Introduction
Immune checkpoint inhibitors have recently attracted
attention as an innovative cancer therapy (1). Programmed death 1 (PD-1)/programmed
death ligand-1 (PD-L1) checkpoint inhibitors have been shown to
have a continuous clinical effect and low toxicity in some
responder patients with several types of recurrent and metastatic
diseases (2). Among these, the anti
PD-1 monoclonal antibody nivolumab has led to a good clinical
response in several cancer patients with lung cancer, melanoma, and
renal cell carcinoma (3–6). However, many cancer patients do not gain
sufficient benefits even with the anti-PD-1 antibody treatment.
Some researchers have proposed that the level of PD-L1 expression
and the DNA mismatch-repair status in a tumor are biomarker
candidates for predicting the sensitivity to immune checkpoint
blockade (3,7). However, invasive tumor sampling is
required to determine the protein expression and DNA status in
tumor cells. Therefore, further research is needed globally to
identify a new blood biomarker that does not require invasive
sampling.
In cancer patients, circulating tumor cells (CTCs)
in peripheral blood have been identified as a reliable blood tumor
marker (8,9). The evaluation of CTCs generally includes
an enrichment step and detection processes depending on the CTC
characteristics, such as tumor size, density, and cell surface
antigen expression, which are conducted using a cytometric-based or
polymerase chain reaction (PCR)-based method (10). Nagrath et al (11), were the first to report on a new
microfluid device known as a CTC chip that was coated with
epithelial cell adhesion molecule (EpCAM) antibody, which could be
used to collect CTC-expressing EpCAM from whole blood samples of
cancer patients. Because CTCs are known to be larger than nearly
all normal blood cells (12,13), the use of only the large cells in
blood samples as PCR templates will enable CTCs to be detected more
easily using a highly sensitive PCR-based method against the CTC
markers carcinoembryonic antigen (CEA), human Telomerase
Reverse Transcriptase (hTERT), cytokeratin 19 (CK19),
and PD-L1 (9,14–16).
Therefore, in the present study, we used a modified CTC chip that
was based on a continuous particle separation method (17) to enrich CTCs according to cell
size.
The purpose of the present study was to utilize this
new CTC chip to collect the large cell fraction from whole blood
samples and to find a blood sensitivity marker for nivolumab in
advanced, pre-treatment lung cancer patients. To do this, we first
examined the sorting efficacy of the new CTC chip using the lung
cancer cell line PC-9 and then evaluated the mRNA expression of the
CTC markers CEA, hTERT, CK19, and PD-L1
in the large cell fraction of clinical lung cancer patients' blood
samples collected by this chip.
Materials and methods
Preparation of a polymeric CTC chip
for size sorting
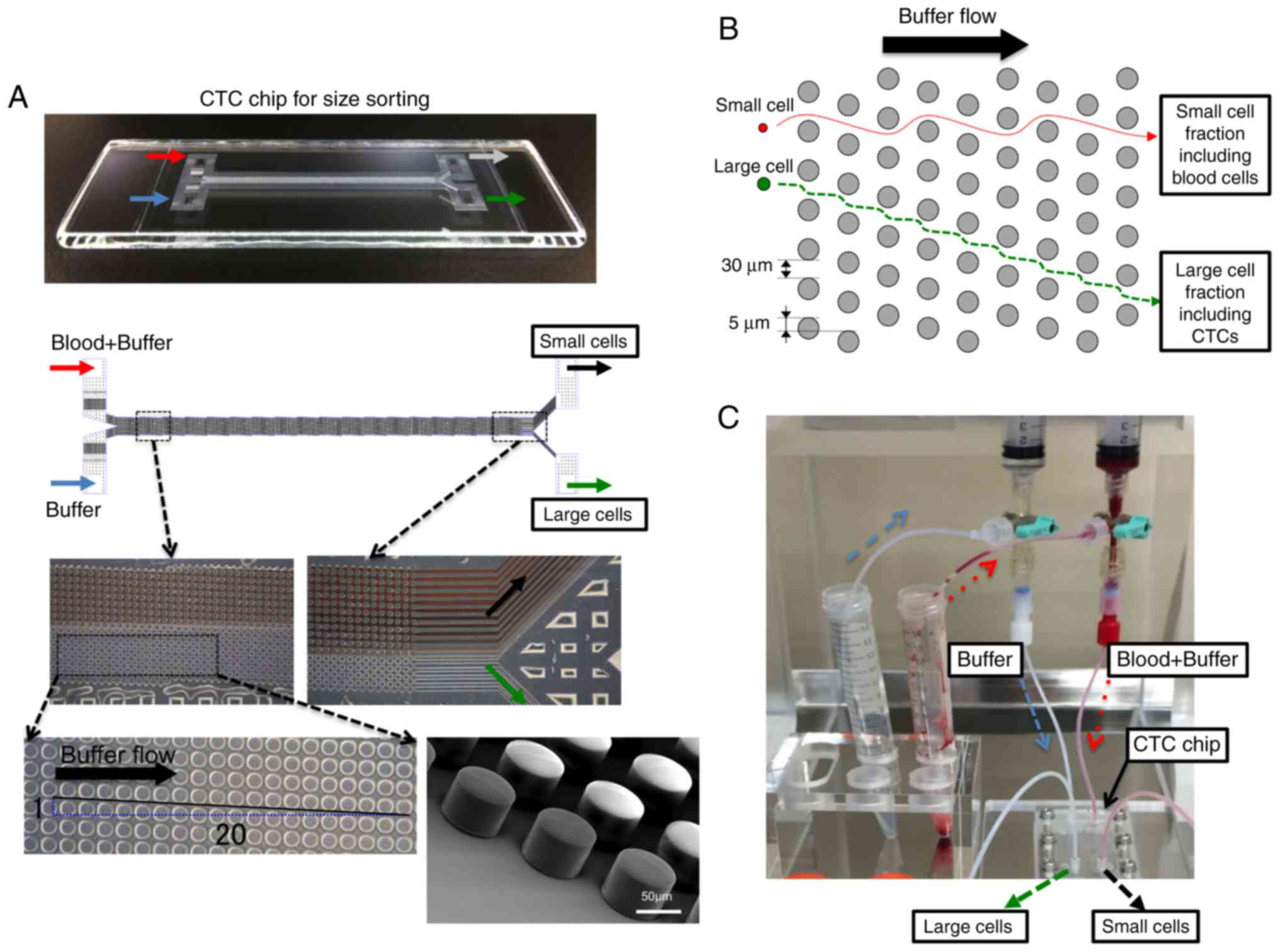
A polymeric CTC chip for size sorting was produced
as previously described (18). The
chip was set in a holder to enable liquid samples to flow from two
inlets to two outlets. Cell suspension samples and mere buffer were
sent from each inlet tube into the CTC chip at 0.2 ml/min using a
syringe pump (Fig. 1). Two outlets
were used to allow the large cell fraction including CTCs to be
enriched and blood cells in the small cell fraction to be collected
separately. Since the diagonally-arranged microposts cannot
influence the flow of small cells, these are carried into the small
cell fraction simply as a result of the buffer flow. By contrast,
large cells, which include CTCs, are sorted by the microposts based
on their size (Fig. 1B and C). Blood
samples from advanced lung cancer patients were obtained before
nivolumab treatment and collected into ethylenediaminetetraacetic
acid (EDTA) blood collection tubes. A 1-ml aliquot of these blood
samples was then diluted two times with phosphate-buffered saline
(PBS) before size sorting using the CTC chip. Once the large cell
fraction had been collected, it was used for further analysis.
Clinical samples and cell lines
This study was prospectively conducted on 15
patients at Gunma University Hospital, Japan, who had advanced lung
cancer and were candidates for nivolumab treatment (Table I). The inclusion criteria for this
study were as follows: pathologically proven lung cancer; recurrent
lung cancer candidate for nivolumab as a result of progressive
disease (PD) following chemotherapy; Eastern Cooperative Oncology
Group (ECOG) performance status of 0–2,
2-[18F]-fluoro-2-deoxy-D-glucose (18F-FDG)
positron emission tomography/computed tomography (PET/CT) scheduled
before and after the first cycle of nivolumab therapy; no evidence
of concurrent cancer; no uncontrolled diabetes mellitus; no
interstitial pneumonia or pulmonary fibrosis; and adequate organ
function.
 | Table I.Clinicopathological characteristics
including genetic backgrounds and circulating tumor cell marker
expression in 15 patients with lung cancer. |
Table I.
Clinicopathological characteristics
including genetic backgrounds and circulating tumor cell marker
expression in 15 patients with lung cancer.
| Age | Gender | Histology | EGFR gene
mutation | ALK fusion
gene | CEA in serum
(ng/ml) | CEA | hTERT | CK19 | PDL1 | Response |
|---|
| 74 | Female | SqCC | Unknown | Unknown | 7.9 | Low | Low | Negative | Negative | PR |
| 48 | Male | AdenoCa | No | No | 1.2 | Low | Low | Negative | Negative | PR |
| 64 | Male | AdenoCa | No | No | 4.3 | Low | Low | Negative | Negative | PR |
| 66 | Male | AdenoCa | No | No | 8.8 | Low | Low | Negative | Negative | PR |
| 82 | Male | AdenoCa | No | No | 9.3 | High | High | Negative | Negative | SD |
| 73 | Male | SqCC | No | No | Unknown | High | High | Negative | Negative | SD |
| 73 | Male | SqCC | Unknown | Unknown | Unknown | Low | High | Negative | Negative | SD |
| 47 | Male | AdenoCa | No | No | 11342 | Low | High | Negative | Positive | SD |
| 57 | Male | AdenoCa | Yes (T790M) | No | 70.6 | Low | High | Negative | Positive | SD |
| 52 | Male | AdenoCa | No | No | 99.7 | High | Low | Negative | Negative | SD |
| 62 | Male | AdenoCa | No | No | 3.7 | Low | Low | Positive | Negative | SD |
| 78 | Male | AdenoCa | Yes | No | 1.8 | High | High | Negative | Negative | PD |
| 63 | Male | AdenoCa | No | No | 41.6 | High | High | Negative | Negative | PD |
| 70 | Male | AdenoCa | No | No | 4.4 | High | Low | Negative | Negative | PD |
| 72 | Female | AdenoCa | No | Yes | 5.5 | High | Low | Negative | Negative | PD |
A pre-treatment 18F-FDG-PET/CT study and
blood sampling for CTC sorting were performed as part of the
disease evaluation workup prior to the administration of nivolumab.
If the tumor size was successfully suppressed by 3 months after the
initiation of nivolumab, a subsequent post-treatment PET/CT was
considered at around this time at the discretion of the
investigator. The clinical response to nivolumab was assessed from
the 18F-FDG uptake and chest CT of these patients, and
categorized as a partial response (PR), stable disease (SD), or PD
according to the Response Evaluation Criteria in Solid Tumors
(RECIST). All blood samples were prospectively collected and used
in accordance with the Helsinki Declaration and the guidelines of
the Gunma University Ethical Review Board for Medical Research
Involving Human Subjects after obtaining written informed consent
from each patient (approval no. 1404).
The human lung cancer cell line PC-9 and breast
cancer cell lines MCF7 and MDA-MB-231 were used to examine the
efficacy of the CTC chip for sorting cancer cells according to
size. These cell lines were provided by the RIKEN BioResource
Center and the American Type Culture Collection. The cells were
cultured in Roswell Park Memorial Institute (RPMI) 1640 medium
supplemented with 100 units/ml penicillin, 100 units/ml
streptomycin, and 10% fetal bovine serum (FBS) in a humidified
5%-CO2 incubator at 37°C. Before cell sorting, these
cells were labeled using the CellTrace™
Carboxyfluorescein Diacetate Succinimidyl Ester (CFSE) Cell
Proliferation kit (Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocol. The labeled cells
were then spiked into the CTC chip and separated into the small or
large cell fraction (Fig. 2A),
following which the sorted cancer cells in each fraction were
counted microscopically.
Reverse transcription-polymerase chain
reaction (RT-PCR)
To discover a useful biomarker for predicting
patients' sensitivity to the immune checkpoint inhibitor nivolumab,
we examined the relationships between the clinical response to
nivolumab and several genetic and CTC markers, including epidermal
growth factor receptor (EGFR) gene mutation, anaplastic
lymphoma kinase (ALK) fusion gene mutation, CEA levels in
the serum, and the expressions of existing CTC markers in the large
cell fraction including CEA, hTERT, CK19, and
PD-L1 in advanced lung cancer patients. RNA extraction and
reverse transcription were performed using NucleoSpin RNA XS kit
and PrimeScript RT reagent kit with gDNA Eraser (Takara Bio Inc.,
Tokyo, Japan) according to the manufacturer's protocol. The
following gene-specific oligonucleotide primers were designed for
PCR: The CEA (66 bp) sense primer 5′-ACCACAGTCACGACGATCAC-3′
and antisense primer 5′-GGAGTTGTTGCTGGTGATG-3′; the hTERT
(61 bp) sense primer 5′-GCCTTCAAGAGCCACGTC-3′ and antisense primer
5′-CCACGAACTGTCGCATGT-3′; the CK19 (126 bp) sense primer
5′-GCCACTACTACACGACCATCC-3′ and antisense primer
5′-CAAACTTGGTTCGGAAGTCAT-3′; the PD-L1 (124 bp) sense primer
5′-CTACTGGCATTTGCTGAACG-3′ and antisense primer
5′-TGCAGCCAGGTCTAATTGTTT-3′; and the 18S ribosomal RNA (rRNA) sense
primer 5′-GATGGTAGTCGCCGTGCC-3′ and antisense primer
5′-GCCTGCTGCCTTCCTTGG-3′. PCR amplification was performed in a
LightCycler® system (Roche Diagnostics, Basel,
Switzerland) using the LightCycler 480 SYBR Green I Master kit, as
previously described (19). Each of
the 40 amplification cycles comprised initial denaturation at 95°C
for 10 min, followed by denaturation at 95°C for 10 sec, annealing
at 62°C for 10 sec, and elongation at 67°C for 10 sec. The relative
expression levels of these genes were obtained by normalizing the
amount of mitochondrial RNA (mRNA) to that of 18S rRNA as an
endogenous control in each sample.
Fluorescent immunohistochemistry
Sorted cells were seeded on glass coverslips and
incubated for 12 h at 37°C. After washing with PBS to exclude
non-attached circulating cells such as lymphocytes, the cells were
fixed with 100% methanol at −20°C for 15 min, and then incubated
with mouse CEA antibody (1:100) (Kyowa Medex Co., Ltd., Tokyo,
Japan) and rabbit hTERT antibody (Abcam, Cambridge, CA, USA) for 1
h at room temperature. To detect antibodies against CEA and hTERT,
fluorophore-labeled antibodies with anti-mouse fluorescein
isothiocyanate (FITC) and anti-rabbit Cy3 specificities (Thermo
Fisher Scientific, Inc.) were used for 1 h at room temperature at a
dilution of 1:2,000. All sections were then counterstained with
4′,6-diamidino-2-phenylindole (DAPI) and examined under an
All-in-One BZ-X710 Fluorescence Microscope (Keyence Corporation,
Osaka, Japan). Negative control sections were stained as described
above but without the primary antibodies.
Statistical analysis
The relationship between the number of patients with
positive marker expression in the large cell fraction and the
clinical response to nivolumab was analyzed using Pearson's
chi-square test. All statistical analyses were performed using the
JMP software package (SAS Institute, Inc., Cary, NC, USA).
Results
The new polymeric microfluid CTC chip
effectively collected lung cancer cell line PC-9 into the large
cell fraction
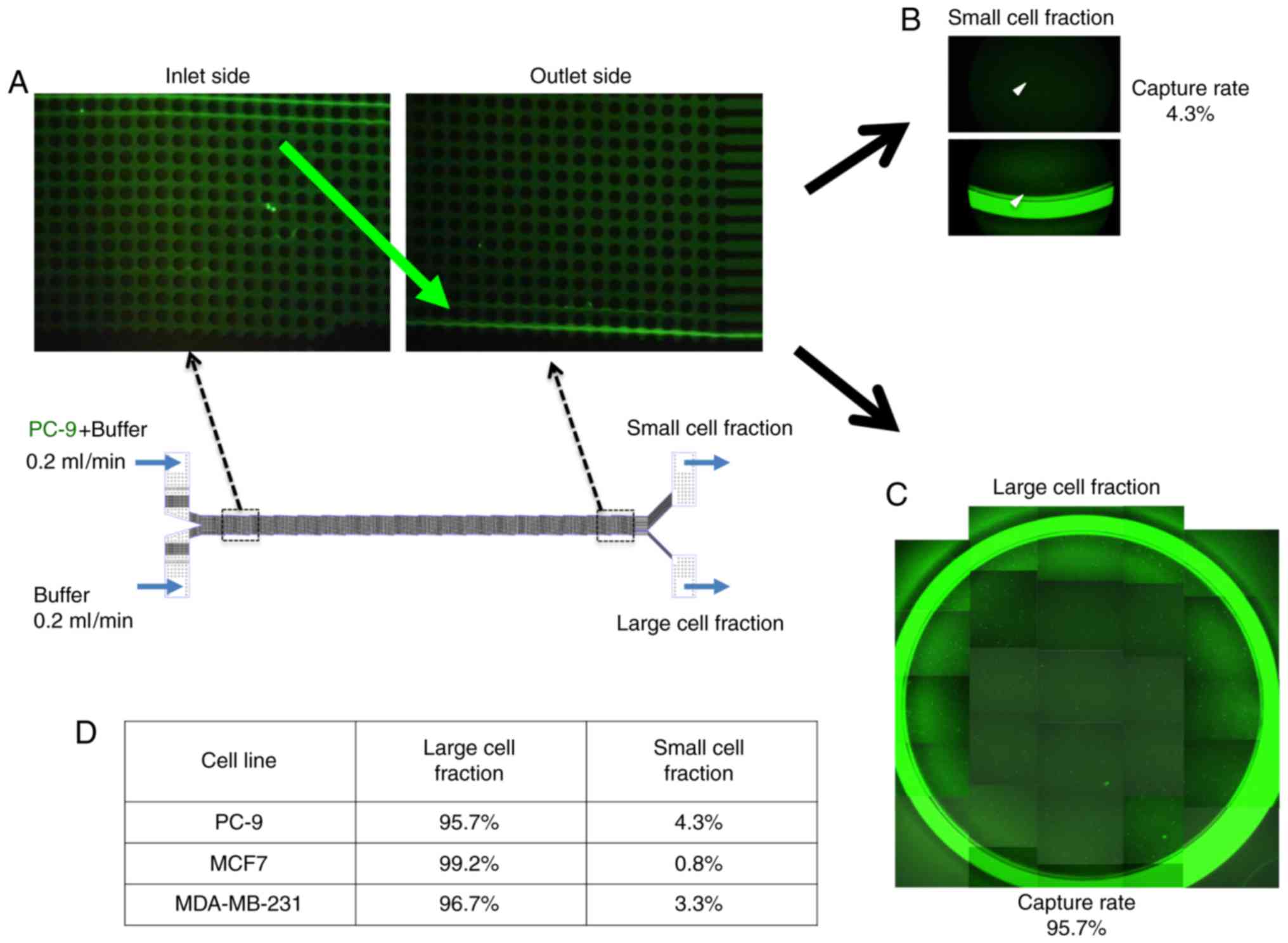
To examine the capture efficacy of lung cancer cells
using the size-sorting CTC chip device, we spiked the CTC chip with
CFSE-labeled PC-9 cells and counted how many entered the small and
large cell fractions (Fig. 2). We
found that nearly all of the PC-9 cells were moved into the large
cell fraction side as they approached the outlet (Fig. 2B and C). Moreover we could validate
the sorting efficacy of our CTC chip using breast cancer cell lines
MCF7 and MDA-MB-231 similar to PC-9 cells (Fig. 2D).
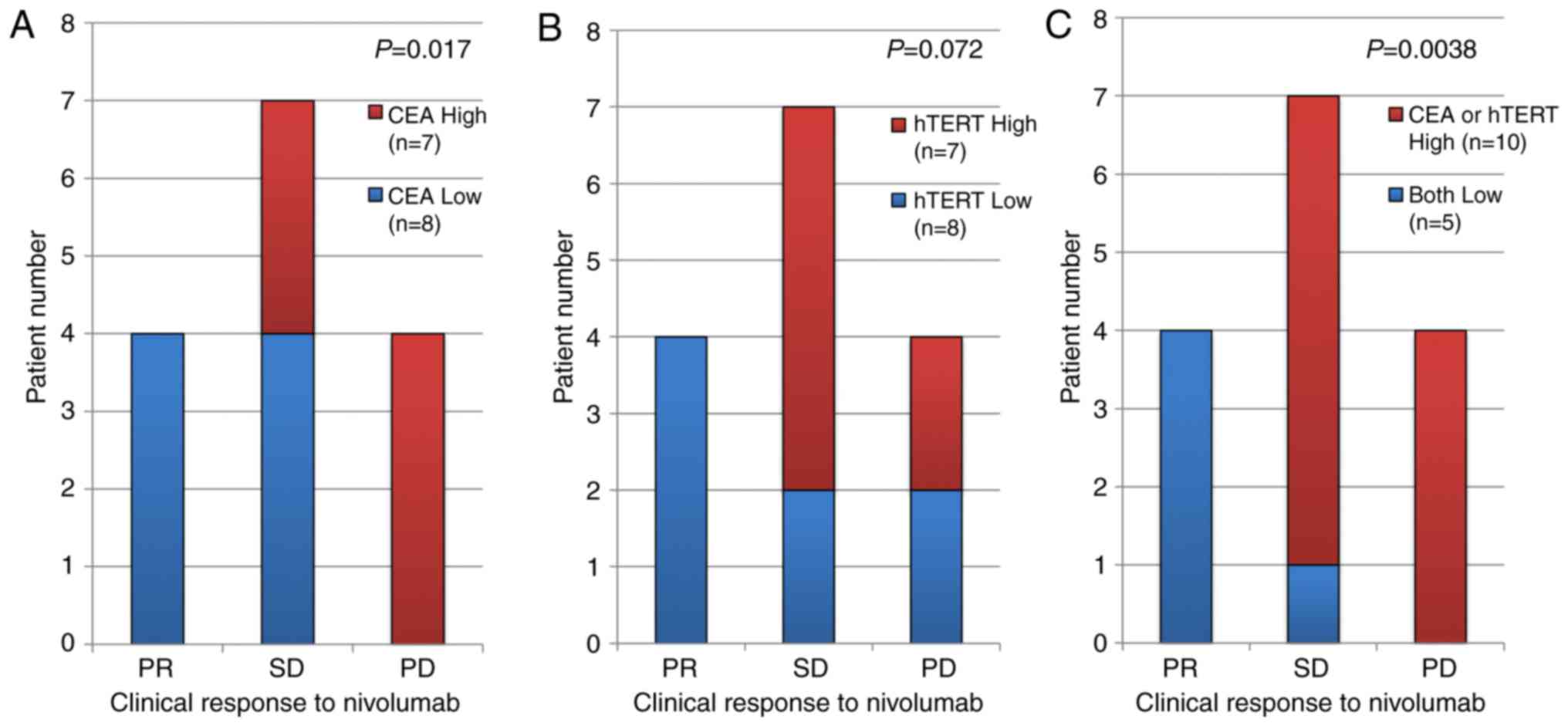
High expression of CEA and hTERT in
the sorted CTCs were associated with poor clinical response to the
anti-PD-1 antibody nivolumab
We found that the large cell fraction collected by
the CTC chip exhibited high expression of CEA and
hTERT. Furthermore, the high expression of CEA was
significantly correlated with a poor clinical response to nivolumab
and there was also a weak negative correlation between the
expression of hTERT and the clinical response, although this
was not significant (Fig. 3A and B).
Interestingly, both low expressions were strongly correlated with
PR clinical response (Fig. 3C).
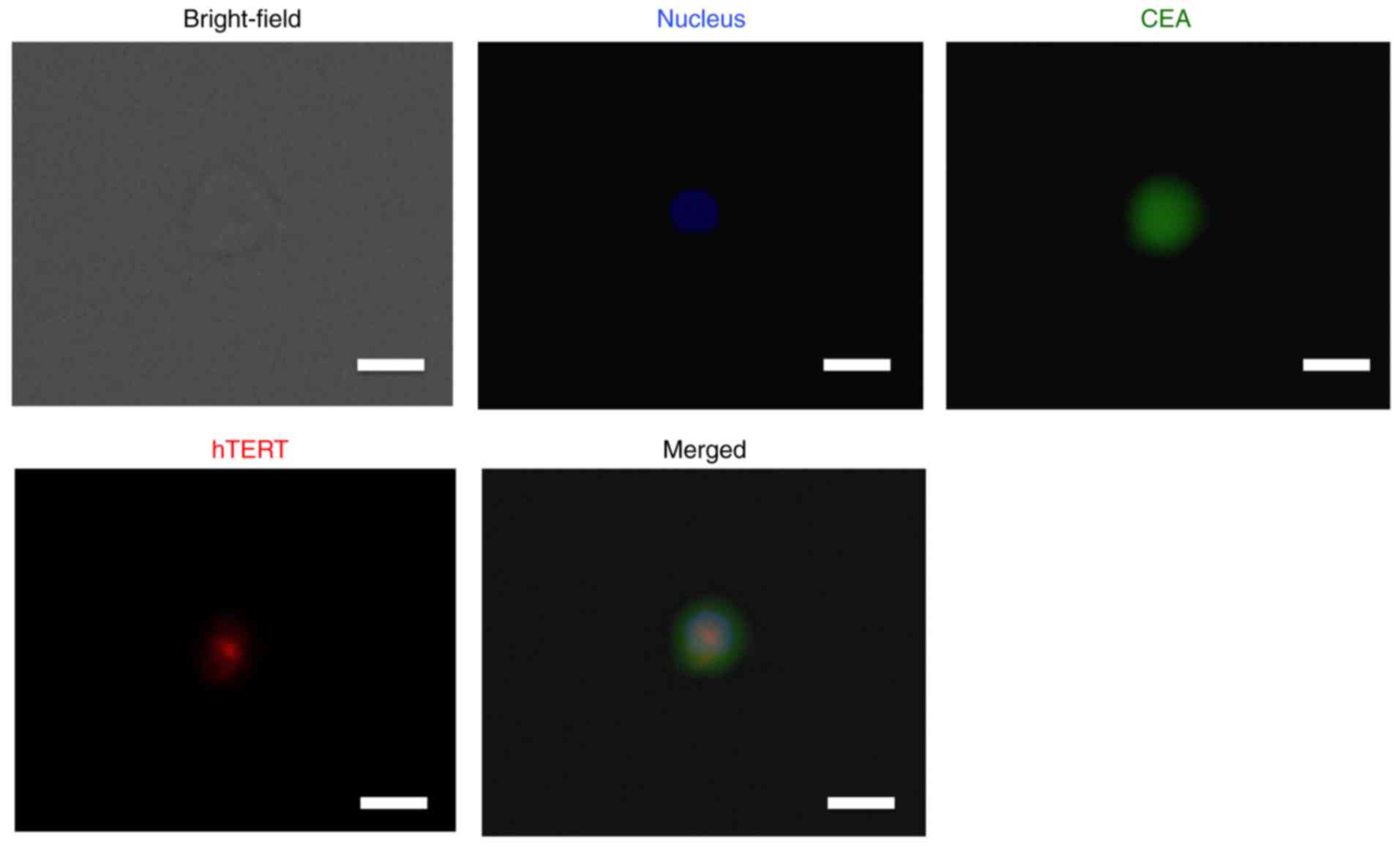
Immunohistochemical analysis of CEA
and hTERT expression in CTCs collected by the size-sorting CTC
chip
Once large cells had been collected by the CTC chip,
we used fluorescent immunohistochemistry to validate the expression
of CEA and hTERT in CTCs. This showed that CTCs of lung cancer
expressed the CTC markers CEA and hTERT (Fig. 4). These findings are consistent with
the above-mentioned PCR data.
Discussion
In the present study, we designed a new CTC chip
that could effectively sort PC-9 cells from a cell suspension in
vitro depending on their size. Moreover, we demonstrated that
this CTC chip was able to collect the large cells including CTCs
from whole blood and that a high expression level of CEA in
this fraction was significantly correlated with a poor clinical
response in clinical lung cancer patients treated with the
anti-PD-1 antibody nivolumab. By contrast, serum CEA levels and the
expression of other markers in the large cell fraction, such as the
representative epithelial marker CK19 and PD-L1, were
not associated with the clinical response to nivolumab.
In general, it is important for cancer cells to
avoid immune surveillance not only at the primary site but also in
the bloodstream to allow them to survive. However, cancers also
have several other hallmarks that help with their survival,
including the ability to sustain proliferative signaling; evade
growth suppressors; enable replicative immortality and
tumor-promoting inflammation; activate invasion and metastasis;
induce angiogenesis, genome instability, and mutation; resist cell
death; and deregulate cellular energetics (20). It has previously been reported that
CEA and hTERT may help in the proliferation,
inflammation, angiogenesis, metastasis, resistance to apoptosis,
DNA damage repair, and replicative immortality of cancers, which
are expected to be important for their survival (20–22).
Consequently, it has been suggested that if cancers depend on
avoiding immune destruction for their survival, the immune
checkpoint inhibitor nivolumab would be an effective treatment,
whereas if they are not depending on this, nivolumab may not induce
an anti-cancer effect against the cancer cells with other hallmark
characteristics. Thus, the use of the method outlined here for
detecting CEA and hTERT in CTCs in combination with
nivolumab treatment may prove useful for evaluating whether the
cancers in advanced lung cancer patients are depending on avoiding
immune destruction for their survival.
Several methods for enriching and detecting CTCs in
blood samples from clinical cancer patients have previously been
reported (8,9,23). Here,
we used a CTC chip to enrich CTCs, which is a morphology-based
isolation technique that related to the isolation by size of
epithelial tumor cells (ISET) method (24), Ficoll isolation for collecting CTC and
mononuclear cells, and RosetteSep™ (25). This size-sorting CTC chip was able to
enrich lung cancer cells in vitro and in clinical samples.
We chose to use a PCR-based method with high sensitivity to detect
CTCs from the large cell fraction because CTCs are known to be rare
in the blood (one CTC per 106-107 mononuclear
cells; [10]). This combination of a size-sorting CTC chip and
high-sensitivity PCR-based CTC detection technology may be more
suitable for evaluating sensitivity to the immune checkpoint
inhibitor nivolumab than the mere detection of CTCs in blood
samples.
Commercially available CTC chip using epithelial
antibody-based detection methods could capture the cancer cell
lines expressing epithelial markers such as MCF7. However,
epithelial mesenchymal transition (EMT) induced cancer cells
including MDA-MB-231 with highly aggressive phenotypes were not
captured by these types of CTC chips (26,27). In
contrast, our CTC chip is based on cell size, not epithelial
markers. We could demonstrate the high capture ratio of EMT-induced
cancer cell line MDA-MB-231 using our CTC chip. From these data, it
was suggested that our CTC chip may be more effective in sorting
the EMT-induced aggressive CTC than epithelial marker dependent CTC
detection.
There is a need to develop a sensitivity marker for
immune checkpoint inhibitors including nivolumab in the clinic
because these agents are very expensive and have characteristic
side effects including an autoimmune response against several
organs. It has previously been reported that the overexpression of
PD-L1 and somatic mutations that encode immunogenic neoantigens are
significantly related to a better response to PD-1/PD-L1 blockade
therapy in several cancers (3).
However, we are in urgent need of a useful predictive biomarker
that uses blood because invasive tumor sampling is currently
required to detect PD-L1 expression and mutations for neoantigens
in tumor cells. The method outlined in the present study could
predict nivolumab sensitivity before treatment using only 1 ml of
blood from a cancer patient, making it very promising. However, our
data have some limitations: We only validated the immunostaining of
CTC markers, CEA, hTERT in a few clinical lung cancer patients in
this study, and the limited number of patients that were used in
this study; therefore, more patients need to be involved in future
studies to elucidate the clinical potential of these methods.
In conclusion, we successfully developed a new
polymeric CTC chip that can sort CTCs from blood samples based on
their size. The CTCs in the large cell fraction expressed the
existing CTC markers CEA and hTERT, the high
expression of which was associated with a poor clinical response to
the immune checkpoint inhibitor nivolumab in advanced lung cancer
patients. Evaluation of CEA and hTERT in CTCs may be
a predictive blood marker candidate for patient sensitivity to
nivolumab.
Acknowledgements
The present study was supported by JSS Young
Researcher Award from Japan Surgical Society, Gunma University
Clinical Biobank, and grants-in-Aid for Scientific Research from
the Japan Society for the Promotion of Science (JSPS) (grant nos.
JP 26461969, JP15K10129, JP15K10085, and JP26350557).
References
|
1
|
Sharma P and Allison JP: The future of
immune checkpoint therapy. Science. 348:56–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ma W, Gilligan BM, Yuan J and Li T:
Current status and perspectives in translational biomarker research
for PD-1/PD-L1 immune checkpoint blockade therapy. J Hematol Oncol.
9:472016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meng X, Huang Z, Teng F, Xing L and Yu J:
Predictive biomarkers in PD-1/PD-L1 checkpoint blockade
immunotherapy. Cancer Treat Rev. 41:868–876. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Borghaei H, Paz-Ares L, Horn L, Spigel DR,
Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al:
Nivolumab versus docetaxel in advanced nonsquamous non-small-cell
lung cancer. N Engl J Med. 373:1627–1639. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Robert C, Long GV, Brady B, Dutriaux C,
Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C,
Kalinka-Warzocha E, et al: Nivolumab in previously untreated
melanoma without BRAF mutation. N Engl J Med. 372:320–330. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Motzer RJ, Escudier B, McDermott DF,
George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G,
Plimack ER, et al: Nivolumab versus everolimus in advanced
renal-cell carcinoma. N Engl J Med. 373:1803–1813. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD-1 Blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pantel K and Alix-Panabières C:
Circulating tumour cells in cancer patients: Challenges and
perspectives. Trends Mol Med. 16:398–406. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alix-Panabières C and Pantel K: Challenges
in circulating tumour cell research. Nat Rev Cancer. 14:623–631.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun YF, Yang XR, Zhou J, Qiu SJ, Fan J and
Xu Y: Circulating tumor cells: Advances in detection methods,
biological issues, and clinical relevance. J Cancer Res Clin Oncol.
137:1151–1173. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nagrath S, Sequist LV, Maheswaran S, Bell
DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky
A, et al: Isolation of rare circulating tumour cells in cancer
patients by microchip technology. Nature. 450:1235–1239. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dong Y, Skelley AM, Merdek KD, Sprott KM,
Jiang C, Pierceall WE, Lin J, Stocum M, Carney WP and Smirnov DA:
Microfluidics and circulating tumor cells. J Mol Diagn. 15:149–157.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Seal SH: A sieve for the isolation of
cancer cells and other large cells from the blood. Cancer.
17:637–642. 1964. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu Y, Xu G, Cao J, Jin S, Man Y and Shang
L: Combination of four gene markers to detect circulating tumor
cells in the peripheral blood of patients with advanced lung
adenocarcinoma using real-time PCR. Oncol Lett. 5:1400–1406. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tanaka F, Yoneda K and Hasegawa S:
Circulating tumor cells (CTCs) in lung cancer: Current status and
future perspectives. Lung Cancer (Auckl). 1:77–84. 2010.PubMed/NCBI
|
|
16
|
Mazel M, Jacot W, Pantel K, Bartkowiak K,
Topart D, Cayrefourcq L, Rossille D, Maudelonde T, Fest T and
Alix-Panabières C: Frequent expression of PD-L1 on circulating
breast cancer cells. Mol Oncol. 9:1773–1782. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang LR, Cox EC, Austin RH and Sturm JC:
Continuous particle separation through deterministic lateral
displacement. Science. 304:987–990. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ohnaga T, Shimada Y, Moriyama M, Kishi H,
Obata T, Takata K, Okumura T, Nagata T, Muraguchi A and Tsukada K:
Polymeric microfluidic devices exhibiting sufficient capture of
cancer cell line for isolation of circulating tumor cells. Biomed
Microdevices. 15:611–616. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yokobori T, Iinuma H, Shimamura T, Imoto
S, Sugimachi K, Ishii H, Iwatsuki M, Ota D, Ohkuma M, Iwaya T, et
al: Plastin3 is a novel marker for circulating tumor cells
undergoing the epithelial-mesenchymal transition and is associated
with colorectal cancer prognosis. Cancer Res. 73:2059–2069. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Beauchemin N and Arabzadeh A:
Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs)
in cancer progression and metastasis. Cancer Metastasis Rev.
32:643–671. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cong Y and Shay JW: Actions of human
telomerase beyond telomeres. Cell Res. 18:725–732. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alix-Panabières C and Pantel K:
Technologies for detection of circulating tumor cells: Facts and
vision. Lab Chip. 14:57–62. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vona G, Sabile A, Louha M, Sitruk V,
Romana S, Schütze K, Capron F, Franco D, Pazzagli M, Vekemans M, et
al: Isolation by size of epithelial tumor cells: A new method for
the immunomorphological and molecular characterization of
circulating tumor cells. Am J Pathol. 156:57–63. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Busch R, Cesar D, Higuera-Alhino D, Gee T,
Hellerstein MK and McCune JM: Isolation of peripheral blood CD4(+)
T cells using RosetteSep and MACS for studies of DNA turnover by
deuterium labeling. J Immunol Methods. 286:97–109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Polyak K and Weinberg RA: Transitions
between epithelial and mesenchymal states: Acquisition of malignant
and stem cell traits. Nat Rev Cancer. 9:265–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ohnaga T, Shimada Y, Takata K, Obata T,
Okumura T, Nagata T, Kishi H, Muraguchi A and Tsukada K: Capture of
esophageal and breast cancer cells with polymeric microfluidic
devices for CTC isolation. Mol Clin Oncol. 4:599–602. 2016.
View Article : Google Scholar : PubMed/NCBI
|