Introduction
Hepatocellular carcinoma (HCC), the most common type
of liver cancer in adults, occurs more frequently in males compared
with females (1). The incidence of
HCC is increasing worldwide due to an increased rate of hepatitis
virus infection (1). Chronic liver
inflammation, due to hepatitis infection or exposure to toxins, is
thought to be the leading cause of HCC (2,3). Many
previous studies suggest that microRNAs (miRNAs/miRs), small
non-coding RNA molecules, ~22 nucleotides in length (4), serve an essential role in the
progression of HCC (4–7). Notably, recent studies of serum from
patients with liver diseases have indicated that circulating miRNAs
are a novel class of biomarkers of HCC and other liver diseases
(6,7).
These studies suggest serum miRNAs may be potential biomarkers for
HCC. Therefore, it is critical to identify novel miRNA biomarkers
to help predict the progression of HCC.
Human miR-760 is involved in the pathogenesis of
various human diseases and its expression varies dramatically in
numerous pathological conditions. In patients with oral submucous
fibrosis (OSF) the expression of miR-760 is decreased in OSF tissue
compared with normal tissue (8). In T
cells of patients with rheumatoid arthritis the expression of
miR-760 is increased compared with that in T cells from healthy
individuals (9). In addition, miR-760
has been identified as a novel biomarker to predict the progression
of various cancer types, including gastric cancer (GC) (10), breast cancer (11–13),
colorectal cancer (CRC) (14), colon
cancer (CC) and ovarian cancer (OC) (15). Based on an RNA-seq analysis of bone
marrow cells from patients with GC, the expression of miR-760 in
bone marrow and tumor tissue is significantly lower in stage IV
patients in comparison with stage I patients (10). A higher miR-760 expression was also
associated with better overall survival (OS) in patients with GC
(10). This may be due to the direct
interaction of miR-760 with histone mRNA, resulting in the
decreased expression of histone cluster 1 H3 family member D
(10). Furthermore, an in
vitro study indicated that miR-760 regulates breast cancer stem
cell metastasis and gene expression by targeting Nanog homeobox
dependent pathways (16). miR-760 is
also significantly downregulated in breast cancer cells that
exhibit chemoresistance (12). By
contrast, overexpression of miR-760 induces the resistance of human
breast cancer cells to doxorubicin (11). In human CC cells and replicatively
senescent human lung fibroblast cells, miR-760 expression is
upregulated and regulates protein kinase CK2 α subunit expression
(17). In Egyptian patients with CRC,
miR-760 is significantly downregulated in CRC tissue compared with
healthy tissue and the expression of miR-760 is inversely
associated with advanced CRC stages (18). Previously, miR-760 has been detected
in the plasma of patients with CRC and further investigation
demonstrated that plasma miR-760 may be a potential novel biomarker
for the early stages of CRC (14). A
systematic review and meta-analysis in gastrointestinal cancer
types has also indicated that miR-760 is downregulated and
associated with poor prognosis (19).
By contrast, a quantitative polymerase chain reaction assay for
miR-760 in OC tissues revealed a markedly higher expression of
miR-760 in tumor tissue compared with healthy tissue. Additionally,
this assay demonstrated that miR-760 directly regulates the
expression of PH domain and leucine rich repeat protein phosphatase
2 (PHLPP2), which subsequently results in tumor cell proliferation
(15). The low expression of miR-760
is strongly associated with a better phenotype and prognosis for
patients with OC, suggesting that miR-760 serves an oncogenic role
in human OC progression (15).
However, to the best of our knowledge, the role of miR-760 in
patients with HCC has not been studied.
The Kaplan-Meier (KM) plotter miRpower analysis tool
has been reported to analyze the prognostic role of miRNAs in
breast cancer (20). The current
study used the KM plotter database to investigate the prognostic
role of miR-760 expression in HCC patients.
Materials and methods
To analyze the prognostic role of miRNA expression
in HCC, the current study used the KM plotter database and miRpower
analysis tool to study the association of miR-760 expression with
OS in different analysis selections (http://kmplot.com/analysis/index.php?p=service&cancer=liver_mirna).
The KM plotter database has combined the published miRNA
expression, OS and clinical data from The Cancer Genome Atlas
(http://cancergenome.nih.gov), Gene
Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), European
Genome-Phenome Archive (https://www.ebi.ac.uk/ega/home) and PubMed (http://www.pubmed.com) (21). The database included the miRNA
expression and OS rates (with a 10 year follow-up) of 614 patients
with HCC.
The KM plotter analysis tool was used to obtain
Kaplan-Meier plots for the OS rate of patients with HCC.
hsa-miR-760 was entered into the database and the number-at-risk
was displayed under the main panel of the OS. Based on the
expression of miR-760, patients were divided into two groups.
Patients with miR-760 expression higher than the median were pooled
into the group with high expression, while the patients with
miR-760 expression lower than the median were pooled into the group
with low expression. Other statistical outcomes calculated from the
database, including hazard ratio (HR), 95% confidence intervals and
log rank P-values, were also included in the figures and table in
the current study. P<0.05 was considered to indicate a
statistically significant difference.
Results
The current study used the KM plotter database and
analysis tool to determine the prognostic value of miR-760 for HCC.
Survival curves were generated for patients with HCC from the
RNA-seq platform (n=372; Fig. 1A) and
from the non-commercial spotted microarray (n=164; Fig. 1B). Compared with high expression, low
expression of miR-760 was associated with higher OS for all
patients with HCC from both platforms (Fig. 1). In the RNA-seq platform, the HR for
miR-760 was 2.04 [95% confidence interval (CI)=1.44–2.89] and
P=4.9×10−5 (Fig. 1A), and
in the non-commercial spotted platform the HR for miR-760 was 1.71
(95% CI=1.05–2.76) and P=0.028 (Fig.
1B).
Subsequently, the current study analyzed the
expression of miR-760 in patients with HCC of different sex
(Table I and Fig. 2), different clinical stages (Table I and Fig.
3) and different ethnicities (Table
I and Fig. 4) using the data from
the RNA-Seq platform. As demonstrated in Fig. 2 and Table
I, compared with high miR-760 expression, low expression of
miR-760 was associated with a higher OS in male patients with HCC
[hazard ratio (HR)=2.36 (1.51–3.67), P=9.2×10−5;
Fig. 2A], but not in female patients
with HCC [HR=1.58 (CI=0.89–2.82), P=0.11; Fig. 2B]. As demonstrated in Fig. 3 and Table
I, compared with high miR-760 expression, low expression of
miR-760 was associated with a higher OS in stage I patients
[HR=2.19 (CI=1.18–4.05), P=0.01; Fig.
3A], stage II patients [HR=3.44 (CI=1.49–7.93), P=0.002;
Fig. 3B] and stage III patients
[HR=2.29 (CI=1.18–4.44), P=0.01; Fig.
3C]. Stage IV patients was excluded due to a low population.
Additionally, compared with high expression, low miR-760 expression
was associated with a higher OS in Asian patients and Caucasian
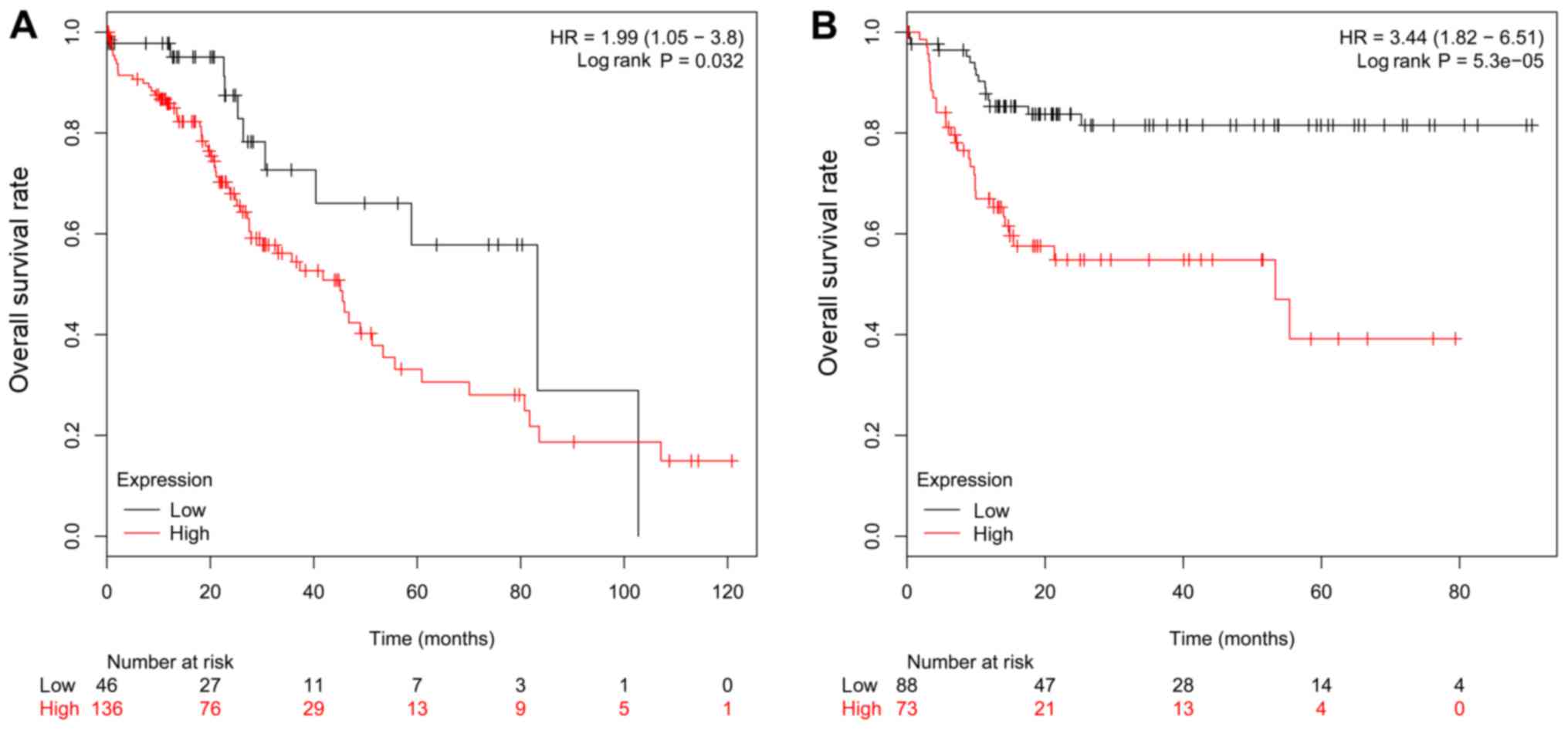
patients (Fig. 4 and Table I). As demonstrated in Fig. 4 and Table
I, in the 393 patients with complete ethnicity information (29
patients were excluded in this study due to the missing of
ethnicity information), the patients with low miR-760 expression
was marginally associated with higher OS in Caucasian patients
[HR=1.99 (CI-1.05–3.8), P=0.032; Fig.
4A] and was strongly associated with higher OS in Asian
patients [HR=3.44 (CI=1.82–6.51), P=5.3×10−5; Fig. 4B].
 | Table I.Association of miR-760 expression and
patients with HCC with different clinical variables. |
Table I.
Association of miR-760 expression and
patients with HCC with different clinical variables.
| Clinical
variables | Cases, n | HR | 95% CI | P-value |
|---|
| Sex |
|
|
|
|
|
Male | 253 | 2.36 | 1.51–3.67 | <0.001 |
|
Female | 119 | 1.58 | 0.89–2.82 | 0.115 |
| Clinical stage |
|
|
|
|
| I | 172 | 2.19 | 1.18–4.05 | 0.010 |
| II | 86 | 3.44 | 1.49–7.93 | 0.002 |
|
III | 85 | 2.29 | 1.18–4.44 | 0.011 |
| Ethnicity |
|
|
|
|
|
Caucasian | 182 | 1.99 | 1.05–3.80 | 0.032 |
|
Asian | 161 | 3.44 | 1.82–6.51 | <0.001 |
Discussion
As the primary cancer of the liver, HCC is one of
the most prevalent cancer types and affects more than
5×105 individuals globally every year (22). HCC is associated with a poor prognosis
rate; the five-year survival rate of patients with <10%
(23). This may be due to various
risk factors and limited approaches for early diagnosis (22,23). Liver
transplantation and resection are commonly used as the most
effective clinical therapies for HCC worldwide (24). However, the worldwide incidence rate
of HCC and HCC-associated cases of mortality are very high,
particularly in developing countries, including China and countries
in Africa (2). Previously, it has
been identified that various protein-coding genes serve
indispensable roles in the onset and development of HCC in humans
(25). HCC is commonly not detected
until it is in the advanced stage (2,23,24), therefore identifying more sensitive
biomarkers for early stage HCC may be beneficial for earlier
diagnosis.
Recently, many fundamental and clinical studies have
identified that miRNAs, including miR-21, −122, −221/222, −520 and
−657, are involved in the progression of HCC (5,26–28). These studies may assist in the
identification of novel biomarkers for early stage HCC and may also
be beneficial in the development of precision therapy for patients
with HCC. Notably, miRNAs have also been detected in the serum of
patients with HCC (29,30). Thus, monitoring serum miRNA profiles
may be a convenient approach to identify early changes in the
pathological functions of the liver during the onset and
development of HCC.
The current study investigated the clinical value of
miR-760 in predicting the development of HCC and the OS rate. The
current data indicated that low expression of miR-760, compared
with high expression, was associated with higher OS for patients
with HCC and suggested that miR-760 may be a novel biomarker for
HCC that serves as an oncogene in HCC development. In the
development of non-small cell lung cancer (NSCLC), miR-760
suppresses the proliferation and metastasis of NSCLC by directly
decreasing the expression of the proto-oncogene, tyrosine-protein
kinase ROS (31). Furthermore, in
human CRC, miR-760 attenuates tumor progression by targeting the
basic leucine zipper transcriptional factor ATF-like 3 dependent
signaling pathways (32). Notably,
miR-760 has been demonstrated to serve different roles in the
development of OC. The expression of miR-760 is increased in the
tumor tissue of patients with OC compared with that in healthy
tissue from patients with OC and miR-670 promotes proliferation of
OC cells via the downregulation of PHLPP2 expression (15). Similar to OC development, miR-760 may
negatively regulate the expression of cancer suppressing genes,
including PHLPP2, to promote the progression of HCC. This potential
function of miR-760 requires further investigation to provide a
better understanding of the mechanistic details involved. The
current detailed analysis of miR-760 expression in different sexes
and ethnicities suggested that miR-760 would be more sensitive for
predicting prognosis in male patients compared with female patients
and more sensitive in Asian patients compared with Caucasian
patients. Notably, compared with high miR-760 expression, low
expression of miR-760 was strongly associated with a higher OS in
patients with HCC in stage I, II and III.
In conclusion, the current study demonstrated the
distinct prognostic role of miR-760 for HCC. Compared with high
expression, low miR-760 expression was associated with better OS
for all patients with HCC, particularly for male patients and Asian
patients. Therefore, the current results suggest that miR-760 may
be a novel biomarker for patients with HCC both in the early and
advanced stages. Furthermore, by targeting miR-760, a novel
approach for earlier diagnosis of HCC may be achieved, which is
essential to improve the survival rate of patients with HCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Key Research
Project of Shandong Province (grant no. 2017GSF218087).
Availability of data and materials
All data generated or analyzed during the present
study are included in this article.
Authors' contributions
DS, JL, CH, QZ, XW, ZZ and SH designed the
experiments and collected the data; DS, JL, ZZ and SH contributed
to the data analysis and wrote the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
HR
|
hazard ratio
|
|
KM
|
Kaplan-Meier
|
|
OS
|
overall survival
|
|
miR
|
microRNA
|
|
OSF
|
oral submucous fibrosis
|
|
GC
|
gastric cancer
|
|
CRC
|
colorectal cancer
|
|
CC
|
colon cancer
|
|
OC
|
ovarian cancer
|
|
PHLPP2
|
PH domain and leucine rich repeat
protein phosphatase 2
|
|
NSCLC
|
non-small cell lung cancer
|
References
|
1
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273 e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gomaa AI, Khan SA, Toledano MB, Waked I
and Taylor-Robinson SD: Hepatocellular carcinoma: Epidemiology,
risk factors and pathogenesis. World J Gastroenterol. 14:4300–4308.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rapisarda V, Loreto C, Malaguarnera M,
Ardiri A, Proiti M, Rigano G, Frazzetto E, Ruggeri MI, Malaguarnera
G, Bertino N, et al: Hepatocellular carcinoma and the risk of
occupational exposure. World J Hepatol. 8:573–590. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Felekkis K, Touvana E, Stefanou CH and
Deltas C: microRNAs: A newly described class of encoded molecules
that play a role in health and disease. Hippokratia. 14:236–240.
2010.PubMed/NCBI
|
|
5
|
Hung CH, Chiu YC, Chen CH and Hu TH:
MicroRNAs in hepatocellular carcinoma: Carcinogenesis, progression,
and therapeutic target. Biomed Res Int. 2014:4864072014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ding Y, Yan JL, Fang AN, Zhou WF and Huang
L: Circulating miRNAs as novel diagnostic biomarkers in
hepatocellular carcinoma detection: A meta-analysis based on 24
articles. Oncotarget. 8:66402–66413. 2017.PubMed/NCBI
|
|
7
|
Loosen SH, Schueller F, Trautwein C, Roy S
and Roderburg C: Role of circulating microRNAs in liver diseases.
World J Hepatol. 9:586–594. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chickooree D, Zhu K, Ram V, Wu HJ, He ZJ
and Zhang S: A preliminary microarray assay of the miRNA expression
signatures in buccal mucosa of oral submucous fibrosis patients. J
Oral Pathol Med. 45:691–697. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lai NS, Yu HC, Tung CH, Huang KY, Huang HB
and Lu MC: The role of aberrant expression of T cell miRNAs
affected by TNF-α in the immunopathogenesis of rheumatoid
arthritis. Arthritis Res Ther. 19:2612017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iwaya T, Fukagawa T, Suzuki Y, Takahashi
Y, Sawada G, Ishibashi M, Kurashige J, Sudo T, Tanaka F, Shibata K,
et al: Contrasting expression patterns of histone mRNA and microRNA
760 in patients with gastric cancer. Clin Cancer Res. 19:6438–6449.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu SH, Wang CH, Huang ZJ, Liu F, Xu CW, Li
XL and Chen GQ: miR-760 mediates chemoresistance through inhibition
of epithelial mesenchymal transition in breast cancer cells. Eur
Rev Med Pharmacol Sci. 20:5002–5008. 2016.PubMed/NCBI
|
|
12
|
Lv J, Fu Z, Shi M, Xia K, Ji C, Xu P, Lv
M, Pan B, Dai L and Xie H: Systematic analysis of gene expression
pattern in has-miR-760 overexpressed resistance of the MCF-7 human
breast cancer cell to doxorubicin. Biomed Pharmacother. 69:162–169.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lv J, Xia K and Xu P: miRNA expression
patterns in chemoresistant breast cancer tissues. Biomedi
Pharmacother. 68:935–942. 2014. View Article : Google Scholar
|
|
14
|
Wang Q, Huang Z, Ni S, Xiao X, Xu Q, Wang
L, Huang D, Tan C, Sheng W and Du X: Plasma miR-601 and miR-760 are
novel biomarkers for the early detection of colorectal cancer. PLoS
One. 7:e443982012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liao Y, Deng Y, Liu J, Ye Z, You Z, Yao S
and He S: MiR-760 overexpression promotes proliferation in ovarian
cancer by downregulation of PHLPP2 expression. Gynecol Oncol.
143:655–663. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han ML, Wang F, Gu YT, Pei XH, Ge X, Guo
GC, Li L, Duan X, Zhu MZ and Wang YM: MicroR-760 suppresses cancer
stem cell subpopulation and breast cancer cell proliferation and
metastasis: By down-regulating NANOG. Biomed Pharmacother.
80:304–310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee YH, Kim SY and Bae YS: Upregulation of
miR-760 and miR-186 is associated with replicative senescence in
human lung fibroblast cells. Mol Cells. 620–627. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Elshafei A, Shaker O, Abd El-Motaal O and
Salman T: The expression profiling of serum miR-92a, miR-375 and
miR-760 in colorectal cancer: An Egyptian Study. Tumour Biol.
39:10104283177057652017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng Q, Chen C, Guan H, Kang W and Yu C:
Prognostic role of microRNAs in human gastrointestinal cancer: A
systematic review and meta-analysis. Oncotarget. 8:46611–46623.
2017.PubMed/NCBI
|
|
20
|
Lánczky A, Nagy Á, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: miRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2178 breast cancer patients. 160:439–446. 2016.
|
|
21
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ananthakrishnan A, Gogineni V and Saeian
K: Epidemiology of primary and secondary liver cancers. Semin
Intervent Radiol. 23:47–63. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee JG, Kang CM, Park JS, Kim KS, Yoon DS,
Choi JS, Lee WJ and Kim BR: The actual five-year survival rate of
hepatocellular carcinoma patients after curative resection. Yonsei
Med J. 47:105–112. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Waghray A, Murali AR and Menon KN:
Hepatocellular carcinoma: From diagnosis to treatment. World J
Hepatol. 7:1020–1029. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wan DW, Tzimas D, Smith JA, Kim S, Araujo
J, David R, Lobach I and Sarpel U: Risk factors for early-onset and
late-onset hepatocellular carcinoma in Asian immigrants with
hepatitis B in the United States. Am J Gastroenterol.
106:1994–2000. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mehra M and Chauhan R: Long noncoding RNAs
as a key player in hepatocellular carcinoma. Biomark Cancer.
9:1179299X177373012017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gramantieri L, Fornari F, Callegari E,
Sabbioni S, Lanza G, Croce CM, Bolondi L and Negrini M: MicroRNA
involvement in hepatocellular carcinoma. J Cell Mol Med.
12:2189–2204. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang CS, Yu W, Cui H, Wang YJ, Zhang L,
Han F and Huang T: Increased expression of miR-21 predicts poor
prognosis in patients with hepatocellular carcinoma. Int J Clin Exp
Pathol. 8:7234–7238. 2015.PubMed/NCBI
|
|
29
|
Liu AM, Yao TJ, Wang W, Wong KF, Lee NP,
Fan ST, Poon RT, Gao C and Luk JM: Circulating miR-15b and miR-130b
in serum as potential markers for detecting hepatocellular
carcinoma: A retrospective cohort study. BMJ Open. 2:e0008252012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qi P, Cheng SQ, Wang H, Li N, Chen YF and
Gao CF: Serum microRNAs as biomarkers for hepatocellular carcinoma
in Chinese patients with chronic hepatitis B virus infection. PLoS
One. 6:e284862011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan C, Zhang W, Shi X, Zheng J, Jin X and
Huo J: MiR-760 suppresses non-small cell lung cancer proliferation
and metastasis by targeting ROS1. Environ Sci Pollut Res Int.
25:18385–18391. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao L, Liu Y, Wang D, Huang L, Li F, Liu
J, Zhang C, Shen Z, Gao Q, Yuan W and Zhang Y: MiR-760 suppresses
human colorectal cancer growth by targeting BATF3/AP-1/cyclinD1
signaling. J Exp Clin Cancer Res. 37:832018. View Article : Google Scholar : PubMed/NCBI
|