Introduction
Hepatocellular carcinoma (HCC) is one of the most
prevalent and mortality-associated cancer types in developing and
developed countries and accounts for 70–90% of primary liver cancer
cases (1). Despite numerous years of
basic and clinical research on HCC, the 5-year survival rate still
remains at ~7% (2). An alternative
approach for addressing the poor survival problem may rely on
discovering novel targets for treatment. Hepatocarcinogenesis is a
slow and complicated process that includes genomic changes that
progressively alter the hepatocellular phenotype to produce
abnormal cellular intermediates, finally resulting in HCC (3); however, the understanding of the
underlying molecular mechanisms that drive hepatocarcinogenesis is
still in its infancy. In the past 15 years, non-coding RNAs,
particularly microRNAs (miRNAs), have received considerable
attention regarding an elucidation of the molecular pathogenesis of
cancer (4). Through recognizing the
seed sequences in the 3′-untranslated region (3′-UTR) of target
mRNAs, each miRNA has the ability to regulate the expression of
numerous genes (5); therefore, miRNAs
are frequently considered to efficiently coordinate and regulate
multiple signaling pathways and biological processes in human
diseases, particularly in cancers (5). Thus far, accumulating evidence has
indicated that an abnormal miRNA expression profile is a hallmark
of malignancies, including HCC (6,7). The
previous study demonstrated that miR-139 was downregulated in HCC
and could serve as a diagnostic and prognostic marker for HCC
(8); however, the major targets and
precise signaling pathways that miR-139 participates in in HCC are
not fully understood. A number of studies determined that
overexpression of miR-139 suppresses the proliferation, invasion
and metastasis of HCC cell lines in vitro (9–11). miR-139
is also associated with the functions of particular genes; it is
reported that miR-139 may target transcription factor 4 (TCF-4)
3′-UTR, regulate the expression of TCF-4 and inhibit the
β-catenin/TCF-4 pathway in HCC cell lines (9). Wong et al (11) reported that miR-139 reduces the
expression of Rho-kinase 2 (ROCK2) in HCC cell lines. c-fos
may be another downstream gene responsible for the metastatic
effect in HCC cell lines. Furthermore, miR-139 is also identified
as one of the post-hepatectomy recurrence-associated miRNAs
(12). The expression of zinc finger
E-box binding homeobox 1 (ZEB1) and ZEB2 was also inhibited by
miR-139 through recognizing the 3′-UTR of these two genes (13). Considering that miRNAs serve a crucial
role in multiple genes' expression and transcription regulation, it
was hypothesized that miR-139 may have a major functional target
gene and possibly acts as a key regulator of HCC progression.
In the present study, a combinational analysis of
the data from four miRNA target prediction tools and biological
experiments was applied to explore potential targets of
tumor-suppressive miR-139 in HCC. It was demonstrated that
Topoisomerase I (TOP1) is a proven, direct target of miR-139
in HCC. Overexpression of miR-139 inhibits HCC cell proliferation
and migration, largely due to TOP1 downregulation. The
present study indicated that miR-139 exerts a tumor-suppressive
effect during hepatocarcinogenesis via suppressing the expression
of TOP1; therefore, miR-139 is not only a biomarker for
diagnosis and prognosis but also a promising target for the
biological treatment of HCC.
Materials and methods
Bioinformatics analysis of miRNA
target prediction
The majority of model organisms have an miRNA target
gene prediction database, including TargetScan (http://www.targetscan.org/) (14), miRanda (http://www.microrna.org/) (15), miRDB (http://www.mirdb.org/) (16) and CLIP-Seq (http://www.starbase.sysu.edu.cn/) (17). Through these databases, an analysis of
miR-139 was performed using bioinformatics in HCC. Using the
combinational analysis of the data from four miRNA target
prediction tools, four groups of genes were selected from the
database and were identified as screening objects.
Cell culture
Human liver cancer cell lines BEL-7404 and SMMC-7721
were obtained from the Type Culture Collection of the Chinese
Academy of Sciences (Shanghai, China). BEL-7404 and SMMC-7721 were
maintained in RPMI-1640 medium (GE Healthcare Life Sciences, Logan,
UT, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone; GE
Healthcare Life Sciences). HEK-293 was maintained in DMEM (GE
Healthcare Life Sciences) supplemented with 10% FBS. The cells were
incubated at 37°C in a humidified atmosphere containing 5%
CO2.
Cell transfection
BEL-7404 and SMMC-7721 cells were transfected with
either miRNA mimics/inhibitors or plasmids using HiPerFect
Transfection Reagent (Qiagen GmbH, Hilden, Germany), following the
manufacturer's protocol. The miRNA mimics were chemically
synthesized, double-stranded RNAs that mimic mature endogenous
miRNAs following transfection into cells, whereas the miRNA
inhibitors were chemically modified antisense RNA oligonucleotides
optimized to specifically target specific miRNA molecules in cells.
miRNA mimics, inhibitors and negative control (NC) sequences were
chemically synthesized by Shanghai GenePharma Co., Ltd., (Shanghai,
China). miR-139-NC: 5′-ACGUGACACGUUCGGAGAAUU3′, miR-139 mimics:
5′-UCUACAGUGCACGUGUCUCCAGU-3′ and miR-139 inhibitors:
5′ACUGGAGACACGUGCACUGUAGA-3′.
The reference miR-139 ID was MIMAT0000250 and the
gene sequence was 5′-UCUACAGUGCACGUGUCUCCAGU-3′. miRNAs and/or DNA
plasmids were diluted in Opti-MEM I reduced serum medium (Thermo
Fisher Scientific, Inc., Waltham, MA, USA).
Briefly, the day prior to transfection, BEL-7404 and
SMMC-7721 cells (2×104) were plated with medium of 100
µl DMEM medium containing 10% FBS, placed in an incubator with 5%
CO2 at 37°C. Transfection were performed when the cells
were at 70–80% confluency and recorded as time 0. Recombination
plasmid TOP1 (Shanghai GenePharma Co., Ltd.) (0.1 µg) was added,
and/or 0.5 µl 20 µM miR-139 mimics or inhibitors were added to
Opti-MEM for a final volume of 10 µl. Subsequently, 0.4 µl
Lipofectamine 2000 was added and the mixture were kept at room
temperature for 15 min. Following this, the transfection mixture
was added to each cell medium and mixed. The media were changed to
RPMI-1640 medium supplemented with 10% FBS following incubation in
the incubator with 5% CO2 at 37°C for 5 h. Subsequently,
the supernatant medium was removed following another incubation in
an atmosphere containing 5% CO2 at 37°C for 48 h.
Following this, the Passive Lysis Buffer (100 µl; Promega
Corporation, Madison, WI, USA) were added and the cells were lysed
and collected following the mixture being shaken gently at room
temperature for 15 min. The cells were then collected for other
subsequent experiments.
Immunoblotting
Total cell lysates were obtained using a Triton
X-100 lysis buffer [20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1
mM EGTA, 1% Triton X-100, 2.5 mM Sodium pyrophosphate, 1 mM
β-glycerophosphate and 1 mM Na3VO4],
supplemented with a protease inhibitor cocktail (Roche Applied
Science, Penzberg, Germany) and phenylmethane sulfonyl fluoride (1
mM), and determined with the BCA Protein Quantification kit (cat.
no. BL521A; BioSharp, Hefei, China), according to the
manufacturer's protocols.
Protein samples (20 µg) were separated respectively
by 12, 10 and 8% SDS-PAGE, according to the following groups, which
were divided by the protein molecular weight: BTG family member 3
(BTG3; 29 kDa); Casitas B-lineage lymphoma-transforming
sequence-like protein 1 (CBLL1; 55 kDa); H2A Histone Family Member
V (H2AFV; 14 kDa); Heterogeneous Nuclear Ribonucleoprotein F
(HNRNPF; 46 kDa); Ligand Dependent Nuclear Receptor Corepressor
(LCOR; 47 kDa); LIM Domain Only 4 (LMO4; 18 kDa); Protein
Phosphatase 2 Catalytic Subunit Alpha (PPP2CA; 34 kDa), and β-actin
(42 kDa) were separated by 12% SDS-PAGE; Mannosyl
(Alpha-1,3-)-Glycoprotein Beta-1,4-N-Acetylglucosaminyltransferase,
Isozyme A (MGAT4A; 62 kDa); Discoidin, CUB And LCCL Domain
Containing 2 (DCBLD2; 78 kDa) and Intestinal Cell Kinase (ICK; 71
kDa) were separated by 10% SDS-PAGE; Eukaryotic Translation
Initiation Factor 4 Gamma 2 (EIF4G2; 102 kDa); DNA Topoisomerase I
(TOP1; 91 kDa) and Zinc Finger And BTB Domain Containing 10
(ZBTB10; 95 kDa) were separated by 8% SDS-PAGE) and were
transferred onto polyvinylidene fluoride membranes for western
blotting. The membranes were blocked with 5% BSA (Solarbio Life
Science, Beijing, China) at room temperature for 1 h, then probed
with primary antibodies [anti-BTG3, (dilution, 1:500; cat. no.
bs-7698R; BIOSS, Beijing, China); anti-H2AFV, (dilution, 1:200;
cat. no. bs-17425R; BIOSS); anti-HNRNPF (dilution, 1:500; cat. no.
bs-4205R; BIOSS); anti-ICK, (dilution, 1:1,000; cat. no. bs-15536R;
BIOSS); anti-LCOR, (dilution, 1:200; cat. no. bs-18198R; BIOSS);
anti-ZBTB10, (dilution, 1:1,000; cat. no. bs-13556R; BIOSS);
anti-LMO4, (dilution, 1:1,000; cat. no. bs-5966R; BIOSS);
anti-PPP2CA, (dilution, 1:1,000; cat. no. bs-0029R; BIOSS);
anti-EIF4G2 (dilution, 1:500; cat. no. bs-1350R; BIOSS);
anti-DCBLD2 (dilution, 1:500; cat. no. bs-5834R; BIOSS);
anti-MGAT4A (dilution, 1:1,000; cat. no. bs-18907R; BIOSS);
anti-TOP1 (dilution, 1:1,000; cat. no. bs-10542R; BIOSS);
anti-CBLL1 (dilution, 1:1,000; cat. no. bs-8386R; BIOSS,) and
anti-β-actin, (dilution, 1:2,000; cat. no. 60008–1-Ig; Proteintech,
Thermo Fisher Scientific, Inc.) overnight at 4°C. The supernatants
were removed and the membranes were washed by TBS with 0.1% Tween
20 (TBST) for 5 min three times. Subsequently, the anti-rabbit
secondary antibodies (1:5,000; cat. no. ZB-2301; OriGene
Technologies, Inc., Beijing, China) labeled with horseradish
peroxidase (HRP) were added, and incubated at room temperature for
1 h. Following this, the supernatants were removed and the
membranes were washed with TBST three times, 5 mins per time.
Finally, the blots were visualized by Trident femto Western HRP
Substrate (GeneTex Inc.) and the images were captured using
ChemiDocEQ detection system (Bio-Rad Laboratories, Hercules,
California, USA.
Luciferase reporter assay
Cells of 50–90% confluence in 48-well plates were
transfected using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). 3′-UTR of TOP1 gene was
transfected with the vector psicheck 2.0 (Shanghai GenePharma Co.,
Ltd.) contained luciferase reporter gene. A total of 100 ng of
TOP1-transfected plasmid and a Renilla luciferase construct (5 ng;
for normalization) were co-transfected in to the cells. Cell
extracts were prepared 48 h after transfection, and the luciferase
activity was measured using the Dual-Luciferase Reporter Assay
System (Promega Corporation).
Cell proliferation assay
Cell proliferation assays were performed using the
Cell Counting Kit-8 (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany). Cells transfected with miR-139-NC were set as the
negative control. The experiments were repeated three times.
BEL-7404 and SMMC-7721 were seeded in 96-well plates at
~5×103 cells/well and cultured in RPMI-1640 medium (GE
Healthcare Life Sciences) supplemented with 10% FBS (Beyotime
Institute of Biotechnology, Shanghai, China). After 24 h, 15 µl
CCK-8 solution was added to each well and they were incubated at
37°C in 5% CO2 for 3 h. The absorbance was quantified at
492 nm by a microplate spectrophotometer (Thermo MK3; Thermo Fisher
Scientific, Inc.).
Wound-healing assay
BEL-7404 and SMMC-7721 were maintained in RPMI-1640
medium (GE Healthcare Life Sciences) supplemented with 10% FBS
(Beyotime Institute of Biotechnology). The cells were seeded onto
six-well dishes at 2×105 cells/well. A single scratch
wound was created in confluent cells using a p10 micropipette tip.
Cells were washed twice with PBS to remove cell debris. Monitored
images were captured by fluorescence microscopy (magnification,
×400) (IX71; Olympus Corporation, Tokyo, Japan) at 0 and 24 h
following wounding.
miRNA target prediction
To define potential downstream targets of miR-139,
candidate genes that were commonly predicted were matched by four
publicly available algorithms: TargetScan version 7.0, miRanda
version 2010, miRDB and CLIP-Seq version 2.
Statistical analysis
Statistical analysis was performed using SPSS 11.0
for Windows (SPSS, Inc., Chicago, IL, USA). All data are presented
as the mean ± standard deviation. A two-tailed Student's t-test was
used to evaluate the statistical significance of differences
between two groups of data in the luciferase, cell proliferation
and wound-healing assays. P<0.05 was considered to
indicate a statistically significant difference.
Results
Prediction of miR-139 targets
To identify the potential targets of
tumor-suppressive miR-139 in HCC cells, a bioinformatics analysis
was performed using miRNA target prediction tools. Computational
predictions indicate that all human genes may be regulated by
microRNAs, with each microRNA possibly targeting thousands of genes
(18). As depicted in Fig. 1A, the four frequently used algorithms
[TargetScan (14), miRanda (15), miRDB (16) and CLIP-Seq (17)] produced divergent sets of predicted
targets of miR-139. To reduce bias caused by one method, the
results predicted by the different algorithms were intersected and
it was determined that a group of 28 genes are jointly identified
by all four algorithms (Fig. 1). Some
of these genes include: BTG3; CBLL1; DCBLD2;
EIF4G2; H2AFV; HNRNPF; ICK;
LCOR; LMO4; MGAT4A; PPP2CA;
TOP1; and ZBTB10. These genes are reported to be
aberrantly expressed in various cancer types and thus became a
focus (Fig. 1B).
miR-139 target screening by western
blot analysis
To validate the potential targets of miR-139 in HCC
cells, western blot analysis was performed to screen the predicted
genes that are dysregulated in cancer. Two frequently used HCC cell
lines, BEL-7404 and SMMC-7721, were selected and miR-139 mimics,
miR-139 inhibitors or a negative control were transfected into
these cells. Following 48 h, cells were lysed, and all samples were
analyzed via semi-quantitative immunoblotting. As depicted in
Figs. 2 and 3, increased miR-139 expression notably
reduced TOP1 protein levels in both HCC cell lines, whilst
miR-139 inhibitors had an opposite effect on TOP1
expression. These results indicated that miR-139 could negatively
regulate TOP1 expression in HCC cells.
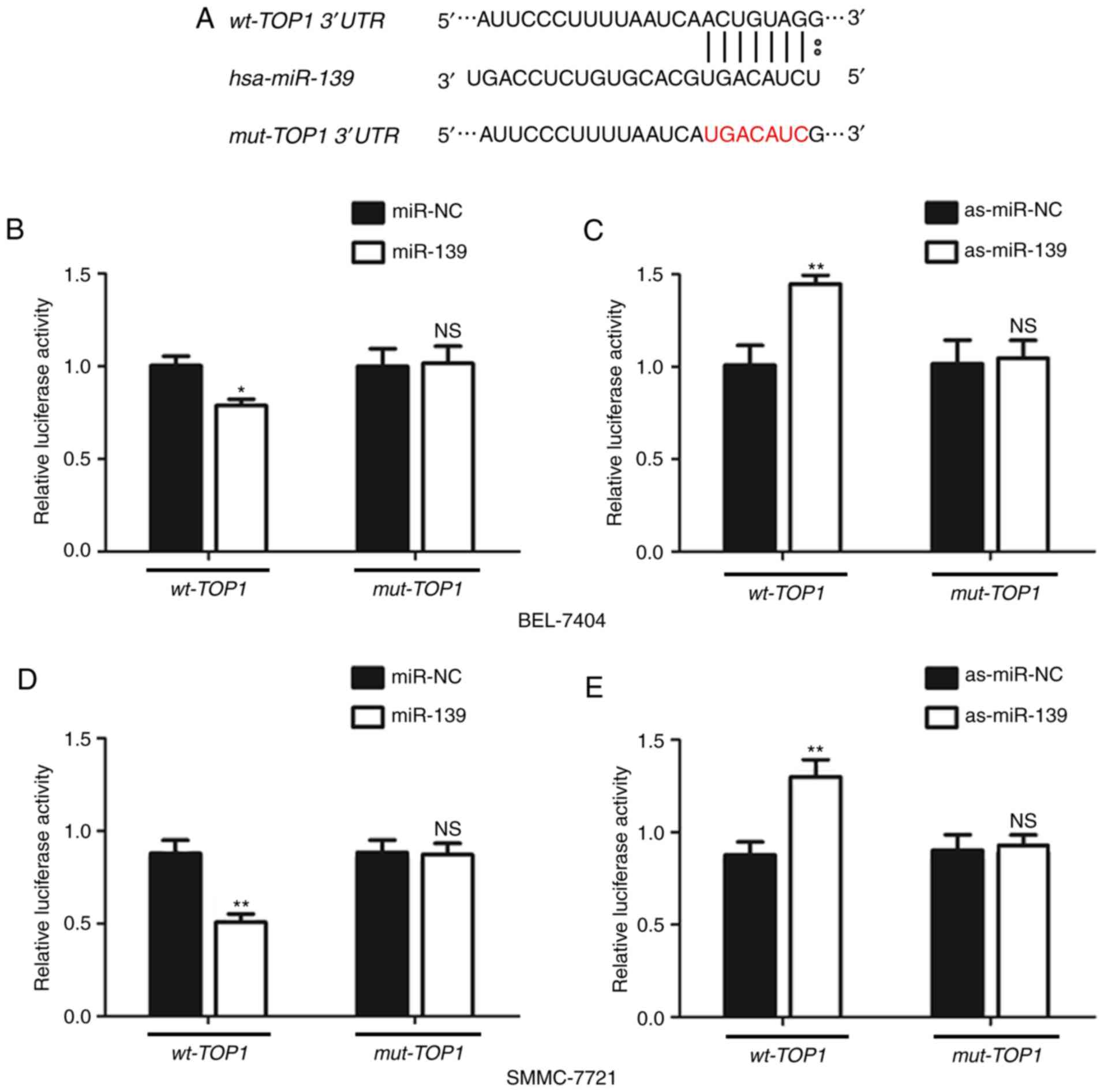
miR-139 directly targets and inhibits
TOP1 expression
As western blot analyses could not discriminate
between direct and indirect effects of miR-139 on TOP1
expression, a Dual-Luciferase reporter analysis was performed to
determine if miR-139 targets TOP1 mRNA directly. The results
demonstrated that miR-139 significantly repressed the luciferase
activity of a reporter vector harboring the wild-type 3′-UTR of
TOP1, whereas mutation of the putative miR-139-binding site
in the 3′-UTR region abrogated the inhibitory effect of miR-139
(Fig. 4A, B and C). Similarly,
inhibition of miR-139 significantly enhanced luciferase activity of
the reporter vector harboring the wild-type 3′-UTR of TOP1,
and this effect could be completely abolished by mutation of the
miR-139-binding site (Fig. 4D and E).
Taken together, these results indicate that miR-139 directly binds
to the 3′-UTR region of TOP1 and inhibits its expression in
HCC cells.
miR-139 suppresses HCC cell
proliferation and migration through downregulation of TOP1
The previous study demonstrated that miR-139 is
significantly downregulated in HCC tissues and is an independent
risk factor for reduced survival (8);
however, the biological function of this tumor-suppressive miRNA is
largely unknown. To determine if miR-139 affects HCC cell
proliferation, BEL-7404 and SMMC-7721 cells were treated with a
negative control, miR-139 mimics or miR-139 inhibitors for 48 h. A
CCK-8 assay demonstrated that enforced miR-139 expression
significantly reduced the proliferation rate of both cell lines,
whilst overexpression of TOP1 lacking the endogenous 3′-UTR
completely abrogated the inhibitory effect of miR-139 (Fig. 5). Furthermore, a wound-healing assay
was performed to test whether miR-139 had any effect on HCC cell
migration. As depicted in Fig. 6,
miR-139 overexpression notably repressed HCC cell migration, and
miR-139-induced migration inhibition could be rescued by exogenous
TOP1 expression. Overall, these data indicated that
tumor-suppressive miR-139 inhibits cell proliferation and migration
through downregulation of TOP1 in HCC.
Discussion
Since the beginning of this century, accumulating
evidence has demonstrated that miRNAs serve pivotal roles in the
process of tumorigenesis (19). An
individual tumor type is characterized by a globally distinctive
expression pattern of miRNAs (20–23) It has
been demonstrated that miRNA expression patterns are closely
associated with a number of important clinical events, including
tumor diagnosis, treatment responses and prognosis (24,25).
Furthermore, novel evidence has revealed more specific roles of
miRNA in tumorigenesis (26). A
number of functional studies have demonstrated that miRNAs serve an
oncogenic or tumor suppressor role in different malignancies, in
vitro and in vivo (27).
MiRNA-139 was determined to be dysregulated in various cancer
types, including breast and colon cancer (28,29), but
the precise function of this miRNA still requires further
exploration. Wong et al (11)
determined that low expression of miR-139 is associated with
metastatic HCC and overexpression of miR-139 suppresses metastasis
and the progression of HCC by downregulating ROCK2 (11). In addition, a low-expression of
miR-139 was also determined in colorectal cancer and breast cancer,
which indicates that miR-139 may be a key regulator in malignancies
(11,28). The previous study demonstrated that
miR-139 is significantly downregulated in HCC tissues and could be
used as an independent risk factor for predicting prognosis in
patients with HCC (8).
Development of dysplastic hepatocytes in point foci
and nodules dysplasia and formation emergence in HCC are associated
with the build-up of an accumulation of irreversible structural
alterations in genes and chromosomes (3); therefore, identification of key genes
that promote genomic instability is of great importance to cancer
gene therapy. In the present study, TOP1 was identified as a
direct target of miR-139 in HCC. DNA topoisomerases are vital
enzymes that solve DNA topological problems that result from strand
separation during replication and transcription. TOP1 is a
nuclear enzyme that cuts one of the two strands of DNA, relaxes the
strand and reanneals the strand, thereby allowing moving DNA
supercoils during DNA replication or gene transcription (30). Based on this function, topoisomerases
are emerging as important factors in a wide range of fundamental
biological processes in nuclear and mitochondrial genomes (31). Topoisomerases introduce transient DNA
breaks using a transesterification mechanism, which is highly
reversible and minimizes the risks of genome stability that would
otherwise occur due to strand breakage (32); therefore, formation of a
DNA-TOP1 complex is a crucial intermediate step in the
transesterification mechanism. However, aberrant expression of
TOP1 is potentially hazardous to the cell due to it
mediating an illegitimate recombination that may lead to genomic
instability and oncogenesis (33).
Therefore, it is now established that topoisomerases can ensure and
endanger genome integrity. Kim et al (32) indicated that TOP1 could provoke
genome instability by action at sites of endogenous and exogenous
DNA damage. The risks associated with strand breakage by
topoisomerases indicate that there are aspects of fundamental
processes, including transcription, that pose unique topological
challenges and that cells require a wide repertoire of responses
and specific repair pathways to safeguard the dangerous process of
introducing transient DNA breaks. The importance of topoisomerases
in genomic maintenance may also explain why cancer cells, which are
high replicative and undergo transcriptional stress, frequently
overexpress nuclear and mitochondrial topoisomerases (34). Furthermore, it has been determined
that TOP1 is highly expressed in a number of malignancy
types, including colon cancer and breast cancer (35,36),
indicating its potential role in tumorigenesis. The major effect of
TOP1-induced DNA lesions on cell survival has resulted in
this enzyme being a prime target for cancer therapies to kill
fast-growing cancer cells (37). To
date, a number of TOP1 inhibitors have been developed,
including camptothecins, irinotecan and topotecan. However,
camptothecins and its water-soluble derivatives have several
limitations. For example, camptothecins produce side effects
(including leucopenia) that limit the dose that can be safely
administered and, therefore, its anti-tumor efficacy (38). The diarrhea induced by irinotecan can
be severe and is possibly due to ‘off-target’ effects that are
associated with the bis-piperidine that confers water-solubility
(38); therefore, novel therapeutic
strategies targeting TOP1 are required to be developed by
further increasing their antitumor activity and decreasing the side
effects. However, miR-139 may be an alternative target for the same
molecular signal pathway.
Due to the potential ability of miRNAs to influence
multiple cellular behaviors, therapeutic strategies based on
modulation of miRNA expression levels have demonstrated great
promise (39). Recent studies
indicated that enforced overexpression of individual miRNA exhibits
a powerful antitumorigenic effect in lymphoma cells transformed by
key oncogenes, including c-Myc and Bcl-2 (40). Furthermore, Kumar et al
(40) demonstrated that systemic
administration of viral vectors expressing let-7 miRNAs impaired
tumor growth in a mouse model with lung adenocarcinoma. In the
present study, it was reported that TOP1 is inhibited by a
tumor-suppressive miRNA miR-139 and increased expression of miR-139
impairs HCC cell proliferation and migration. The present study
indicated that miR-139 could be a promising novel therapeutic
option for targeting TOP1 and holds great potential in the
treatment of HCC. It was speculated that miR-139 may be a safe and
effective TOP1 inhibitor with fewer side effects.
Acknowledgements
Not applicable.
Funding
This work was supported by Shaanxi Science and
Technology Department (grant no. 2015KTCL03-05).
Availability of data and materials
The datasets used during the current study are
available from the corresponding author on reasona-ble request. The
raw data required to reproduce these findings cannot be shared at
this time as the data also forms part of an ongoing study.
Authors' contributions
PZ and JY proposed the study and wrote the first
draft and analyzed the data. LY, QW, XD, RD, GZ and JL conceived
and improved the manuscript. CW, QB and LJ performed the
experiments. GZ and JL analyzed the data and approved the final
version to be published. All authors read and approved the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carr BI: Hepatocellular carcinoma: Current
management and future trends. Gastroenterology. 127:S218–S224.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thorgeirsson SS and Grisham JW: Molecular
pathogenesis of human hepatocellular carcinoma. Nat Genet.
31:339–346. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in Cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li T, Yin J, Yuan L, Wang S, Yang L, Du X
and Lu J: Downregulation of microRNA-139 is associated with
hepatocellular carcinoma risk and short-term survival. Oncol Rep.
31:1699–1706. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gu W, Li X and Wang J: miR-139 regulates
the proliferation and invasion of hepatocellular carcinoma through
the WNT/TCF-4 pathway. Oncol Rep. 31:397–404. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hua S, Lei L, Deng L, Weng X, Liu C, Qi X,
Wang S, Zhang D, Zou X, Cao C, et al: miR-139-5p inhibits aerobic
glycolysis, cell proliferation, migration, and invasion in
hepatocellular carcinoma via a reciprocal regulatory interaction
with ETS1. Oncogene. 37:1624–1636. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wong CC, Wong CM, Tung EK, Au SL, Lee JM,
Poon RT, Man K and Ng IO: The microRNA miR-139 suppresses
metastasis and progression of hepatocellular carcinoma by
down-regulating Rho-kinase 2. Gastroenterology. 140:322–331. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang L, Liu M, Zhu H, Rong W, Wu F, An S,
Liu F, Feng L, Wu J and Xu N: Identification of recurrence-related
serum microRNAs in hepatocellular carcinoma following hepatectomy.
Cancer Biol Ther. 16:1445–1452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qiu G, Lin Y, Zhang H and Wu D: miR-139-5p
inhibits epithelial-mesenchymal transition, migration and invasion
of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2.
Biochem Biophys Res Commun. 463:315–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ritchie W, Rasko JE and Flamant S:
MicroRNA target prediction and validation. Adv Exp Med Biol.
774:39–53. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie X, Tan W, Chen B, Huang X, Peng C, Yan
S, Yang L, Song C, Wang J, Zheng W, et al: Preoperative prediction
nomogram based on primary tumor miRNAs signature and
clinical-related features for axillary lymph node metastasis in
early-stage invasive breast cancer. Int J Cancer. 142:1901–1910.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Slattery ML, Lee FY, Pellatt AJ, Mullany
LE, Stevens JR, Samowitz WS, Wolff RK and Herrick JS: Infrequently
expressed miRNAs in colorectal cancer tissue and tumor molecular
phenotype. Mod Pathol. 30:1152–1169. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mannavola F, Tucci M, Felici C, Stucci S
and Silvestris F: miRNAs in melanoma: A defined role in tumor
progression and metastasis. Exp Rev Clin Immunol. 12:79–89. 2016.
View Article : Google Scholar
|
|
23
|
Tian F, Li R, Chen Z, Shen Y, Lu J, Xie X
and Ge Q: Differentially expressed miRNAs in tumor, adjacent, and
normal tissues of lung adenocarcinoma. Biomed Res Int.
2016:14282712016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang L, Zhang X, Jia LT, Hu SJ, Zhao J,
Yang JD, Wen WH, Wang Z, Wang T, Zhao J, et al: c-Myc-mediated
epigenetic silencing of MicroRNA-101 contributes to dysregulation
of multiple pathways in hepatocellular carcinoma. Hepatology.
59:1850–1863. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong
C, Huang Y, Hu X, Su F, Lieberman J and Song E: let-7 regulates
self renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen K, Liang Q, Xu K, Cui D, Jiang L, Yin
P, Lu Y, Li Q and Liu J: MiR-139 inhibits invasion and metastasis
of colorectal cancer by targeting the type I insulin-like growth
factor receptor. Biochem Pharmacol. 84:320–330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krishnan K, Steptoe AL, Martin HC,
Pattabiraman DR, Nones K, Waddell N, Mariasegaram M, Simpson PT,
Lakhani SR, Vlassov A, et al: miR-139-5p is a regulator of
metastatic pathways in breast cancer. RNA. 19:1767–1780. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ashour ME, Atteya R and El-Khamisy SF:
Topoisomerase-mediated chromosomal break repair: An emerging player
in many games. Nat Rev Cancer. 15:137–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stewart L, Redinbo MR, Qiu X, Hol WG and
Champoux JJ: A model for the mechanism of human topoisomerase I.
Science. 279:1534–1541. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pommier Y, Sun Y, Huang SN and Nitiss JL:
Roles of eukaryotic topoisomerases in transcription, replication
and genomic stability. Nat Rev Mol Cell Biol. 17:703–721. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim N, Huang SN, Williams JS, Li YC, Clark
AB, Cho JE, Kunkel TA, Pommier Y and Jinks-Robertson S: Mutagenic
processing of ribonucleotides in DNA by yeast topoisomerase I.
Science. 332:1561–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Larsen AK and Gobert C: DNA topoisomerase
I in oncology: Dr Jekyll or Mr Hyde? Pathol Oncol Res. 5:171–178.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rahman M, Jackson LK, Johnson WE, Li DY,
Bild AH and Piccolo SR: Alternative preprocessing of RNA-sequencing
data in the cancer genome atlas leads to improved analysis results.
Bioinformatics. 31:3666–3672. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kümler I, Balslev E, Stenvang J, Brünner N
and Nielsen D: A phase II study of weekly irinotecan in patients
with locally advanced or metastatic HER2-negative breast cancer and
increased copy numbers of the topoisomerase 1 (TOP1) gene: A study
protocol. BMC Cancer. 15:782015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Giovanella BC, Stehlin JS, Wall ME, Wani
MC, Nicholas AW, Liu LF, Silber R and Potmesil M: DNA topoisomerase
I-targeted chemotherapy of human colon cancer in xenografts.
Science. 246:1046–1048. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li M and Liu Y: Topoisomerase I in human
disease pathogenesis and treatments. Genomics Proteomics
Bioinformatics. 14:166–171. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pommier Y: Topoisomerase I inhibitors:
Camptothecins and beyond. Nat Rev Cancer. 6:789–802. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sampson VB, Rong NH, Han J, Yang Q, Aris
V, Soteropoulos P, Petrelli NJ, Dunn SP and Krueger LJ: MicroRNA
let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt
lymphoma cells. Cancer Res. 67:9762–9770. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kumar MS, Erkeland SJ, Pester RE, Chen CY,
Ebert MS, Sharp PA and Jacks T: Suppression of non-small cell lung
tumor development by the let-7 microRNA family. Proc Natl Acad Sci
USA. 105:3903–3908. 2008. View Article : Google Scholar : PubMed/NCBI
|