Oncol Lett 18: [Related article:] 814-821, 2019; DOI:
10.3892/ol.2019.10342
Subsequently to the publication of this article, the
authors have realized that the article contained several errors
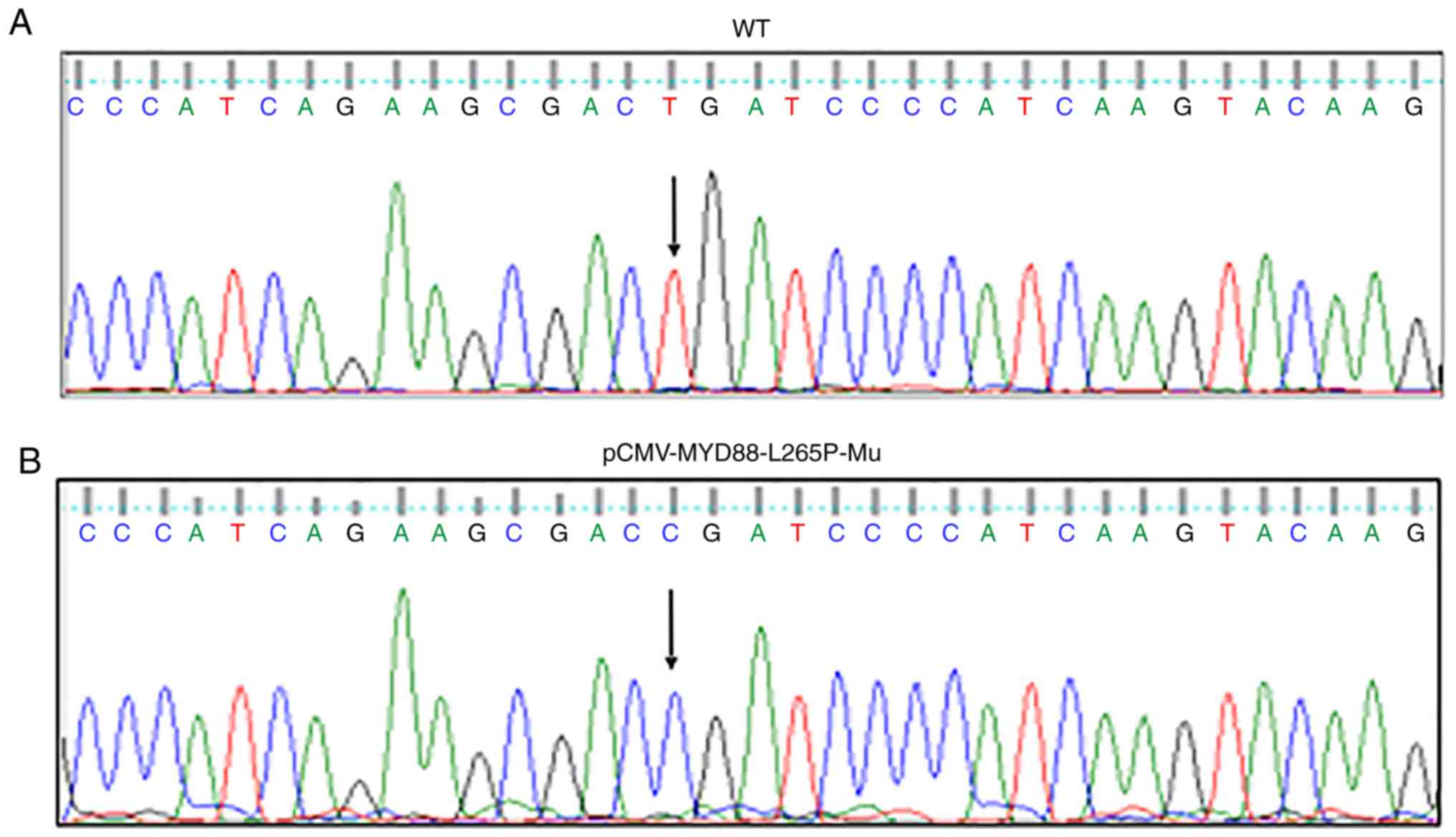
that were not corrected. First, an error was introduced into
Fig. 1; essentially, the mutation of
T>C at position 794 of the nucleotide sequence of the wild-type
MYD88 plasmid was not portrayed correctly (as cytosine) in
panel (B). In addition, the sequencing result in the lowest panel
of Fig. 4 shows the wild-type
sequence, and therefore the nucleotide at position 794 should have
been presented as T, not as C. Corrected versions of Figs. 1 and 4
are shown opposite.
Otherwise, textual errors remained in the paper.
Some textual errors were featured in the subsection of the
Materials and methods entitled “Sensitivity evaluation by HRM
assay and direct sequencing”, in the final paragraph in the
right-hand column of p. 816. The sentence commencing 6 lines from
the bottom of the page should have read as: “The melting curve
obtained using 1% negative control template clearly differed
from the positive template.” (“negative control” instead of
“mutated”, and “positive” instead of “negative”). The subsequent
sentence should have read as: “However, the mutation was detectable
by direct sequencing when the negative control template
frequency was >10% (Fig. 4).”
(“negative control template” instead of “mutant”). Finally, some
errors remained uncorrected in the legend for Fig. 4 on p. 819. The first sentence in the
legend should have read as follows: “Direct sequencing results of
negative control template at serial dilutions (100, 50, 20,
10, 5, 1 and 0%) with positive control.” (“negative control
template” instead of “MYD88 p.L265P mutations”, and
“positive” instead of “negative”).
The authors and the Editor apologize to the
readership of the Journal for any inconvenience caused.