Being a part of the system that specifies pattern
formation during animal embryogenesis, hedgehog (Hh) signalling was
initially discovered to regulate cuticle patterns in
Drosophila more than 30 years ago (1); its existence in mammals suggested the
strictly conservative roles of the Hh pathway in animal
embryogenesis during evolution (2–4).
Considering that development and tumourigenesis are essentially
inextricable processes and that tumourigenesis may be the product
of abnormal development under undesired circumstances, it is quite
reasonable that deregulated Hh signalling was discovered as the
promoter of human cancers. It was initially involved in basal cell
nevus syndrome in 1996 (5,6). Subsequently, aberrantly active Hh
pathways are closely linked to various human cancers (7), including ovarian (8), gastric (9), hepatocellular (10) and bladder cancers (11,12),
indicating its significant roles in adult tissue homeostasis
maintenance. Therefore, Hh pathway-targeted inhibitors have been
developed for cancer therapy (13).
Though highly conserved, the Hh pathway is much more complex in
vertebrates than in Drosophila, likely due to adaptations for the
functional complexity of higher multicellular organisms throughout
evolution. For instance, the mammalian counterpart of the
Drosophila Hh ligand is the Hedgehog family of secreted
proteins, which contains three members: Sonic Hedgehog (Shh),
Indian Hedgehog (Ihh), and Desert Hedgehog (Dhh); these proteins
perform overlapping but quite distinctive functions. Similarly, the
function of the terminal transcription factor Cubitus interruptus
(Ci) in Drosophila is assigned to three glioma-associated
factors (GLI)-GLI1, GLI2 and GLI3-in mammals (14). In addition, the protein suppressor of
fused (Sufu), a negative regulator of Hh signalling pathway, seems
to display a more important function in vertebrates, where Sufu
deletion led to embryo lethality in mice (15) but no visible phenotypic alterations in
Drosophila (16).
Among the three Hh signalling pathways, the most
intensively studied at present is the Shh pathway, which is
generally delineated as follows. The Hh ligand binding to the
twelve-span transmembrane protein Ptc1 (Patched1), relieves the
suppressive effect of Ptc1 on another seven-span transmembrane
protein, smoothened (Smo). Active Smo transduces Hh signalling
across the cytomembrane to activate GLI through incompletely
clarified mechanisms that likely involve dissociating GLI from a
suppressive complex containing Sufu. Finally, activated GLI enters
the nucleus to transcriptionally activate a specific set of genes
that contribute to certain cellular activities. Furthermore,
alternative Hh signalling cascades, through which Hh signalling is
transduced through Ptc1 and Smo but are not mediated by GLI, were
identified to regulate pain perception (17) and cellular metabolism (18). These alternative cascades comprise
non-canonical Hh signalling pathways, which will not be described
herein (19). This review focuses
within the realm of the canonical mammalian Shh signalling pathway,
which will be simply described as the Hh pathway in the following
sections.
As described above, Sufu acts as a negative
regulator of the Hh signalling pathway and plays essential roles in
both embryogenesis and adult tissue homeostasis in species ranging
from invertebrates to vertebrates. Loss-of-function of Sufu is
sufficient to activate the Hh signalling pathway irrespective of
the presence of the Hh ligand (32).
In addition, the embryonic lethality in mice due to Sufu deletion
(15) implies important functions of
Sufu in mammals. These findings strongly suggest a central role of
Sufu in the mammalian Hh pathway, which, along with our observation
of loss of Sufu in colorectal cancer specimens, intrigued us
greatly.
After the Hh pathway was implicated in human cancers
by Ptc1 inactivation associated with basal cell carcinomas
(5,6),
substantial studies have been conducted to explore the
tumour-promoting effects and the underlying mechanisms of a
hyperactive Hh pathway. Presently, multiple Sufu mutations have ben
identified (shown in Table I), which
are named according to the standard nomenclature recommendations of
the HGVS (http://www.HGVS.org/mutnomen/) (33). These mutations determine abnormal
forms of the Sufu protein among various human cancers, with the
medulloblastoma of desmoplastic/nodular subtype being most
abundantly studied, which is one of the five histologic subtypes of
medulloblastoma and is characteristic of aberrant Hh pathway
(34).
In both familial and sporadic desmoplastic
medulloblastoma, multiple somatic and germline Sufu mutations that
are likely to predispose individuals to tumourigenesis were
identified in a series of studies (35–39), with
some mutations, such as c.71del and c.71dul, showing incomplete
penetrance of the predisposition to medulloblastoma for unknown
reasons (40). Brugières et al
reported that germline Sufu mutations are responsible for over 50%
of seemingly sporadic desmoplastic/nodular medulloblastoma cases,
especially in patients diagnosed at younger than 3 years of age
(35). Although abnormal protein
products speculated from the mutated Sufu genes are thought
to lose their capacity to suppress GLI, only a few of them have
been studied with respect to their functional consequences. Studies
taking advantage of genetically modified mice were performed to
confirm the tumour-stimulating effects of the loss of Sufu. While
Sufu-/- mice present embryonic lethality with cephalic and neural
tube defects, indicating a pivotal role of Sufu in mouse embryonic
development, Sufu+/− mice displayed characteristic features of
human Gorlin syndrome due to enhanced Hh signalling but were less
prone to harbour tumour medulloblastoma and rhabdomyosarcoma
compared with ptch+/− individuals (15,41).
However, the absence of the p53 gene could facilitate
tumourigenesis in Sufu+/− mice. It was shown that Sufu+/− p53-/-
mice but not p53-/- individuals frequently developed
medulloblastoma and rhabdomyosarcoma similar to those observed in
Ptc1+/− mice (32,42), suggesting that abrogation of Sufu
activity remains a critical step to initiate tumours in the context
of p53 inactivation. To some extent, this finding is in agreement
with the "two-hit" theory of tumor initiation. The observation that
Sufu+/− mice are less prone than Ptc1+/− individuals to develop
tumours may be due not only to the differences in functions of Sufu
and Ptc1 but also to the genetic background of the experimental
mouse model. Taken together, these studies indicate a stimulatory
role of Sufu inactivation in medulloblastoma. However, in sharp
contrast to the observations of Sufu mutations in the studies
above, no somatic or germ line mutations or altered expression in
Sufu was revealed in a screen of 145 primitive neuroectodermal
tumours, suggesting that genetic alteration of Sufu is a relatively
rare event in human medulloblastoma (43). In addition, in a study to discover
critical mutational targets in region 10q23.3–10q25.3, which
contains Sufu and displays deletion in medulloblastoma, no
tumour-specific sequence variations or DNA hypermethylation of
promoter CpG islands of Sufu and other known genes in that
region were discovered (44). One
explanation for these observed disparities in Sufu mutations among
medulloblastoma tumours includes the heterogeneous characteristics
of tumours. Moreover, it is also possible that in cancers
harbouring hyperactive Hh pathways with intact Sufu loci, the
function of Sufu is debilitated through epigenetic modifications of
some unidentified regulatory sequences or translational and
post-translational modifications of the Sufu protein. Nonetheless,
these modifications cannot be detected with techniques involving
the screening of only genetic mutations or by simply analysing
putative regulatory genomic sequence (45,46) or
protein expression levels.
In addition to medulloblastoma, Sufu mutations have
also been found in other human cancers. Applying genome-wide
linkage analysis and exome sequencing, Aavikko et al
revealed a germline Sufu mutation (c.367C>T) in meningiomas
encoding the protein mutant p.Arg123Cys. As a result, an altered
tertiary structure and compromised function of Sufu occurred
(47). Genetic mutations in Sufu and
other Hh pathway components were also detected in mesothelioma
(48) and rhabdomyoma (49). Some other studies have linked Sufu
mutations to basal cell carcinoma (BCC) (50–52) and
skin hamartomata (53). It was also
reported that the Hh pathway hyperactivation attributed to the loss
of Sufu or that overexpression of the Hh ligand may participate in
the progression and metastases of prostate cancer (54), while the germline splicing mutation
c.1022+1 G>A generated a Sufu protein with a deletion of exon 8
instead of the general Ptc1 mutation that was found in a family
with atypical Gorlin syndrome (55).
In summary, consistent with the fact that Sufu is a
critical suppressor of the Hh pathway, which promotes
tumourigenesis when aberrantly activated, Sufu inactivation has
been implicated in various human cancers. Subsequently, the
molecular mechanisms underlying the regulation of Hh signalling
activity by Sufu will be illustrated in detail.
Within the realm of the Hh signalling pathway,
substantial research focusing on Sufu has revealed multiple
mechanisms by which it regulates the transcription activity of GLI,
supporting its critical regulatory role in the Hh pathway. As a
suppressor of the Hh pathway in both invertebrates and vertebrates,
the regulation modes of GLI by Sufu in vertebrates are much more
intricate than the regulation of Ci by Sufu in Drosophila
(56), in part due to different
regulation patterns of GLI1, GLI2 and GLI3 because of differences
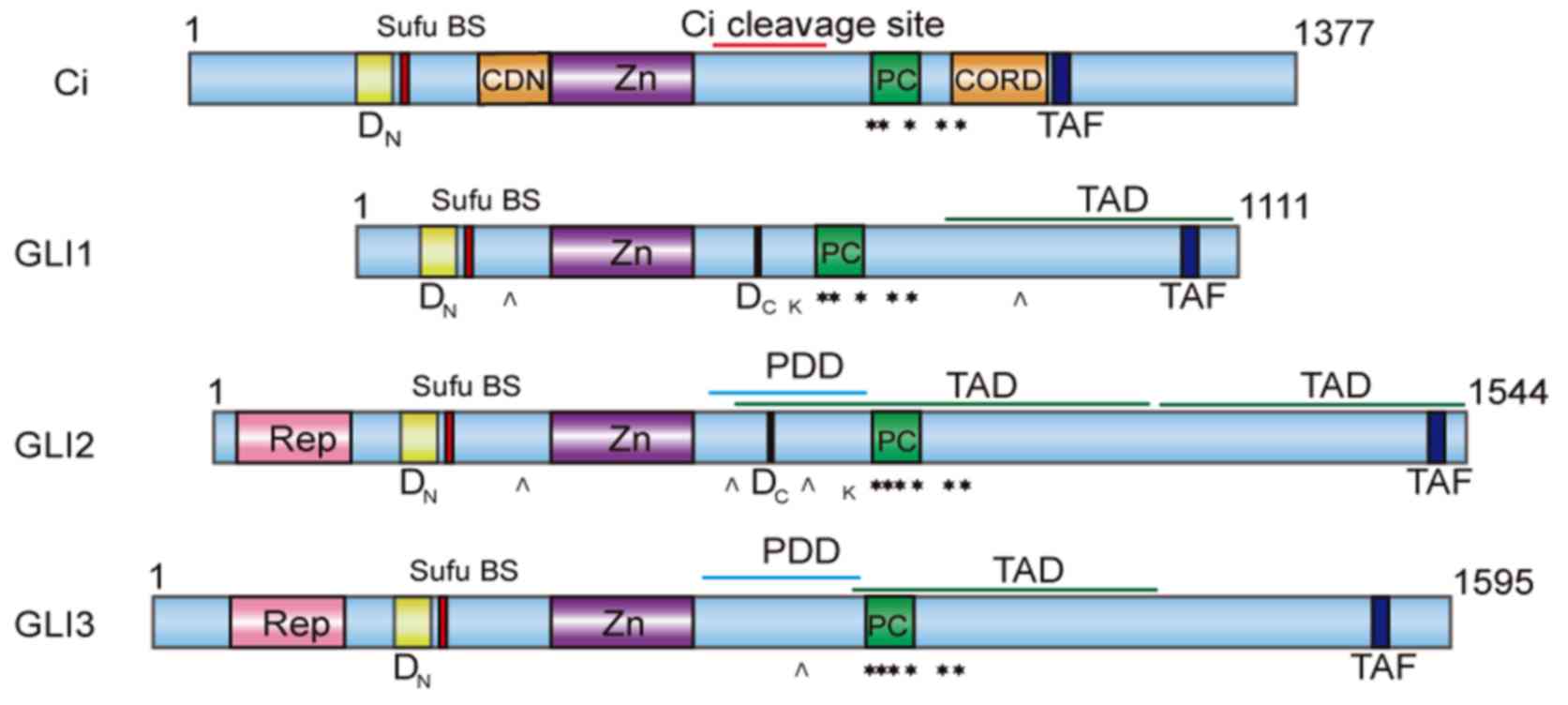
in structure (as shown in Fig. 1) and
functionality among GLI proteins (14). As primary mediators of the Hh pathway,
GLI2 mainly functions as a potential activator of the pathway,
while GLI3 acts mainly as a suppressor (57,58),
although they both cooperate to modulate the expression of GLI1 and
other target genes. Once generated, GLI1 functions as a
constitutive activator to amplify Hh signalling due to the lack of
an N-terminal suppressor domain in GLI, which exists in GLI2 and
GLI3 (14).
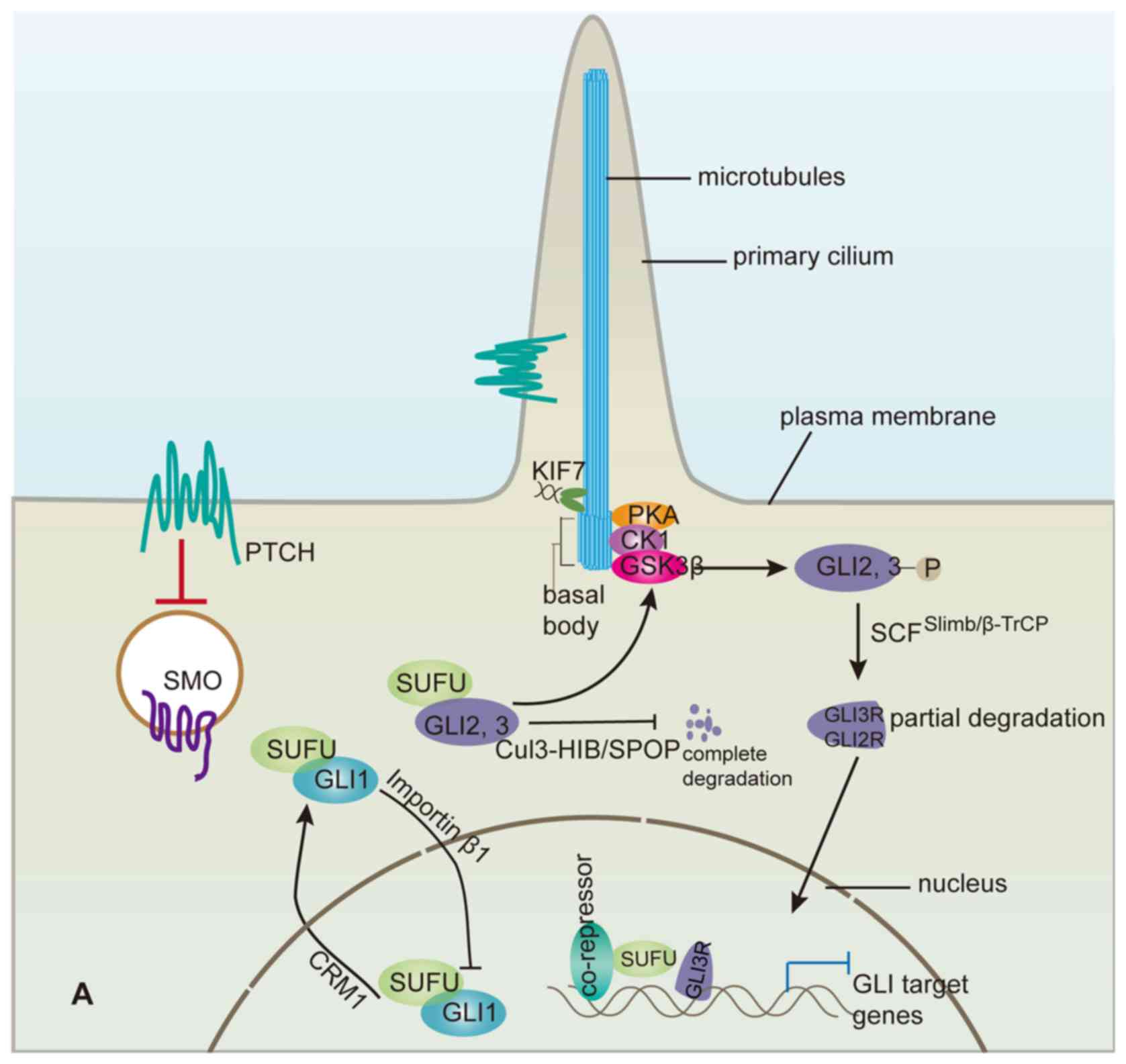
Hh signalling transduction is realized by
alterations in the distributions of Hh pathway components, such as
Ptc1, Smo, GLI and Sufu, within the primary cilium, a
microtubule-based projection of the cell membrane that generally
functions in vertebrates as a sensor of various extracellular
signal pathways, including the Hh pathway (59). When the Hh signalling pathway is
quiescent in the absence of functional Hh ligand, the binding of
Sufu to GLI2 and GLI3 sequesters them in a cytoplasmic complex
containing a cilium-associated kinesin Kif7 (60–62), known
as the vertebrate homologue of Cos2 in Drosophila. This
sequestration directly prohibits full-length GLI proteins from
entering the nucleus and functioning as transcriptional activators.
A study indicated that the binding of Sufu to GLI1 impeded the
nuclear accumulation of GLI1 (63),
consequently inhibiting proliferation, invasion and vascular
mimicry of glioma cancer cells, while Sufu knockdown reversed these
suppressive effects. Later, Sufu was reported to compete with
importin β1 to bind to the same site on GLI1, thus retarding the
importin β1-mediated nuclear translocation of GLI1 (64). Similarly, by masking the NLS (nuclear
localization sequence), Sufu precludes Transportin-mediated nuclear
import of Ci in Drosophila, as well as the nuclear accumulation of
GLI2 and GLI3 mediated by Kapβ2 (65), the mammalian homologue of Transportin.
Recently, a study revealed that Sufu can sequester Ci/GLI proteins
in the cytoplasm by binding to their N-terminal regions, while in
the nucleus, the suppression of Ci/GLI activity is dependent on
their C-terminal regions (66).
Moreover, binding of Sufu to GLI2/3 in the cytoplasmic complex
facilitates the production of repressor GLI3 (GLI3R) and, to a
lesser extent, GLI2R, which result from the partial degradation of
the full-length GLI2/3 and function as transcription suppressors
(67). The hyper-phosphorylated
full-length GLI2/3 resulting from sequential phosphorylation by
protein kinase A (PKA), GSK3β and CK1 within the complex are
partially degraded through the ubiquitin-proteasome pathway
mediated by SCFβTrCP ubiquitin E3 ligase, truncating the
C-terminal transcriptional activator domain of GLI2/3 proteins
(68,69). In Drosophila, a cytoplasmic complex
composed of fu, Sufu, Cos2 and Ci similarly regulates Ci activity,
suggesting some conserved scenario of Hh pathway regulation by Sufu
during evolution. GLI2R and GLI3R likely compete only with
full-length GLI2/3 for the same regulatory sequences to inhibit the
otherwise activated transcription or may cooperate with
co-repressors recruited in a GLI2/3R-dependent manner.
Nevertheless, the inhibitory cytoplasmic complex described above
would be transformed by Hh signalling (70,71).
Following Smo activation, the cytoplasmic complex containing Sufu
and GLI2/3 is recruited to the primary cilium tip, where
dissociation of the complex releases full-length GLI2 and GLI3
(72), which are subsequently
translocated into the nucleus as potential transcriptional
activators to take the place of GLIR, while Sufu is induced to
degrade (73,74). Recently, it was reported that
responding to Hh signalling, Sufu accompanies GLI1 to the primary
cilium, which is coupled to the subsequent nuclear transport of
GLI1 (75). The necessity of the
primary cilium for GLI derepression is supported by the report of a
novel central domain characterized in GLI2 that is essential for
both primary cilium localization and transcriptional activity
(76). However, the specific
mechanisms by which Hh signal is relayed from activated Smo in
primary cilium to finally derepress GLI2/3 are far from fully
understood. Recently, a primary cilium G protein-coupled receptor,
Gpr161 (77), which also regulates
the Wnt signalling pathway, was linked to Hh signalling
transduction via Smo in primary cilium (78). Several studies have determined that
the EVC/EVC2 ciliary complex, whose loss-of-function mutations
contribute to Ellis-van Creveld syndrome, interacts with Smo to
particularly regulate Sufu/GLI3 dissociation in primary cilium for
normal endochondral bone formation, where Evc is needed for Hh
pathway activation even when Sufu is deleted (79–81). In
contrast, PKA is reported to efficiently suppress ciliary
localization of GLI2-Sufu and GLI3-Sufu, while just moderately
affecting GLI1-Sufu (72). While
essential for antagonizing Sufu activity, the primary cilium is not
required for the inhibitory effect of Sufu on the Hh pathway
(82), and localization of Sufu in
primary cilium is dependent on GLI, which alone is capable of
accumulating in primary cilium (83).
However, considering the potently suppressive effect
of Sufu on GLI activity, the finding that the protein levels of
GLI2/3 in mice and Ci in Drosophila were reduced when Sufu was
deleted seems paradoxical (82,84). Sufu
could retard proteasome-dependent degradation of GLI2/GLI3 promoted
by Spop, a substrate-binding subunit for Cul3-based E3 ubiquitin
ligase, by competing with Spop in binding the N- and C-terminal
regions of GLI3 and the C-terminal region of GLI2 (67). Notably, the stabilities of the GLI2R,
GLI3R and GLI1 proteins are not influenced in this manner.
Nevertheless, a recent study reported that Sufu is also essential
for maintaining high protein level and nuclear accumulation of GLI1
in response to Hh signalling, sustaining the proliferation of
granule neuron precursor cells (75).
Mechanistically, Sufu-mediated stabilization of full-length GLI2/3
is of great importance. Without Hh signalling, the binding of Sufu
to GLI2/3 preserves a pool of full-length GLI2/3 by preventing
complete degradation stimulated by Spop. It also facilitates
generation of GLI3R and, to a lesser extent, GLI2R mediated by
SCFSlimb/β-TrCP, as mentioned previously (68,69), which
restrains gene transcription. As soon as Hh signalling is present,
the preserved full-length GLI2/3 are derepressed to produce a
transcriptional activator, which counteracts GLI2R and GLI3R,
resulting in the rapid response of Hh signalling. Understandably,
the activated full-length GLI2/3 are subjected to Spop-promoted
degradation and are thus labile and unable to activate the Hh
pathway constitutively, which is similar to the situation of Ci
seen in Drosophila (85).
As in the cytoplasm, Sufu also regulates GLI
proteins within the nucleus. For instance, it can recruit the
SAP18-mSin3-HDAC co-repressor complex to suppress GLI
transcriptional activity (86,87). SAP18
is recruited to GLI3R-Sufu to form a suppressive complex occupying
the GLI binding site when Hh signalling is quiescent, and upon Hh
pathway activation this suppressive complex will be replaced by
GLI1-Sufu (75) to activate the
transcription of target genes. Other nuclear proteins mainly
involved in basal transcription machinery were identified to
interact with Sufu in a yeast two-hybrid screen using mouse Sufu as
a bait, indicating crucial functions of Sufu in the nucleus. Sufu
was revealed to interact with those other nuclear proteins and GLI
proteins through the N- and C-termini, respectively, to form the
suppressive complex (86). Similarly,
p66β is the co-repressor that is recruited by Sufu to suppress
GLI2/3 activity in the nucleus, whereas Hh signalling can induce
Sufu/p66β dissociation from GLI and stimulate GLI activity
(88). In addition, binding of Sufu
in the nucleus also promotes GLI1 and GLI2 to export from the
nucleus to the cytoplasm (89,90). In
C3H10T1/2 cells, treatment with leptomycin B, which inhibits
nuclear export by binding to the nuclear export receptor CRM1,
dramatically shifts the localization of co-expressed GLI1/Sufu from
the cytoplasm to the nucleus, suggesting that Sufu stimulates GLI1
export from the nucleus in a CRM1-dependent manner. However, CRM1
is not involved in Sufu-promoted export of GLI2 from the nucleus.
It is possible that the inhibition of GLIs in the nucleus by Sufu
is a remedy for its failure to sequester them in the cytoplasm.
The integrated regulations of GLI by Sufu are at
least partially responsible for further modulating the Hh pathway
to maintain graded Hh pathway activity (14,91,92), which
is primarily represented by the GLIA/GLIR ratio and is essential
for realizing the complex physiological functions of the Hh pathway
within multicellular organisms. For example, deletion of Sufu
disrupts the gradient of Ptc1 expression, which displays a
high-to-low gradient from ventral to dorsal areas of the wild-type
mouse neural tube, similar to the Hh level gradient (67). Furthermore, Sufu deletion compromised
Hh pathway activity in the ventral area of neural tube in Sufu-/-
mice because more full-length GLI2/3 proteins are completely
degraded compared with the situation in wild-type mice; conversely,
in the dorsal area of the neural tube, Hh pathway activity is
enhanced because GLI3R expression is reduced due to the lack of
full-length GLI3 in the absence of Sufu (67). Thus, in combination with other
factors, such as Cul3-HIB/SPOP and SCFSlimb/β-TrCP, Sufu
functions as a sort of a buffer for regulating GLI proteins (as
shown in Fig. 2) that makes it
possible to alter Hh pathway activity with high sensitivity in
response to Hh signalling. Furthermore, Sufu is helpful for the
interpretation of the Hh level gradient into graded Hh pathway
activity.
Another important aspect of studies of GLI
regulation by Sufu is the exploration of the protein structure of
Sufu since it is helpful for explaining the physical interactions
between Sufu and GLI (93), which is
the basis for the regulations described above. As early as 2003, it
was reported that a 19-aa C-terminal fragment in Sufu was essential
for the interaction with a novel motif SYGH within residues 116–125
in GLI1, which is consistent with the observation that among the
three alternatively spliced transcripts of Sufu, two variants
lacking the C-terminal fragment failed to interact with and
suppress GLI1 (94). In addition,
GLI1 with mutations in the SYGH motif is no longer suppressed by
Sufu. Following arduous exploration, great progress has been made
in the study of the Sufu protein crystal structure (30,31), which
in both humans and Drosophila presents as a monomer
consisting of two globular domains-NTD (N-terminal domain) and CTD
(C-terminal domain)-with a short linker in between. An
‘open-to-closed’ transition mode of Sufu conformation during
interaction with GLI was proposed in both humans and
Drosophila. In the analysis of the crystal structure of
hSufu (with 286–345 aa deleted) in a complex with hGLI1 (112-128aa)
containing the SYGHLS motif, Sufu bound to the GLI1 peptide via
clamping to the NTD and the CTD, showing the closed conformation of
Sufu. This complex could be induced by Hh signalling to dissociate,
which most likely occurs by transformation of Sufu from a closed to
an open conformation, while the specific mechanism remains to be
defined. Moreover, the key residues mediating the association
between GLI and Sufu were further investigated through the analysis
of the crystal structure in combination with site-specific
mutagenesis.
To recapitulate the findings above, depending on the
physical interaction between Sufu and GLI in combination with other
factors, Sufu is engaged in regulating GLI protein along with the
signalling transduction from the cytoplasm into the nucleus,
modulating both the subcellular localization and stability of GLI
to exquisitely realize the graded activity of GLI proteins in
response to the Hh signalling gradient. These sophisticated
regulations of GLI by Sufu indicate a central position of GLI-Sufu
correlation downstream of Smo in the canonical mammalian Hh
signalling pathway.
In contrast to the relatively extensive
accomplishments in the regulation of GLI by Sufu, how Sufu itself
is regulated by other factors remains obscure, although there have
been some achievements in the study of Sufu regulation at the
post-transcriptional level. Given the critical suppressive role of
Sufu in the Hh pathway, it is conceivable that Sufu deactivation
should be one of the prerequisites of Hh pathway activation, as was
previously reported. Hh signalling, especially Shh, is capable of
destabilizing the Sufu protein through inducing Sufu
polyubiquitination at K257 and its subsequent degradation by the
proteasome. In contrast, a K257R mutant of Sufu is, to a large
extent, resistant to Hh signalling-induced degradation and exhibits
a more potent suppressive function against cell growth (73), suggesting that the much more reduced
stability of the Sufu protein is likely responsible for cancer
development when there is an excess of Hh ligand. Though
ubiquitin-proteasome pathway-mediated degradation is of great
importance for Sufu regulation at the post-translational level, the
exact ubiquitin ligase involved in this process remains unknown.
Further studies revealed that phosphorylation also plays an
important role in regulating Sufu stability. When Hh binds to Ptc1,
following recruitment and dissociation of the Sufu/GLI complex in
primary cilium, GLIs are released to enter the nucleus, while Sufu
was transported out of primary cilium and degraded via the
proteasome-dependent pathway. Dual phosphorylation at Ser-346 and
Ser-342 by PKA and GSK3β can prolong the stay of Sufu in primary
cilium and stabilize the protein (95). Recently, a study showed that the E3
ubiquitin ligase Skp1-Cul1-F-box protein Fbxl17 (F-box and
leucine-rich repeat protein 17) targets Sufu proteolysis in the
nucleus and promotes medulloblastoma (74), thus bridging the above ubiquitination
and phosphorylation modifications in Sufu protein regulation. In
response to Hh signalling, phosphorylation at Ser-346 and Ser-342
by PKA and GSK3β is debilitated, while Sufu dephosphorylation
facilitates its polyubiquitination at K257 by SCFFbxl17
and subsequent degradation, leading to Hh pathway activation.
Given the crucial modulatory effects of Sufu on GLI
in both the cytoplasm and the nucleus, and the existence of
different ubiquitin ligases responsible for GLI ubiquitination,
such as Cul3-HIB/SPOP and SCFSlimb/β-TrCP, other
ubiquitin ligases for Sufu ubiquitination may exist in the
cytoplasm that are distinct from SCFFbxl17, which
ubiquitinates nuclear Sufu. Additionally, our team recently
identified NIMA-related expressed kinase 2A (Nek2A) as a
Sufu-interacting protein by a yeast two-hybrid screen; the enzyme
could phosphorylate and stabilize Sufu, consequently dampening
Hh/GLI2 signalling (96,97). It is likely that different
post-translational modifications, including phosphorylation and
ubiquitination, are orchestrated to modulate the stability of the
Sufu protein, although additional studies are required to unveil
other regulations and the relationships between them.
In addition to regulating the stability of the Sufu
protein, Hh signalling also attenuates Sufu activity through
suppressing the maturation of Sufu mRNA. In Drosophila, the target
gene HIB (Hh-induced protein) of the Hh pathway is a substrate
recognition component of Cul3-based ubiquitin ligase. In response
to Hh signalling, the ubiquitin ligase Cul3-HIB, through poorly
clarified mechanisms, stimulates enrichment of the spliceosome
factor crooked neck (Crn) in the nucleus, which likely inhibits the
formation of functional sufu mRNA and ultimately weakens Sufu
activity. Moreover, a similar mechanism may also exist in mammals.
Spop, the mammalian homologue of HIB, is also capable of
downregulating Sufu levels in Drosophila (98). Other factors discovered to impact Sufu
mRNA include micro-RNAs. MicroRNA-378 can promote cell survival,
tumour growth and angiogenesis through attenuating the translation
of both Sufu and Fus-1 (99).
Furthermore, microRNA-214 can directly target Sufu mRNA and
compromise its function, which facilitates lung adenocarcinoma
metastasis and the epithelial-mesenchymal transition (EMT), which
are most likely attributed to deregulated Hh signalling (100). Shown here, microRNA-214 and
microRNA-378 display tumour-promoting roles, whereas some studies
reported tumour-suppressive roles in cervical and breast cancer
cells (101,102), implying the intrinsic complexity of
multi-cellular organisms and the considerable influence of the
microenvironment on gene functions.
Although the mechanism underlying Sufu regulation
remains obscure, its exploration is mounting, as the various
post-transcriptional regulations that have been identified have
begun to delineate the regulatory system focusing on Sufu. These
intricate regulatory mechanisms at different levels, through the
overall process of gene expression and at the post-translational
level, remain to be clarified.
While a substantial number of studies have examined
the Hh signalling pathway, some mechanisms within the Hh signalling
cascade remain elusive. For instance, i) how is Smo inhibited by
Ptc1? ii) How does the relay of Hh signalling from activated Smo to
GLI occur? iii) How is the specific transcription by GLI proteins
realized? and iv) Given the pivotal role of Sufu in Hh signalling,
the study on the regulation of Sufu itself seems somewhat
inadequate, with no studies reporting on the transcription of the
Sufu gene. In addition, what has been presented in this
review is merely part of the whole picture, especially since Sufu
is also implicated in the regulation of the Wnt pathway (103,104),
probably coordinating the Wnt and Hh pathways for their proper
functions. In addition, Sufu was first associated with human
immunity by its single-nucleotide polymorphism involvement in
graft-versus-host disease (105).
Furthermore, in a study on non-canonical Hh signalling, Sufu binds
to and stabilizes cellular nucleic acid-binding protein (CNBP) to
promote polyamine biosynthesis, which supports neuronal and
medulloblastoma cell growth (106),
suggesting a tumour-stimulating effect of Sufu. This seeming
contradiction is likely due to our general inclination to assign
definite functions to certain proteins within linear-pattern
cellular signalling pathways, which are in actuality more like
networks with proteins functioning in context-dependent manners.
Perhaps a panoramic view is needed to understand these
interconnected networks.
|
1
|
Nüsslein-Volhard C and Wieschaus E:
Mutations affecting segment number and polarity in Drosophila.
Nature. 287:795–801. 1980. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Briscoe J and Thérond PP: The mechanisms
of Hedgehog signalling and its roles in development and disease.
Nat Rev Mol Cell Biol. 14:416–429. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ingham PW, Nakano Y and Seger C:
Mechanisms and functions of Hedgehog signalling across the metazoa.
Nat Rev Genet. 12:393–406. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Varjosalo M and Taipale J: Hedgehog:
Functions and mechanisms. Genes Dev. 22:2454–2472. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Johnson RL, Rothman AL, Xie J, Goodrich
LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH Jr
and Scott MP: Human homolog of patched, a candidate gene for the
basal cell nevus syndrome. Science. 272:1668–1671. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hahn H, Wicking C, Zaphiropoulous PG,
Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E,
Unden AB, Gillies S, et al: Mutations of the human homolog of
Drosophila patched in the nevoid basal cell carcinoma syndrome.
Cell. 85:841–851. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Barakat MT, Humke EW and Scott MP:
Learning from Jekyll to control Hyde: Hedgehog signaling in
development and cancer. Trends Mol Med. 16:337–348. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Q, Gao G and Luo S: Hedgehog
signaling pathway and ovarian cancer. Chin J Cancer Res.
25:346–353. 2013.PubMed/NCBI
|
|
9
|
Zeng C, Wang Y, Lu Q, Chen J, Zhang J, Liu
T, Lv N and Luo S: SPOP suppresses tumorigenesis by regulating
Hedgehog/Gli2 signaling pathway in gastric cancer. J Exp Clin
Cancer Res. 33:752014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi C, Huang D, Lu N, Chen D, Zhang M, Yan
Y, Deng L, Lu Q, Lu H and Luo S: Aberrantly activated Gli2-KIF20A
axis is crucial for growth of hepatocellular carcinoma and predicts
poor prognosis. Oncotarget. 7:26206–26219. 2016.PubMed/NCBI
|
|
11
|
Shin K, Lim A, Zhao C, Sahoo D, Pan Y,
Spiekerkoetter E, Liao JC and Beachy PA: Hedgehog signaling
restrains bladder cancer progression by eliciting stromal
production of urothelial differentiation factors. Cancer Cell.
26:521–533. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fei DL, Sanchez-Mejias A, Wang Z, Flaveny
C, Long J, Singh S, Rodriguez-Blanco J, Tokhunts R, Giambelli C,
Briegel KJ, et al: Hedgehog signaling regulates bladder cancer
growth and tumorigenicity. Cancer Res. 72:4449–4458. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rimkus TK, Carpenter RL, Qasem S, Chan M
and Lo HW: Targeting the sonic hedgehog signaling pathway: Review
of smoothened and GLI inhibitors. Cancers (Basel). 8:pii: E22.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hui CC and Angers S: Gli proteins in
development and disease. Annu Rev Cell Dev Biol. 27:513–537. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Svärd J, Heby-Henricson K, Persson-Lek M,
Rozell B, Lauth M, Bergström A, Ericson J, Toftgård R and Teglund
S: Genetic elimination of suppressor of fused reveals an essential
repressor function in the mammalian hedgehog signaling pathway. Dev
Cell. 10:187–197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Préat T: Characterization of suppressor of
fused, a complete suppressor of the fused segment polarity gene of
Drosophila melanogaster. Genetics. 132:725–736. 1992.PubMed/NCBI
|
|
17
|
Babcock DT, Shi S, Jo J, Shaw M, Gutstein
HB and Galko MJ: Hedgehog signaling regulates nociceptive
sensitization. Curr Biol. 21:1525–1533. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Teperino R, Amann S, Bayer M, McGee SL,
Loipetzberger A, Connor T, Jaeger C, Kammerer B, Winter L, Wiche G,
et al: Hedgehog partial agonism drives Warburg-like metabolism in
muscle and brown fat. Cell. 151:414–426. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Teperino R, Aberger F, Esterbauer H, Riobo
N and Pospisilik JA: Canonical and non-canonical Hedgehog
signalling and the control of metabolism. Semin Cell Dev Biol.
33:81–92. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hooper JE: Distinct pathways for autocrine
and paracrine Wingless signalling in Drosophila embryos. Nature.
372:461–464. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pham A, Therond P, Alves G, Tournier FB,
Busson D, Lamour-Isnard C, Bouchon BL, Préat T and Tricoire H: The
suppressor of fused gene encodes a novel PEST protein involved in
Drosophila segment polarity establishment. Genetics. 140:587–598.
1995.PubMed/NCBI
|
|
22
|
Monnier V, Dussillol F, Alves G,
Lamour-Isnard C and Plessis A: Suppressor of fused links fused and
Cubitus interruptus on the hedgehog signalling pathway. Curr Biol.
8:583–586. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Delattre M, Briand S, Paces-Fessy M and
Blanchet-Tournier MF: The Suppressor of fused gene, involved in
Hedgehog signal transduction in Drosophila, is conserved in
mammals. Dev Genes Evol. 209:294–300. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pearse RV II, Collier LS, Scott MP and
Tabin CJ: Vertebrate homologs of Drosophila suppressor of fused
interact with the gli family of transcriptional regulators. Dev
Biol. 212:323–336. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ding Q, Fukami Si, Meng X, Nishizaki Y,
Zhang X, Sasaki H, Dlugosz A, Nakafuku M and Hui Cc: Mouse
suppressor of fused is a negative regulator of sonic hedgehog
signaling and alters the subcellular distribution of Gli1. Curr
Biol. 9:1119–1122. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Simon-Chazottes D, Paces-Fessy M,
Lamour-Isnard C, Guénet JL and Blanchet-Tournier MF: Genomic
organization, chromosomal assignment, and expression analysis of
the mouse suppressor of fused gene (Sufu) coding a Gli protein
partner. Mamm Genome. 11:614–621. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Stone DM, Murone M, Luoh S, Ye W, Armanini
MP, Gurney A, Phillips H, Brush J, Goddard A, de Sauvage FJ and
Rosenthal A: Characterization of the human suppressor of fused, a
negative regulator of the zinc-finger transcription factor Gli. J
Cell Sci. 112:4437–4448. 1999.PubMed/NCBI
|
|
28
|
Rasheed BK, McLendon RE, Friedman HS,
Friedman AH, Fuchs HE, Bigner DD and Bigner SH: Chromosome 10
deletion mapping in human gliomas: A common deletion region in
10q25. Oncogene. 10:2243–2246. 1995.PubMed/NCBI
|
|
29
|
Gray IC, Phillips SM, Lee SJ, Neoptolemos
JP, Weissenbach J and Spurr NK: Loss of the chromosomal region
10q23-25 in prostate cancer. Cancer Res. 55:4800–4803.
1995.PubMed/NCBI
|
|
30
|
Cherry AL, Finta C, Karlström M, Jin Q,
Schwend T, Astorga-Wells J, Zubarev RA, Del Campo M, Criswell AR,
de Sanctis D, et al: Structural basis of SUFU-GLI interaction in
human Hedgehog signalling regulation. Acta Crystallogr D Biol
Crystallogr. 69:2563–2579. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Fu L, Qi X, Zhang Z, Xia Y, Jia
J, Jiang J, Zhao Y and Wu G: Structural insight into the mutual
recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun. 4:26082013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee Y, Kawagoe R, Sasai K, Li Y, Russell
HR, Curran T and McKinnon PJ: Loss of suppressor-of-fused function
promotes tumorigenesis. Oncogene. 26:6442–6447. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ogino S, Gulley ML, den Dunnen JT and
Wilson RB: Association for Molecular Patholpogy Training and
Education Committtee: Standard mutation nomenclature in molecular
diagnostics: Practical and educational challenges. J Mol Diagn.
9:1–6. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brugières L, Remenieras A, Pierron G,
Varlet P, Forget S, Byrde V, Bombled J, Puget S, Caron O, Dufour C,
et al: High frequency of germline SUFU mutations in children with
desmoplastic/nodular medulloblastoma younger than 3 years of age. J
Clin Oncol. 30:2087–2093. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ng D, Stavrou T, Liu L, Taylor MD, Gold B,
Dean M, Kelley MJ, Dubovsky EC, Vezina G, Nicholson HS, et al:
Retrospective family study of childhood medulloblastoma. Am J Med
Genet A. 134:399–403. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Taylor MD, Liu L, Raffel C, Hui CC,
Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, et
al: Mutations in SUFU predispose to medulloblastoma. Nat Genet.
31:306–310. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Smith MJ, Beetz C, Williams SG, Bhaskar
SS, O'Sullivan J, Anderson B, Daly SB, Urquhart JE, Bholah Z, Oudit
D, et al: Germline mutations in SUFU cause Gorlin
syndrome-associated childhood medulloblastoma and redefine the risk
associated with PTCH1 mutations. J Clin Oncol. 32:4155–4161. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Slade I, Murray A, Hanks S, Kumar A,
Walker L, Hargrave D, Douglas J, Stiller C, Izatt L and Rahman N:
Heterogeneity of familial medulloblastoma and contribution of
germline PTCH1 and SUFU mutations to sporadic medulloblastoma. Fam
Cancer. 10:337–342. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Brugières L, Pierron G, Chompret A,
Paillerets BB, Di Rocco F, Varlet P, Pierre-Kahn A, Caron O, Grill
J and Delattre O: Incomplete penetrance of the predisposition to
medulloblastoma associated with germ-line SUFU mutations. J Med
Genet. 47:142–144. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Heby-Henricson K, Bergström A, Rozell B,
Toftgård R and Teglund S: Loss of Trp53 promotes medulloblastoma
development but not skin tumorigenesis in Sufu heterozygous mutant
mice. Mol Carcinog. 51:754–760. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Svärd J, Rozell B, Toftgård R and Teglund
S: Tumor suppressor gene co-operativity in compound Patched1 and
suppressor of fused heterozygous mutant mice. Mol Carcinog.
48:408–419. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Koch A, Waha A, Hartmann W, Milde U,
Goodyer CG, Sörensen N, Berthold F, Digon-Söntgerath B, Krätzschmar
J, Wiestler OD and Pietsch T: No evidence for mutations or altered
expression of the Suppressor of Fused gene (SUFU) in primitive
neuroectodermal tumours. Neuropathol Appl Neurobiol. 30:532–539.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Scott DK, Straughton D, Cole M, Bailey S,
Ellison DW and Clifford SC: Identification and analysis of tumor
suppressor loci at chromosome 10q23.3-10q25.3 in medulloblastoma.
Cell Cycle. 5:2381–2389. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zabidi MA and Stark A: Regulatory
enhancer-core-promoter communication via transcription factors and
cofactors. Trends Genet. 32:801–814. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Spurrell CH, Dickel DE and Visel A: The
ties that bind: Mapping the dynamic enhancer-promoter interactome.
Cell. 167:1163–1166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Aavikko M, Li SP, Saarinen S, Alhopuro P,
Kaasinen E, Morgunova E, Li Y, Vesanen K, Smith MJ, Evans DG, et
al: Loss of SUFU function in familial multiple meningioma. Am J Hum
Genet. 91:520–526. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lim CB, Prêle CM, Cheah HM, Cheng YY,
Klebe S, Reid G, Watkins DN, Baltic S, Thompson PJ and Mutsaers SE:
Mutational analysis of hedgehog signaling pathway genes in human
malignant mesothelioma. PLoS One. 8:e666852013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tostar U, Malm CJ, Meis-Kindblom JM,
Kindblom LG, Toftgård R and Undén AB: Deregulation of the hedgehog
signalling pathway: A possible role for the PTCH and SUFU genes in
human rhabdomyoma and rhabdomyosarcoma development. J Pathol.
208:17–25. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yin WC, Li ZJ and Hui CC: BCC or not: Sufu
keeps it in check. Oncoscience. 2:77–78. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Schulman JM, Oh DH, Sanborn JZ, Pincus L,
McCalmont TH and Cho RJ: Multiple hereditary infundibulocystic
basal cell carcinoma syndrome associated with a germline SUFU
mutation. JAMA Dermatol. 152:323–327. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Reifenberger J, Wolter M, Knobbe CB,
Köhler B, Schönicke A, Scharwächter C, Kumar K, Blaschke B, Ruzicka
T and Reifenberger G: Somatic mutations in the PTCH, SMOH, SUFUH
and TP53 genes in sporadic basal cell carcinomas. Br J Dermatol.
152:43–51. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mann K, Magee J, Guillaud-Bataille M,
Blondel C, Bressac-de Paillerets B, Yeatman J and Winship I:
Multiple skin hamartomata: A possible novel clinical presentation
of SUFU neoplasia syndrome. Fam Cancer. 14:151–155. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sheng T, Li C, Zhang X, Chi S, He N, Chen
K, McCormick F, Gatalica Z and Xie J: Activation of the hedgehog
pathway in advanced prostate cancer. Mol Cancer. 3:292004.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pastorino L, Ghiorzo P, Nasti S,
Battistuzzi L, Cusano R, Marzocchi C, Garrè ML, Clementi M and
Scarrà GB: Identification of a SUFU germline mutation in a family
with Gorlin syndrome. Am J Med Genet A. 149A:1539–1543. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ruel L and Thérond PP: Variations in
Hedgehog signaling: Divergence and perpetuation in Sufu regulation
of Gli. Genes Dev. 23:1843–1848. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
McDermott A, Gustafsson M, Elsam T, Hui
CC, Emerson CP Jr and Borycki AG: Gli2 and Gli3 have redundant and
context-dependent function in skeletal muscle formation.
Development. 132:345–357. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pan Y, Wang C and Wang B: Phosphorylation
of Gli2 by protein kinase A is required for Gli2 processing and
degradation and the Sonic Hedgehog-regulated mouse development. Dev
Biol. 326:177–189. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Goetz SC and Anderson KV: The primary
cilium: A signalling centre during vertebrate development. Nat Rev
Genet. 11:331–344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wen X, Lai CK, Evangelista M, Hongo JA, de
Sauvage FJ and Scales SJ: Kinetics of hedgehog-dependent
full-length Gli3 accumulation in primary cilia and subsequent
degradation. Mol Cell Biol. 30:1910–1922. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Endoh-Yamagami S, Evangelista M, Wilson D,
Wen X, Theunissen JW, Phamluong K, Davis M, Scales SJ, Solloway MJ,
de Sauvage FJ and Peterson AS: The mammalian Cos2 homolog Kif7
plays an essential role in modulating Hh signal transduction during
development. Curr Biol. 19:1320–1326. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cheung HO, Zhang X, Ribeiro A, Mo R,
Makino S, Puviindran V, Law KK, Briscoe J and Hui CC: The kinesin
protein Kif7 is a critical regulator of Gli transcription factors
in mammalian hedgehog signaling. Sci Signal. 2:ra292009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu X, Wang X, Du W, Chen L, Wang G, Cui
Y, Liu Y, Dou Z, Wang H, Zhang P, et al: Suppressor of fused (Sufu)
represses Gli1 transcription and nuclear accumulation, inhibits
glioma cell proliferation, invasion and vasculogenic mimicry,
improving glioma chemo-sensitivity and prognosis. Oncotarget.
5:11681–11694. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Szczepny A, Wagstaff KM, Dias M, Gajewska
K, Wang C, Davies RG, Kaur G, Ly-Huynh J, Loveland KL and Jans DA:
Overlapping binding sites for importin b1 and suppressor of fused
(SuFu) on glioma-associated oncogene homologue 1 (Gli1) regulate
its nuclear localization. Biochem J. 461:469–476. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shi Q, Han Y and Jiang J: Suppressor of
fused impedes Ci/Gli nuclear import by opposing Trn/Kapb2 in
Hedgehog signaling. J Cell Sci. 127:1092–1103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Han Y, Shi Q and Jiang J: Multisite
interaction with Sufu regulates Ci/Gli activity through distinct
mechanisms in Hh signal transduction. Proc Natl Acad Sci USA.
112:6383–6388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang C, Pan Y and Wang B: Suppressor of
fused and Spop regulate the stability, processing and function of
Gli2 and Gli3 full-length activators but not their repressors.
Development. 137:2001–2009. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Tempé D, Casas M, Karaz S,
Blanchet-Tournier MF and Concordet JP: Multisite protein kinase A
and glycogen synthase kinase 3beta phosphorylation leads to Gli3
ubiquitination by SCFbetaTrCP. Mol Cell Biol. 26:4316–4326. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang B and Li Y: Evidence for the direct
involvement of {beta}TrCP in Gli3 protein processing. Proc Natl
Acad Sci USA. 103:33–38. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Pan Y, Bai CB, Joyner AL and Wang B: Sonic
hedgehog signaling regulates Gli2 transcriptional activity by
suppressing its processing and degradation. Mol Cell Biol.
26:3365–3377. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang B, Fallon JF and Beachy PA:
Hedgehog-regulated processing of Gli3 produces an
anterior/posterior repressor gradient in the developing vertebrate
limb. Cell. 100:423–434. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tukachinsky H, Lopez LV and Salic A: A
mechanism for vertebrate Hedgehog signaling: Recruitment to cilia
and dissociation of SuFu-Gli protein complexes. J Cell Biol.
191:415–428. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yue S, Chen Y and Cheng SY: Hedgehog
signaling promotes the degradation of tumor suppressor Sufu through
the ubiquitin-proteasome pathway. Oncogene. 28:492–499. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Raducu M, Fung E, Serres S, Infante P,
Barberis A, Fischer R, Bristow C, Thézénas ML, Finta C,
Christianson JC, et al: SCF (Fbxl17) ubiquitylation of Sufu
regulates Hedgehog signaling and medulloblastoma development. EMBO
J. 35:1400–1416. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang Z, Shen L, Law K, Zhang Z, Liu X,
Hua H, Li S, Huang H, Yue S, Hui CC and Cheng SY: Suppressor of
fused chaperones Gli proteins to generate transcriptional responses
to sonic hedgehog signaling. Mol Cell Biol. 37:pii: e00421. –16.
2017. View Article : Google Scholar
|
|
76
|
Santos N and Reiter JF: A central region
of Gli2 regulates its localization to the primary cilium and
transcriptional activity. J Cell Sci. 127:1500–1510. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li BI, Matteson PG, Ababon MF, Nato AQ Jr,
Lin Y, Nanda V, Matise TC and Millonig JH: The orphan GPCR, Gpr161,
regulates the retinoic acid and canonical Wnt pathways during
neurulation. Dev Biol. 402:17–31. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Pal K, Hwang SH, Somatilaka B, Badgandi H,
Jackson PK, DeFea K and Mukhopadhyay S: Smoothened determines
b-arrestin-mediated removal of the G protein-coupled receptor
Gpr161 from the primary cilium. J Cell Biol. 212:861–875. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Blair HJ, Tompson S, Liu YN, Campbell J,
MacArthur K, Ponting CP, Ruiz-Perez VL and Goodship JA: Evc2 is a
positive modulator of Hedgehog signalling that interacts with Evc
at the cilia membrane and is also found in the nucleus. BMC Biol.
9:142011. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yang C, Chen W, Chen Y and Jiang J:
Smoothened transduces Hedgehog signal by forming a complex with
Evc/Evc2. Cell Res. 22:1593–1604. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Caparrós-Martín JA, Valencia M, Reytor E,
Pacheco M, Fernandez M, Perez-Aytes A, Gean E, Lapunzina P, Peters
H, Goodship JA and Ruiz-Perez VL: The ciliary Evc/Evc2 complex
interacts with Smo and controls Hedgehog pathway activity in
chondrocytes by regulating Sufu/Gli3 dissociation and Gli3
trafficking in primary cilia. Hum Mol Genet. 22:124–139. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Jia J, Kolterud A, Zeng H, Hoover A,
Teglund S, Toftgård R and Liu A: Suppressor of Fused inhibits
mammalian Hedgehog signaling in the absence of cilia. Dev Biol.
330:452–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zeng H, Jia J and Liu A: Coordinated
translocation of mammalian Gli proteins and suppressor of fused to
the primary cilium. PLoS One. 5:e159002010. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chen MH, Wilson CW, Li YJ, Law KK, Lu CS,
Gacayan R, Zhang X, Hui CC and Chuang PT: Cilium-independent
regulation of Gli protein function by Sufu in Hedgehog signaling is
evolutionarily conserved. Genes Dev. 23:1910–1928. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ohlmeyer JT and Kalderon D: Hedgehog
stimulates maturation of Cubitus interruptus into a labile
transcriptional activator. Nature. 396:749–753. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Paces-Fessy M, Boucher D, Petit E,
Paute-Briand S and Blanchet-Tournier MF: The negative regulator of
Gli, Suppressor of fused (Sufu), interacts with SAP18, Galectin3
and other nuclear proteins. Biochem J. 378:353–362. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Cheng SY and Bishop JM: Suppressor of
Fused represses Gli-mediated transcription by recruiting the
SAP18-mSin3 corepressor complex. Proc Natl Acad Sci USA.
99:5442–5447. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Lin C, Yao E, Wang K, Nozawa Y, Shimizu H,
Johnson JR, Chen JN, Krogan NJ and Chuang PT: Regulation of Sufu
activity by p66b and Mycbp provides new insight into vertebrate
Hedgehog signaling. Genes Dev. 28:2547–2563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Barnfield PC, Zhang X, Thanabalasingham V,
Yoshida M and Hui CC: Negative regulation of Gli1 and Gli2
activator function by Suppressor of fused through multiple
mechanisms. Differentiation. 73:397–405. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kogerman P, Grimm T, Kogerman L, Krause D,
Undén AB, Sandstedt B, Toftgård R and Zaphiropoulos PG: Mammalian
suppressor-of-fused modulates nuclear-cytoplasmic shuttling of
Gli-1. Nat Cell Biol. 1:312–319. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
91
|
Ruiz i Altaba A, Sánchez P and Dahmane N:
Gli and hedgehog in cancer: Tumours, embryos and stem cells. Nat
Rev Cancer. 2:361–372. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
92
|
Humke EW, Dorn KV, Milenkovic L, Scott MP
and Rohatgi R: The output of Hedgehog signaling is controlled by
the dynamic association between Suppressor of Fused and the Gli
proteins. Genes Dev. 24:670–682. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Merchant M, Vajdos FF, Ultsch M, Maun HR,
Wendt U, Cannon J, Desmarais W, Lazarus RA, de Vos AM and de
Sauvage FJ: Suppressor of fused regulates Gli activity through a
dual binding mechanism. Mol Cell Biol. 24:8627–8641. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Dunaeva M, Michelson P, Kogerman P and
Toftgard R: Characterization of the physical interaction of Gli
proteins with SUFU proteins. J Biol Chem. 278:5116–5122. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Chen Y, Yue S, Xie L, Pu XH, Jin T and
Cheng SY: Dual Phosphorylation of suppressor of fused (Sufu) by PKA
and GSK3beta regulates its stability and localization in the
primary cilium. J Biol Chem. 286:13502–13511. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wang Y, Li Y, Hu G, Huang X, Rao H, Xiong
X, Luo Z, Lu Q and Luo S: Nek2A phosphorylates and stabilizes SuFu:
A new strategy of Gli2/Hedgehog signaling regulatory mechanism.
Cell Signal. 28:1304–1313. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhou F, Huang D, Li Y, Hu G, Rao H, Lu Q,
Luo S and Wang Y: Nek2A/SuFu feedback loop regulates Gli-mediated
Hedgehog signaling pathway. Int J Oncol. 50:373–380. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Liu C, Zhou Z, Yao X, Chen P, Sun M, Su M,
Chang C, Yan J, Jiang J and Zhang Q: Hedgehog signaling
downregulates suppressor of fused through the HIB/SPOP-Crn axis in
Drosophila. Cell Res. 24:595–609. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lee DY, Deng Z, Wang CH and Yang BB:
MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis
by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA.
104:20350–20355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Long H, Wang Z, Chen J, Xiang T, Li Q,
Diao X and Zhu B: microRNA-214 promotes epithelial-mesenchymal
transition and metastasis in lung adenocarcinoma by targeting the
suppressor-of-fused protein (Sufu). Oncotarget. 6:38705–38718.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Peng RQ, Wan HY, Li HF, Liu M, Li X and
Tang H: MicroRNA-214 suppresses growth and invasiveness of cervical
cancer cells by targeting
UDP-N-acetyl-a-D-galactosamine:polypeptide
N-acetylgalactosaminyltransferase 7. J Biol Chem. 287:14301–14309.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Browne G, Dragon JA, Hong D, Messier TL,
Gordon JA, Farina NH, Boyd JR, VanOudenhove JJ, Perez AW, Zaidi SK,
et al: MicroRNA-378-mediated suppression of Runx1 alleviates the
aggressive phenotype of triple-negative MDA-MB-231 human breast
cancer cells. Tumour Biol. 37:8825–8839. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Min TH, Kriebel M, Hou S and Pera EM: The
dual regulator Sufu integrates Hedgehog and Wnt signals in the
early Xenopus embryo. Dev Biol. 358:262–276. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Peng Y, Zhang X, Ma Q, Yan R, Qin Y, Zhao
Y, Cheng Y, Yang M, Wang Q, Feng X, et al: MiRNA-194 activates the
Wnt/β-catenin signaling pathway in gastric cancer by targeting the
negative Wnt regulator, SUFU. Cancer Lett. 385:117–127. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Bari R, Hartford C, Chan WK, Vong Q, Li Y,
Gan K, Zhou Y, Cheng C, Kang G, Shurtleff S, et al: Genome-wide
single-nucleotide polymorphism analysis revealed SUFU suppression
of acute graft-versus-host disease through downregulation of HLA-DR
expression in recipient dendritic cells. Sci Rep. 5:110982015.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
D'Amico D, Antonucci L, Di Magno L, Coni
S, Sdruscia G, Macone A, Miele E, Infante P, Di Marcotullio L, De
Smaele E, et al: Non-canonical Hedgehog/AMPK-mediated control of
polyamine metabolism supports neuronal and medulloblastoma cell
growth. Dev Cell. 35:21–35. 2015. View Article : Google Scholar : PubMed/NCBI
|