AS1 in CRC patients was significantly
correlated with advanced tumor stage (P=0.004) and large tumor size
(P=0.002). In addition, patients with high expression of FAM83H-AS1
were associated with worse overall survival in CRC patients
(P=0.006; Fig. 2). These data
indicated that FAM83H-AS1 might act as a potent biomarker for
predicting prognosis in CRC patients.
 | Table I.Associations between FAM83H antisense
RNA 1 (Head to Head) expression and clinical features. |
Table I.
Associations between FAM83H antisense
RNA 1 (Head to Head) expression and clinical features.
|
| FAM83H-AS1
expression |
|
|---|
|
|
|
|
|---|
| Variable | Low (n) | High (n) | P-value |
|---|
| Sex |
|
| 0.310 |
| Male | 18 | 13 |
|
|
Female | 13 | 18 |
|
| Age (years) |
|
| 0.124 |
|
<60 | 10 | 17 |
|
| ≥60 | 21 | 14 |
|
| Tumor
differentiation |
|
| 0.309 |
|
Well/moderately | 12 | 17 |
|
|
Poorly | 19 | 14 |
|
| Distant
metastasis |
|
| 0.202 |
| No | 14 | 20 |
|
| Yes | 17 | 11 |
|
| TNM stages |
|
| 0.004a |
| I–II | 24 | 12 |
|
|
III–IV | 7 | 19 |
|
| Tumor size |
|
| 0.002a |
| <5
cm | 24 | 11 |
|
| ≥5
cm | 7 | 20 |
|
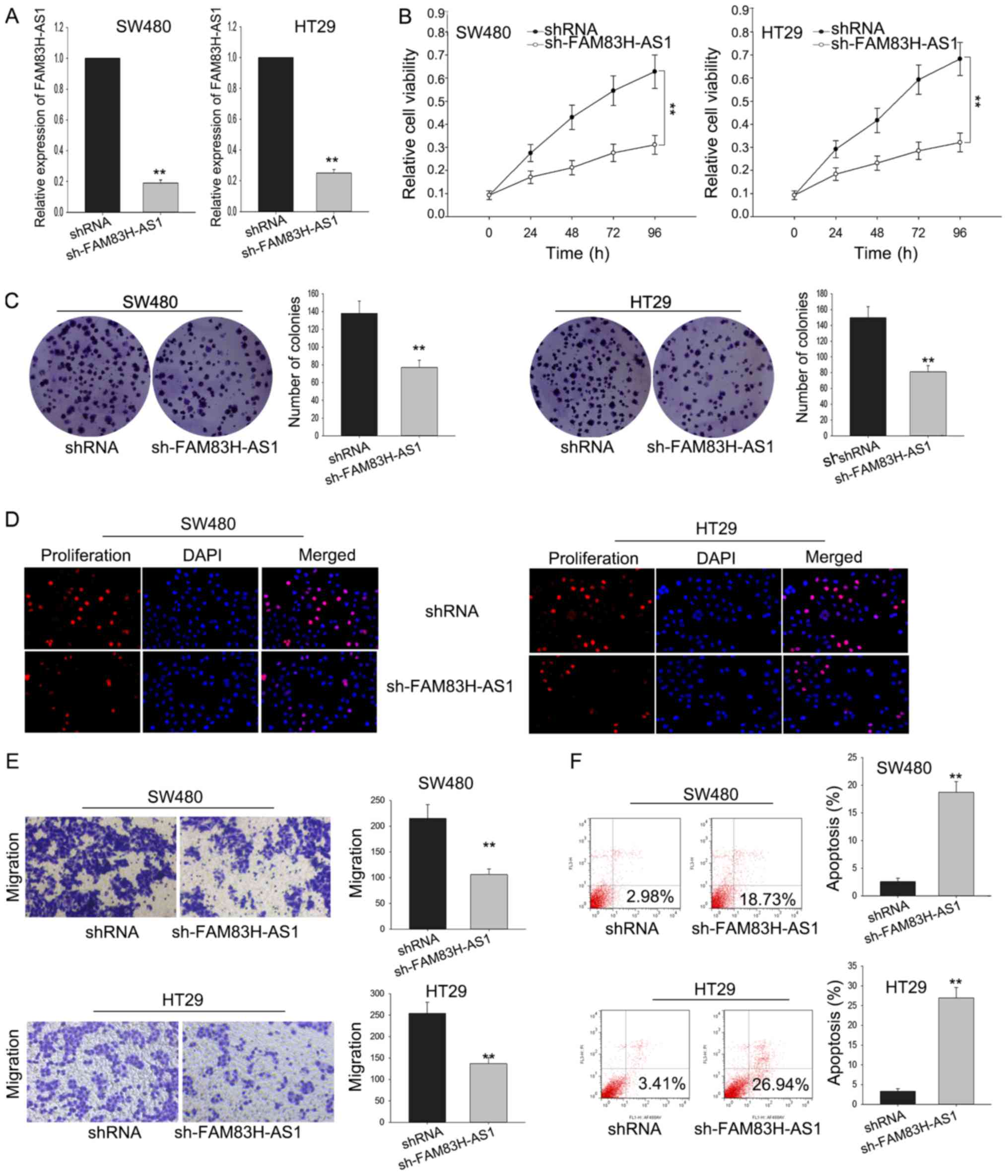
Silenced FAM83H-AS1 inhibited cell
proliferation and migration via affecting cell apoptosis
To investigate the function of FAM83H-AS1 on the
cell proliferation of CRC cells, SW480 and HT29 cells were
transfected with sh-FAM83H-AS1. Satisfied transfection efficiency
was obtained after 48 h (Fig. 3A).
MTT assay was performed to measure the viability of SW480 and HT29
cells transfected with sh-FAM83H-AS1, and results illustrated that
silenced FAM83H-AS1 could significantly inhibit the cell
proliferation ability (Fig. 3B). The
result of colony formation assays indicated that colony formation
was remarkably inhibited by FAM83H-AS1 knockdown (Fig. 3C). Besides, the inhibited cell
proliferation by sh-FAM83H-AS1 was further affirmed by the data
from EdU assay (Fig. 3D).
Furthermore, transwell assay was designed to detect how silenced
FAM83H-AS1 affected cell migration. As diagramed in Fig. 3E, the knockdown of FAM83H-AS1
remarkably impaired migratory cells in comparison with the negative
control. Meanwhile, the silence of FAM83H-AS1 also augmented the
apoptotic cells compared to the negative control (Fig. 3F). These data indicated that knockdown
FAM83H-AS1 suppressed the proliferative and migratory ability of
CRC cells through influencing cell apoptosis.
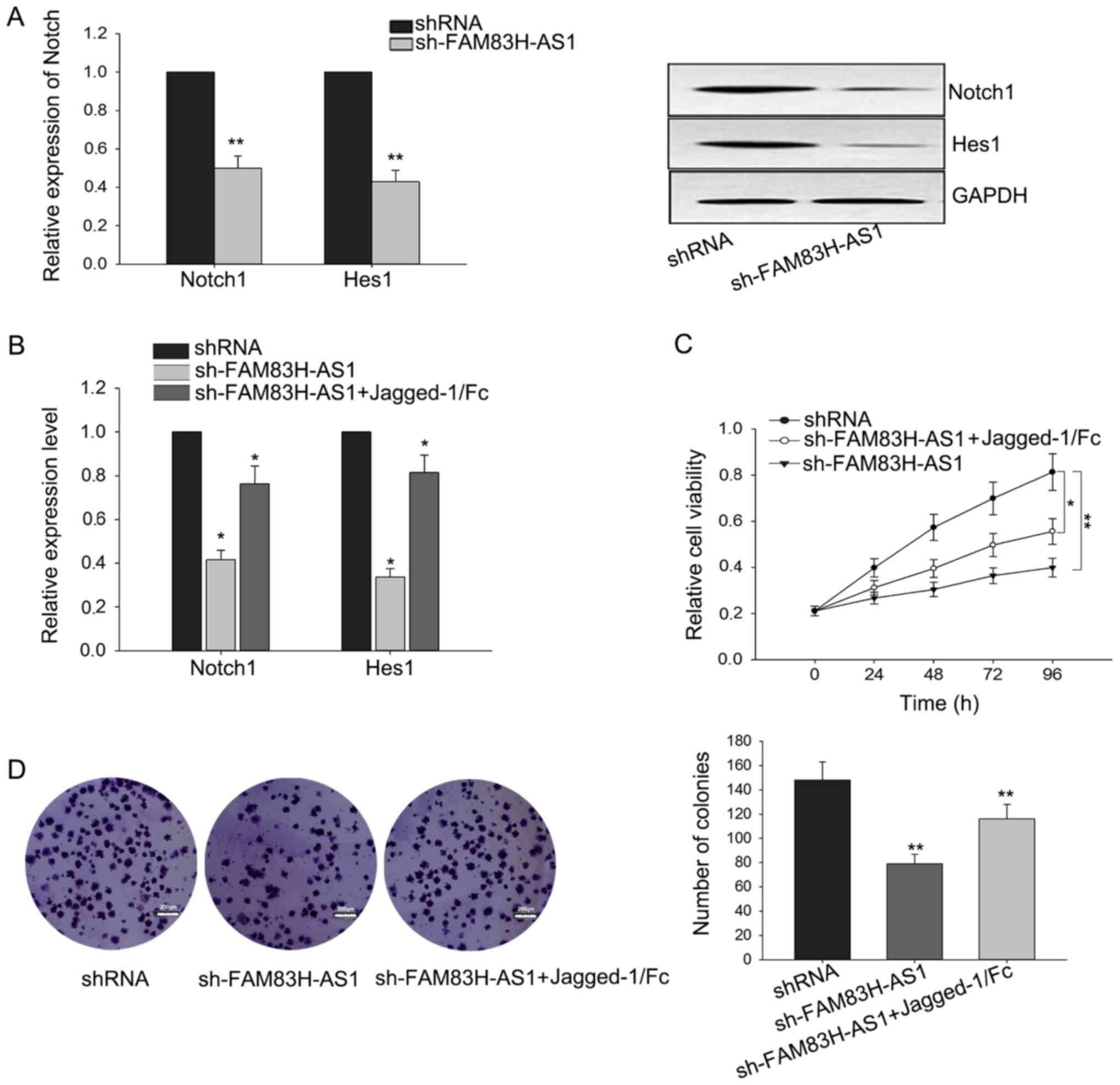
The growth-inhibition in CRC cells
mediated by the knockdown of FAM83H-AS1 could be reversed by Notch1
activator
It has been implicated that Notch signal pathway was
involved in regulating cell proliferation (24,25). Due
to the correlation of FAM83H-AS1 with Notch1 and its downstream
gene Hes1, we hypothesized that the function of FAM83H-AS1 in CRC
might be mediated by Notch signal pathway. To confirm this
hypothesis, we measured the level of Notch in response to the
knockdown of FAM83H-AS1. As illustrated in Fig. 4A, silenced FAM83H-AS1 could suppress
the expressions of Notch1 and Hest1 both in mRNA and protein
levels. To make further confirmation, Jagged-1/Fc, one Notch1
activator, was added to activate the Notch signal pathway. After
Jagged-1/Fc was added, the weakened expressions of Notch1 ans Hes1
triggered by the silence of FAM83H-AS1 were recovered a little
(Fig. 4B). Additionally, as presented
in Fig. 4C and D, results from MTT
and colony formation revealed that the growth-inhibition effect
mediated by silenced FAM83H-AS1 could be reversed by the addition
of Jagged-1/Fc. These findings revealed that the function of
FAM83H-AS1 exerted in CRC was at least partially mediated by Notch
signal pathway.
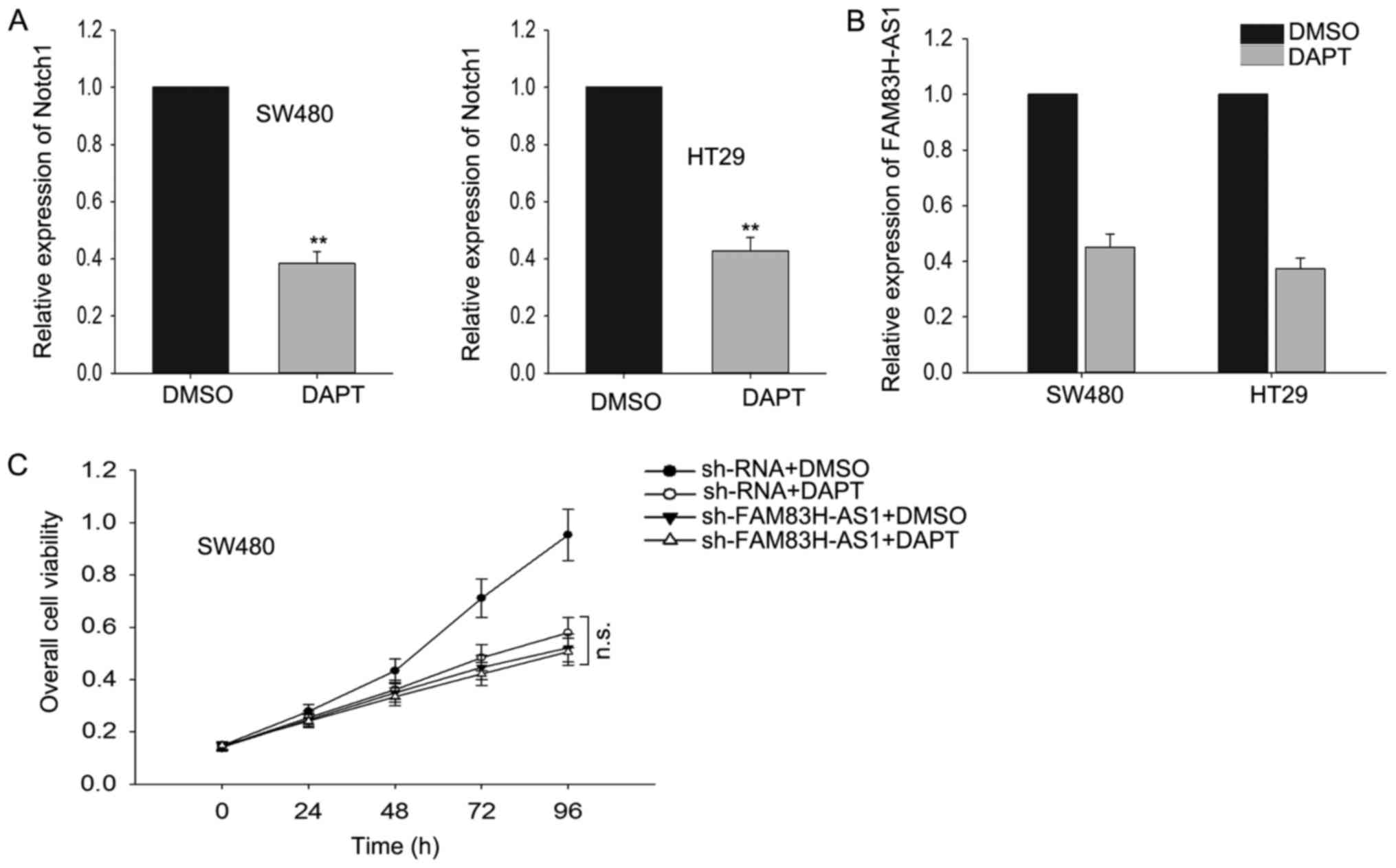
The effect of FAM83H-AS1 on CRC cells
was dependent on Notch pathway
To determine whether Notch signal pathway was
essential for FAM83H-AS, we applied
N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl
ester (DAPT), a widely used γ-secretase inhibitor (GSI) which could
effectively repress the expression of receptors and ligands in
Notch signaling pathway, to inhibit Notch-1 expression in our study
and then we measured the effect of FAM83H-AS1 knockdown on cell
proliferation. As presented in Fig.
5A, after cells were treated with DAPT, the level of Notch-1
was significantly reduced. Subsequently, we observed how the
expression level of FAM83H-AS1 responded to the silenced Notch1 by
DAPT, and discovered that silenced Notch1 resulted in the knockdown
of FAM83H-AS1 (Fig. 5B). Then we
performed MTT assays and found that after Notch-1 was suppressed,
the effect induced by the knockdown of FAM83H-AS1 was vanished
(Fig. 5C). Our results indicated that
FAM83H-AS1 worked through the Notch pathway.
Discussion
lncRNAs, a class of RNAs longer than 200 nucleotides
with limited or no protein-coding ability, have attracting more and
more attention of researchers. Currently, accumulating lncRNAs have
been identified in different biological processes, in which they
act as critical regulators in both physiology and pathology,
including tumorigenesis. Idogawa et al (26) reported that lncRNA NEAT1 was a
transcriptional target of p53 and modulated p53-induced
transactivation and tumor-suppressor function. Zhao et al
(27) revealed that SNHG5/miR-32 axis
regulated cell proliferation and migration of gastric cancer by
targeting KLF4. Malakar et al (28) demonstrated that long noncoding RNA
MALAT1 promoted hepatocellular carcinoma development by
upregulating SRSF1 and activating mTOR. As for CRC, plenty
well-studied lncRNAs have been reported to be dysregulated in CRC
tissues and participate in CRC progression via multiple mechanisms
(29–31). FAM83H-AS1 has been identified to play
critical roles in lung cancer and breast cancer (19,20).
However, its biological function in CRC has not been studied.
In our present study, we demonstrated for the first
time that lncRNA FAM83H-AS1 was upregulated in CRC tissues and cell
lines. Further functional studies revealed that silenced FAM83H-AS1
could inhibit cell proliferation and migration through inducing
more apoptosis. Furthermore, concomitant upregulation of two Notch
signaling molecules, Notch1 and Hes1, was obtained in both CRC
tissues and cell lines, indicating the potential mechanism of
FAM83H-AS1/Notch signal pathway in CRC. Notch signaling pathway has
been reported to involve in the tumorigenesis of various cancer
types (23,32,33). In
our study, we showed the accompanied activation of Notch signaling
in CRC tissues and cells and rescue assays revealed that the growth
inhibition mediated by the knockdown of FAM83H-AS1 was at least in
a Notch signal dependent manner, which was further confirmed by the
effect of DAPT on Notch1 and FAM83H-AS1 as well as the unchanged
cell proliferation after Notch1 was silenced by DAPT. All in all,
despite the fact that we have made progress in studying the
initiation and development of CRC to some extent, our research just
takes a little part in the mechanisms of the tumorigenesis of CRC.
Therefore, this paper has not been involved in the study about the
fact that cadmium prolonged exposure involved malignant progression
of A549 cells. And as for the situation above, we will put much
more attention in the future study upon the research that whether
the hot spicy eating habit of people in Sichuan is related to the
tumorigenesis of CRC.
In conclusion, FAM83H-AS1 was demonstrated to be
upregulated in both CRC tissues and cell lines, wherein acting as
an oncogene via activating Notch signaling. Due to the complicated
cross talk of Notch signaling with various tumorigenesis
regulators, still many studies need to be made to further
investigate the Notch1-mediated mechanism under FAM83H-AS1
dysfunction, for exploring the potential therapeutic value of
FAM83H-AS1 in treating CRC.
References
|
1
|
Booth RA: Minimally invasive biomarkers
for detection and staging of colorectal cancer. Cancer Lett.
249:87–96. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu M, Liu J, Xiao J, Yang L, Cai M, Shen
H, Chen X, Ma Y, Hu S, Wang Z, et al: Lnc-mg is a long non-coding
RNA that promotes myogenesis. Nat Commun. 8:147182017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou X, Yuan P, Liu Q and Liu Z: LncRNA
MEG3 regulates imatinib resistance in chronic myeloid leukemia via
suppressing microRNA-21. Biomol Ther (Seoul). 25:490–496. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou C, Huang C, Wang J, Huang H, Li J,
Xie Q, Liu Y, Zhu J, Li Y, Zhang D, et al: LncRNA MEG3
downregulation mediated by DNMT3b contributes to nickel malignant
transformation of human bronchial epithelial cells via modulating
PHLPP1 transcription and HIF-1α translation. Oncogene.
36:3878–3889. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang H, Xiong Y, Xia R, Wei C, Shi X and
Nie F: The pseudogene-derived long noncoding RNA SFTA1P is
down-regulated and suppresses cell migration and invasion in lung
adenocarcinoma. Tumour Biol. 39:10104283176914182017.PubMed/NCBI
|
|
7
|
Zhang D, Sun G, Zhang H, Tian J and Li Y:
Long non-coding RNA ANRIL indicates a poor prognosis of cervical
cancer and promotes carcinogenesis via PI3K/Akt pathways. Biomed
Pharmacother. 85:511–516. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang CZ: Long non-coding RNA FTH1P3
facilitates oral squamous cell carcinoma progression by acting as a
molecular sponge of miR-224-5p to modulate fizzled 5 expression.
Gene. 607:47–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang CL, Zhu KP and Ma XL: Antisense
lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by
increasing the expression of FOXC2. Cancer Lett. 396:66–75. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu H, Xue Y, Wang P, Liu X, Ma J, Zheng J,
Li Z, Li Z, Cai H and Liu Y: Knockdown of long non-coding RNA XIST
increases blood-tumor barrier permeability and inhibits glioma
angiogenesis by targeting miR-137. Oncogenesis. 6:e3032017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang C, Li Z, Li Y, Xu R, Wang Y, Tian Y
and Chen W: Long non-coding RNA NEAT1 overexpression is associated
with poor prognosis in cancer patients: A systematic review and
meta-analysis. Oncotarget. 8:2672–2680. 2017.PubMed/NCBI
|
|
12
|
Xu J, Zhang R and Zhao J: The novel long
noncoding RNA TUSC7 inhibits proliferation by sponging miR-211 in
colorectal cancer. Cell Physiol Biochem. 41:635–644. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xia S, Ji R and Zhan W: Long noncoding RNA
papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3)
inhibits proliferation and invasion of glioma cells by suppressing
the Wnt/β-catenin signaling pathway. BMC Neurol. 17:302017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Su J, Zhang E, Han L, Yin D, Liu Z, He X,
Zhang Y, Lin F, Lin Q, Mao P, et al: Long noncoding RNA BLACAT1
indicates a poor prognosis of colorectal cancer and affects cell
proliferation by epigenetically silencing of p15. Cell Death Dis.
8:e26652017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pei Z, Du X, Song Y, Fan L, Li F, Gao Y,
Wu R, Chen Y, Li W, Zhou H, et al: Down-regulation of lncRNA CASC2
promotes cell proliferation and metastasis of bladder cancer by
activation of the Wnt/b-catenin signaling pathway. Oncotarget.
8:18145–18153. 2017.PubMed/NCBI
|
|
16
|
Li H, Jiang X and Niu X: Long non-coding
RNA reprogramming (ROR) promotes cell proliferation in colorectal
cancer via affecting p53. Med Sci Monit. 23:919–928. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang ZY, Yang F, Zhang YL, Liu B, Wang M,
Hong X, Yu Y, Zhou YH and Zeng H: LncRNA-ANCR down-regulation
suppresses invasion and migration of colorectal cancer cells by
regulating EZH2 expression. Cancer Biomark. 18:95–104. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang L, Zhao Z, Feng W, Ye Z, Dai W, Zhang
C, Peng J and Wu K: Long non-coding RNA TUG1 promotes colorectal
cancer metastasis via EMT pathway. Oncotarget. 7:51713–51719. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang J, Feng S, Su W, Bai S, Xiao L, Wang
L, Thomas DG, Lin J, Reddy RM, Carrott PW, et al: Overexpression of
FAM83H-AS1 indicates poor patient survival and knockdown impairs
cell proliferation and invasion via MET/EGFR signaling in lung
cancer. Sci Rep. 7:428192017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang F, Lv SX, Lv L, Liu YH, Dong SY, Yao
ZH, Dai XX, Zhang XH and Wang OC: Identification of lncRNA
FAM83H-AS1 as a novel prognostic marker in luminal subtype breast
cancer. Onco Targets Ther. 9:7039–7045. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiao W, Gao Z, Duan Y, Yuan W and Ke Y:
Notch signaling plays a crucial role in cancer stem-like cells
maintaining stemness and mediating chemotaxis in renal cell
carcinoma. J Exp Clin Cancer Res. 36:412017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Teoh SL and Das S: Notch signalling
pathways and their importance in the treatment of cancers. Curr
Drug Targets. Mar 9–2017.(Epub ahead of print).
|
|
23
|
Gao J, Long B and Wang Z: Role of Notch
signaling pathway in pancreatic cancer. Am J Cancer Res. 7:173–186.
2017.PubMed/NCBI
|
|
24
|
Gotte M, Greve B, Kelsch R, Müller-Uthoff
H, Weiss K, Masouleh B Kharabi, Sibrowski W, Kiesel L and Buchweitz
O: The adult stem cell marker Musashi-1 modulates endometrial
carcinoma cell cycle progression and apoptosis via Notch-1 and
p21WAF1/CIP1. Int J Cancer. 129:2042–2049. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo Q, Qian Z, Yan D, Li L and Huang L:
LncRNA-MEG3 inhibits cell proliferation of endometrial carcinoma by
repressing Notch signaling. Biomed Pharmacother. 82:589–594. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Idogawa M, Ohashi T, Sasaki Y, Nakase H
and Tokino T: Long non-coding RNA NEAT1 is a transcriptional target
of p53 and modulates p53-induced transactivation and
tumor-suppressor function. Int J Cancer. 140:2785–2791. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao L, Han T, Li Y, Sun J, Zhang S, Liu
Y, Shan B, Zheng D and Shi J: The lncRNA SNHG5/miR-32 axis
regulates gastric cancer cell proliferation and migration by
targeting KLF4. FASEB J. 31:893–903. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Malakar P, Shilo A, Mogilavsky A, Stein I,
Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV
and Karni R: Long noncoding RNA MALAT1 promotes hepatocellular
carcinoma development by SRSF1 upregulation and mTOR activation.
Cancer Res. 77:1155–1167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Peng W, Wang Z and Fan H: LncRNA NEAT1
impacts cell proliferation and apoptosis of colorectal cancer via
regulation of Akt signaling. Pathol Oncol Res. 23:651–656. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ye Z, Zhou M, Tian B, Wu B and Li J:
Expression of IncRNA-CCAT1, E-cadherin and N-cadherin in colorectal
cancer and its clinical significance. Int J Clin Exp Med.
8:3707–3715. 2015.PubMed/NCBI
|
|
31
|
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang
XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al: Human colorectal
cancer-specific CCAT1-L lncRNA regulates long-range chromatin
interactions at the MYC locus. Cell Res. 24:513–531. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fujiki K, Inamura H, Miyayama T and
Matsuoka M: Involvement of Notch1 signaling in malignant
progression of A549 cells subjected to prolonged cadmium exposure.
J Biol Chem. 292:7942–7953. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Badenes M, Trindade A, Pissarra H,
Lopes-da-Costa L and Duarte A: Erratum to: Delta-like 4/Notch
signaling promotes ApcMin/+ tumor initiation through angiogenic and
non-angiogenic related mechanisms. BMC Cancer. 17:2052017.
View Article : Google Scholar : PubMed/NCBI
|