Introduction
Incidence of colorectal cancer (CRC) was the third
most common type of cancer in certain regions of the world in 2018,
such as the USA (1). CRC is a fatal
disease, resulting in >500,000 mortalities every year worldwide
(2). Patients with early stages of
CRC are treated surgically, followed by adjuvant therapy to inhibit
cancer cell proliferation and metastasis (3). However, a considerable number of
patients with CRC are diagnosed with metastasis to the lymph nodes
and major organs, such as the lungs and liver, leading to poor
treatment outcomes and a low overall 5-year survival rate (4,5).
Therefore, novel therapeutic targets are required to improve the
survival of patients with advanced CRC.
Genome-wide analysis of genetic alterations has
revealed that certain somatic and germline mutations are critical
determinants of tumorigenesis in CRC (6,7). Long
non-coding RNAs (lncRNAs) are RNA transcripts >200 nucleotides
in length with non-protein-coding abilities, and have been
identified as critical players in cancer biology through their
regulatory roles in gene expression at posttranscriptional or
translational levels and through epigenetic pathways (8). In effect, certain lncRNAs are potential
therapeutic targets for the treatment of many types of cancers,
such as lung cancer, liver cancer and CRC (9,10).
However, the application of most lncRNAs in cancer therapies is
limited by their unknown functions. lncRNA DiGeorge syndrome
critical region gene 5 (DGCR5) has been implicated in many types of
cancer, and serves a cancer-specific role as a tumor suppressor in
papillary thyroid carcinoma or as an oncogene lung adenocarcinoma
(11,12). The majority of studies investigating
the role of DGCR5 in cancer biology have revealed its interactions
with microRNAs (miRNAs/miRs), a group of small non-coding RNA
molecules ~22 nucleotides in length which serve important roles in
gene expression regulation through the cleavage of transcripts
(11,12). miR-21 is a well-characterized
oncogenic miRNA in several types of cancer, including CRC (13). miR-21 participates in CRC by
regulating various cancer cell behaviors through the interaction
with multiple downstream tumor suppressive and oncogenic pathways,
such as the phosphatase and tensin homolog (PTEN) and signal
transducer and activator of transcription 3 pathways, respectively
(13). Preliminary data has revealed
that DGCR5 and miR-21 were inversely correlated in human CRC
tissues (data not shown). The present study investigated the role
of DGCR5 in CRC as well as the possible interaction between DGCR5
and miR-21.
Materials and methods
Patients
The present study included 65 patients with CRC
enrolled at The Second Affiliated Hospital of Guangxi University of
Science and Technology between March 2016 and March 2018. The
patients included 35 males and 30 females and their mean age was
51.3±5.7 (range, 34–66 years). The diagnosis of the patients was
confirmed by histopathological examinations. The inclusion criteria
for participation in the current study were as follows: i) patients
diagnosed with CRC for the first time; ii) patients had not been
treated prior to admission; and iii) patients with complete medical
records. The exclusion criteria were as follows: i) patients with
previous history of malignancy; ii) patients who received treatment
within 3 months prior to admission; iii) patients who were
transferred from other hospitals. Based on the American Joint
Committee on Cancer staging criteria (AJCC) used (14), there were 14, 20, 18 and 13 patients
at stage I, II, III and IV, respectively. The Ethics Committee of
The Second Affiliated Hospital of Guangxi University of Science and
Technology approved the current study and all patients signed
informed consent.
Local blast
A local blast analysis was performed using
NCBI-blast software (version 2.2.30; National Center for
Biotechnology Information). In this analysis, human miRNAs
sequences (http://www.mirbase.org) were used as
queries, and DGCR5 was used as a targeted database.
Specimens and cell lines
A biopsy was performed on each patient to obtain
cancer and adjacent healthy tissues (2 cm distance from the tumor
site) from each patient. The weight of the tissues ranged between
0.07 and 0.11 g. Tissues were stored at −70°C until use. All tissue
samples were confirmed by histopathological examination. RKO
(American Type Culture Collection) and CR4 (Sigma-Aldrich; Merck
KGaA) human CRC cell lines were used to perform in vitro
cell experiments. Cells were cultured according to the
manufacturer's protocol using Eagle's Minimum Essential Medium
(Sigma-Aldrich; Merck KGaA) supplemented with 10% fetal bovine
serum (FBS; Sigma-Aldrich; Merck KGaA) at 5% CO2 and 37°C.
Total RNA extraction and
reverse-transcription quantitative polymerase chain reaction
(RT-qPCR)
For the analysis of DGCR5 expression levels,
RNAzol® RT reagent (Sigma-Aldrich; Merck KGaA) was used
to extract total the RNA from CRC and adjacent healthy tissues as
well as the RKO and CR4 cell lines. RNA samples were reverse
transcribed into cDNA using the AMV Reverse Transcriptase kit
(Promega Corporation) at the following thermal conditions: 15 min
at 55°C and 10 min at 80°C. qPCR was subsequently performed using
the SYBR®-Green master mix (Bio-Rad Laboratories, Inc.)
using 18S rRNA as an endogenous control. The following primer
sequences were used: DGCR5 forward, 5′-CCAAGCCTGTCTGTGTGTTC-3′ and
reverse, 5′-GGGAGACACAGACCACAAGA-3′; and 18S forward,
5′-GCTTAATTTGACTCAACACGGGA-3′ and reverse,
5′-AGCTATCAATCTGTCAATCCTGTC-3′. Thermal conditions were: 1 min at
95°C, followed by 40 cycles of 10 sec at 95°C and 40 sec at 95°C.
For the analysis of miR-21 expression levels, extraction of miRNAs
from CRC and adjacent healthy tissues as well as the RKO and CR4
cell lines was performed using a microRNA purification kit (cat.
no. 21300; Norgen Biotek Corp.) according to manufacturer's
protocol. TaqMan microRNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.) was used to perform reverse transcription
according to manufacturer's protocol. TaqMan™ Fast Advanced Master
mix (Thermo Fisher Scientific, Inc.) was used to prepare all qPCR
reaction mixtures using U6 as the endogenous control. The following
primer was used for the reaction: miR-21 forward,
5′-GCCCGCTAGCTTATCAGACTGATG-3′. The miR-21 reverse primer and U6
forward and reverse primers were included in the TaqMan™ Fast
Advanced Master mix kit. The therrmal conditions were: 1 min at
95°C, followed by 40 cycles of 10 sec at 95°C and 20 sec at 95°C.
All RT-qPCR reactions were performed in triplicate.
All data were analyzed using the 2−ΔΔCq
method (15) and each experiment was
performed in triplicate.
Transient transfection
Sequences for the negative control
(5′-CGCGCUUCGGUUUAACUAGC-3′) miRNA and miR-21 mimic
(5′-CAACACCAGUCGAUGGGCUGU-3′) were obtained from Sigma-Aldrich;
Merck KGaA. The DGCR5-expression pcDNA3.1 and empty pcDNA3.1
vectors were obtained from by Sangon Biotech Co., Ltd. A total of
105 CRC or RKO cells were transfected with 40 nM miRNA
or 10 nM vector using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.) according to manufacturer's protocol. Two types
of control cells, including cells without transfection (C) and
cells transfected with empty vectors or negative control miRNAs
(NC) were included. Subsequent experiments were performed 24 h
following transfection.
Cell proliferation assay
CRC and RKO cells were harvested 24 h following
transfection and single cell suspensions were prepared using
Eagle's Minimum Essential Medium supplemented with 10% FBS to a
final cell density of 4×104 cell/ml. Cells suspensions
were cultured at 37°C and 5% CO2 in a 96-well plate containing 0.1
ml per well. A total of 10 µl Cell Counting Kit-8 solution
(Sigma-Aldrich; Merck KGaA) was added into each well every 24 h for
96 h. The optical density was measured at a wavelength of 450 nm to
represent cell proliferation.
Statistical analysis
Three biological replicates were included in each
experiment. Differences between CRC and healthy adjacent tissues
were analyzed by the paired t-test. Differences among different
clinical stages and cell transfection groups were analyzed by the
one-way ANOVA and Tukey test. Linear regression was performed to
analyze the association between the expression levels of DGCR5 and
miR-21 in CRC and healthy adjacent tissues. P<0.05 was
considered to indicate a statistically significant difference.
Results
DGCR5 and miR-21 are dysregulated in
CRC tissues
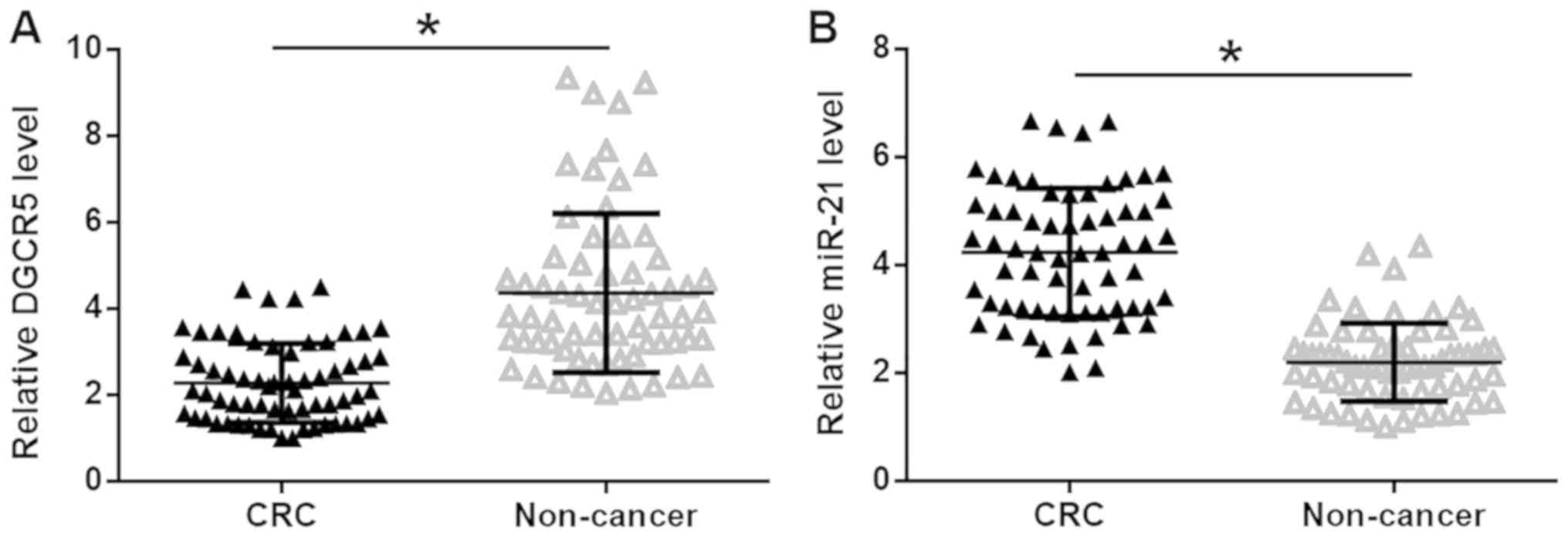
The expression levels of DGCR5 and miR-21 in CRC and
healthy adjacent tissues were quantified by RT-qPCR and the
expression data were analyzed using the paired t-test. It was
observed that DGCR5 was significantly downregulated (Fig. 1A), while miR-21 was significantly
upregulated (Fig. 1B), in CRC
tissues compared with healthy adjacent tissues in patients with CRC
(P<0.05), indicating the involvement of DGCR5 and miR-21 in
CRC.
Expression levels of DGCR5 and miR-21
in CRC tissues are affected by clinical stages
The expression levels of DGCR5 and miR-21 in
patients with different clinical stages of CRC were analyzed using
the one-way ANOVA and Tukey test. The expression levels of DGCR5 in
CRC tissues were significantly decreased (Fig. 2A), while expression levels of miR-21
were significantly increased (Fig.
2B), with advancing clinical stages (P<0.05).
DGCR5 and miR-21 expression levels are
inversely associated in CRC tissues
Linear regression was performed to analyze the
association between the expression levels of DGCR5 and miR-21. It
was observed that DGCR5 and miR-21 were inversely associated in CRC
tissues (Fig. 3A). However, the
association between DGCR5 and miR-21 was not statistically
significant in healthy adjacent tissues (Fig. 3B). It suggested that DGCR5 and miR-21
may interact with each other in CRC.
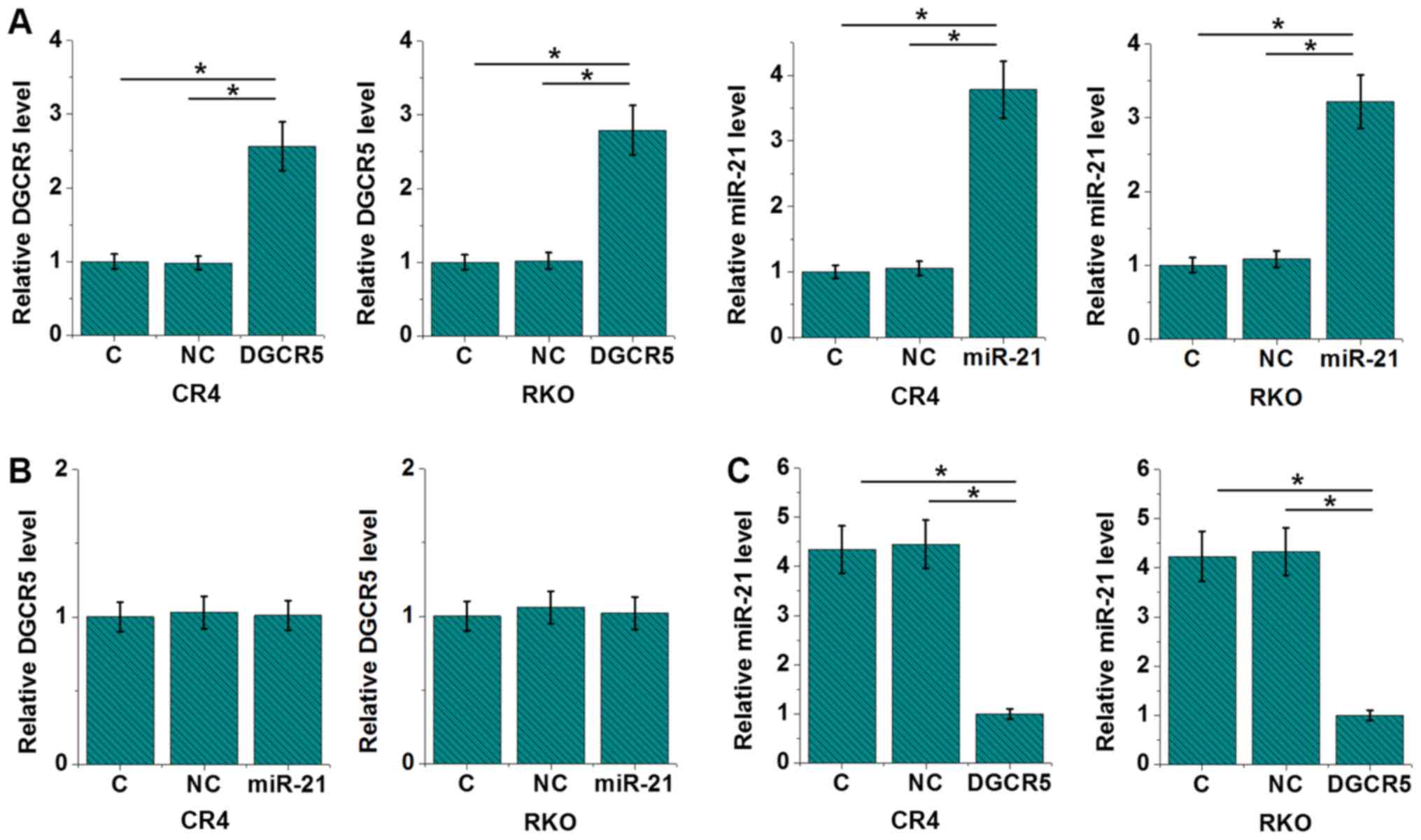
DGCR5 overexpression causes the
downregulation of miR-21 in CRC cells
In order to explore the interaction between DGCR5
and miR-21 in CRC, a DGCR5 expression vector and miR-21 mimic were
transfected into CRC and RKO cell lines. Compared with control (C)
and negative control (NC) groups, expression levels of DGCR5 and
miR-21 were upregulated by more than 2 fold at 24 h after
transfection (Fig. 4A; P<0.05),
indicating the successful transfections. In addition, miR-21
overexpression failed to significantly affect DGCR5 (Fig. 4B), while DGCR5 overexpression led to
reduced expression levels of miR-21 (Fig. 4C; P<0.05).
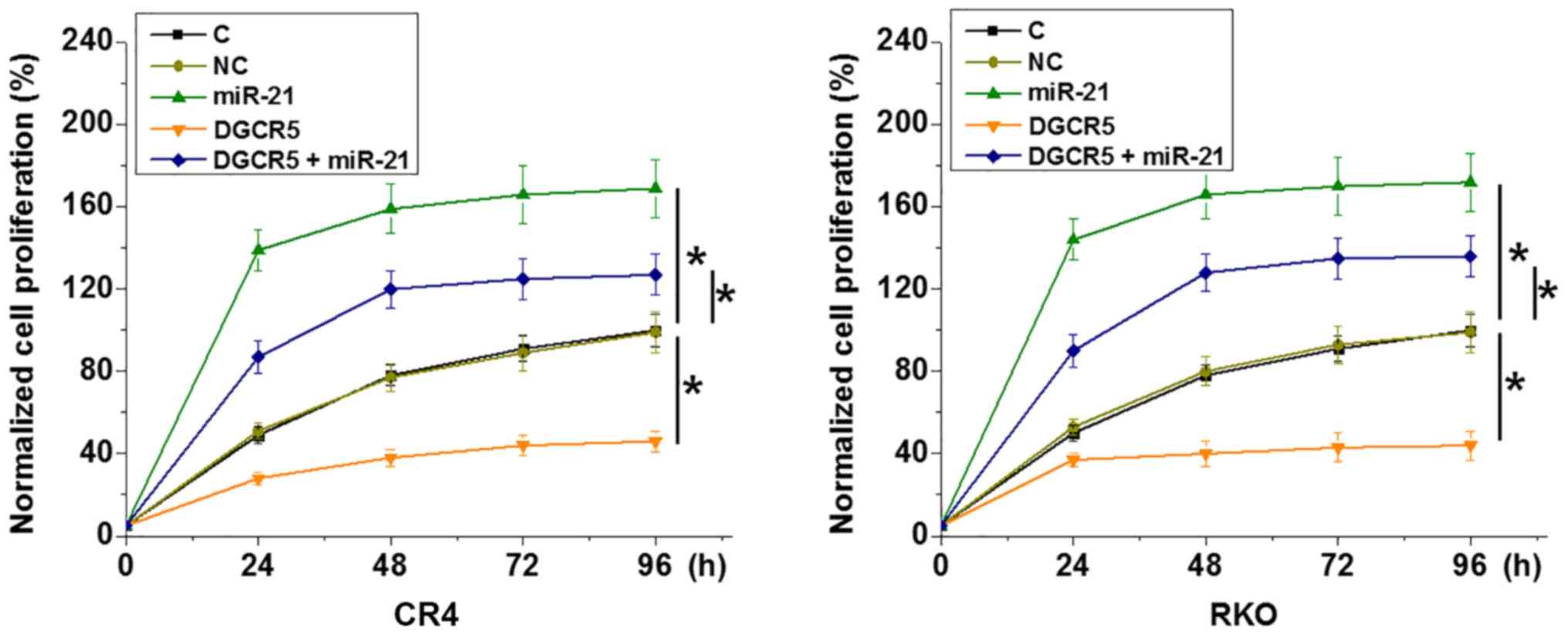
DGCR5 regulates cancer cell
proliferation but not migration and invasion through miR-21
Comparing with the C and NC groups, DGCR5
overexpression showed no significant effects on RKO and CR4 cell
migration and invasion (data not shown; revealed by Transwell
migration and invasion assay). By contrast, DGCR5 overexpression
resulted in inhibited RKO and CR4 cell proliferation compared with
the C and NC groups (Fig. 5;
P<0.05). miR-21 overexpression resulted in increased cancer cell
proliferation and attenuated the effects of DGCR5 overexpression
(Fig. 5; P<0.05).
Discussion
lncRNA DGCR5 plays a role as tumor suppressor in
certain types of cancer, including lung cancer, but serves as an
oncogene in other types of cancer, including papillary thyroid
carcinoma and liver cancer (11,12,16). The
present study revealed the tumor suppressive role of DGCR5 in CRC
and demonstrated that DGCR5 may inhibit the proliferation of CRC
cells by downregulating miR-21, an oncogenic miRNA in CRC (13).
DGCR5 has opposite functions in different types of
cancer. DGCR5 was upregulated during the development of lung
cancer, and DGCR5 overexpression inhibited hsa-mir-22-3p to promote
cancer development (11). By
contrast, DGCR5 inhibits miR-2861 to serve a tumor suppressive role
in papillary thyroid carcinoma (12). The present study revealed that that
DGCR5 was downregulated in CRC tissues compared with healthy
adjacent non-cancer in patients with CRC patients. Additionally,
overexpression of DGCR5 decreased the proliferation of CRC cells
in vitro. Therefore, the results obtained in the present
study suggested that DGCR5 serves a tumor suppressive role in
CRC.
To the best of our knowledge, the present study was
the first to report the inhibitory role of DGCR5 on miR-21, which
as a well-characterized oncogenic miRNA in several types of cancer
(17). miR-21 expression in cancer
development may be regulated by certain lncRNAs (18,19). One
possible mechanism of the interaction between lncRNAs and miRNAs is
that lncRNAs may sponge miRNAs and inhibit their function (20). However, a BLAST (NCBI) search did not
reveal potential binding sites for miR-21 on DGCR5, suggesting that
other molecular mechanisms may mediate interactions between lncRNAs
and miRNAs. Consistently, preliminary luciferase assay data (data
not shown) suggested an indirect interaction between miR-21 and
DGCR5. The relative luciferase signals of miR-21 and negative
control were 100±5.6 vs. 99.7±4.9. It has been shown that both
DGCR5 and miR-21 interact with PTEN (21,22). In
addition, PTEN is downregulated in CRC (23), and PTEN can mediate the interaction
between certain lncRNAs and miRNAs (24). Therefore, PTEN may mediate the
interaction between DGCR5 and miR-21; however, future studies are
required to elucidate the molecular mechanism involved.
DGCR5 overexpression resulted in downregulated
miR-21 but failed to significantly affect CRC cell migration and
invasion (data not shown). miR-21 may regulate in vitro CRC
cell invasion and migration (16).
Therefore, DGCR5 may interact with multiple effectors to regulate
CRC cell behavior. Future studies are required to identify the
effectors involved.
In conclusion, DGCR5 was downregulated in CRC
tissues compared to healthy adjacent samples in patients with CRC
and overexpression of DGCR5 in vitro inhibited CRC cell
proliferation by downregulating miR-21.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
HH and XY designed the experiments. HH, XY, JC and
JF performed the experiments. CC, JW and QM collected and analyzed
the data. XY drafted the manuscript. All authors have read and
approved this manuscript.
Ethics approval and consent to
participate
The Ethics Committee of The Second Affiliated
Hospital of Guangxi University of Science and Technology approved
the current study and all patients signed informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: Incidence, mortality, survival and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Van Cutsem E, Lenz HJ, Köhne CH, Heinemann
V, Tejpar S, Melezínek I, Beier F, Stroh C, Rougier P, van Krieken
JH and Ciardiello F: Fluorouracil, leucovorin, and irinotecan plus
cetuximab treatment and RAS mutations in colorectal cancer. J Clin
Oncol. 33:692–700. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang M, Miao F, Huang R, Liu W, Zhao Y,
Jiao T, Lu Y, Wu F, Wang X, Wang H, et al: RHBDD1 promotes
colorectal cancer metastasis through the Wnt signaling pathway and
its downstream target ZEB1. J Exp Clin Cancer Res. 37:222018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marquez J, Fernandez-Piñeiro I,
Araúzo-Bravo MJ, Poschmann G, Stühler K, Khatib AM, Sanchez A, Unda
F, Ibarretxe G, Bernales I and Badiola I: Targeting liver
sinusoidal endothelial cells with miR-20a-loaded nanoparticles
reduces murine colon cancer metastasis to the liver. Int J Cancer.
143:709–719. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sillars-Hardebol AH, Carvalho B, de Wit M,
Postma C, Delis-van Diemen PM, Mongera S, Ylstra B, van de Wiel MA,
Meijer GA and Fijneman RJ: Identification of key genes for
carcinogenic pathways associated with colorectal
adenoma-to-carcinoma progression. Tumour Biol. 31:89–96. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wahlestedt C: Targeting long non-coding
RNA to therapeutically upregulate gene expression. Nat Rev Drug
Discov. 12:433–446. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dong HX, Wang R, Jin XY, Zeng J and Pan J:
LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via
inhibiting hsa-mir 22-3p. J Cell Physiol. 233:4126–4136. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen F, Yin S, Zhu J, Liu P, Yang C, Feng
Z and Deng Z: lncRNA DGCR5 acts as a tumor suppressor in papillary
thyroid carcinoma via sequestering miR-2861. Exp Ther Med.
17:895–900. 2019.PubMed/NCBI
|
|
13
|
Feiersinger F, Nolte E, Wach S, Rau TT,
Vassos N, Geppert C, Konrad A, Merkel S, Taubert H, Stürzl M and
Croner RS: MiRNA-21 expression decreases from primary tumors to
liver metastases in colorectal carcinoma. PLoS One.
11:e01485802016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hari D M, Leung AM, Lee JH, Sim MS, Vuong
B, Chiu CG and Bilchik AJ: AJCC Cancer Staging Manual 7th edition
criteria for colon cancer: Do the complex modifications improve
prognostic assessment? J Am Coll Surg. 217:181–190. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang R, Wang X, Zhang W, Zhangyuan G, Jin
K, Yu W, Xie Y, Xu X, Wang H and Sun B: Down-regulation of LncRNA
DGCR5 correlates with poor prognosis in hepatocellular carcinoma.
Cell Physiol Biochem. 40:707–715. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pan X, Wang ZX and Wang R: MicroRNA-21: A
novel therapeutic target in human cancer. Cancer Biol Ther.
10:1224–1232. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu L, Ye H, Huang G, Luo F, Liu Y, Liu Y,
Yang X, Shen J, Liu Q and Zhang J: Long noncoding RNA GAS5
suppresses the migration and invasion of hepatocellular carcinoma
cells via miR-21. Tumuor Biol. 37:2691–2702. 2016. View Article : Google Scholar
|
|
19
|
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY,
Gong W and Quan ZW: Long non-coding RNA CCAT1 promotes gallbladder
cancer development via negative modulation of miRNA-218-5p. Cell
Death Dis. 6:e15832015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Q, Chen F, Zhao J, Li B, Liang Y, Pan
W, Zhang S, Wang X and Zheng D: Long non-coding RNA PVT1 promotes
osteosarcoma development by acting as a molecular sponge to
regulate miR-195. Oncotarget. 7:82620–82633. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu Y, Zhang G, Zou C, Gong Z, Wang S, Liu
J, Ma G, Liu X, Zhang W and Jiang P: Long noncoding RNA DGCR5
suppresses gastric cancer progression by acting as a competing
endogenous RNA of PTEN and BTG1. J Cell Physiol. 234:11999–12010.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song L, Liu S, Zhang L, Yao H, Gao F, Xu D
and Li Q: MiR-21 modulates radiosensitivity of cervical cancer
through inhibiting autophagy via the PTEN/Akt/HIF-1α feedback loop
and the Akt-mTOR signaling pathway. Tumuor Biol. 37:12161–12168.
2016. View Article : Google Scholar
|
|
23
|
Rana C, Piplani H, Vaish V, Nehru B and
Sanyal SN: Downregulation of PI3-K/Akt/PTEN pathway and activation
of mitochondrial intrinsic apoptosis by Diclofenac and Curcumin in
colon cancer. Mol Cell Biochem. 402:225–241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang N, Chen J, Zhang H, Wang X, Yao H,
Peng Y and Zhang W: LncRNA OIP5-AS1 loss-induced microRNA-410
accumulation regulates cell proliferation and apoptosis by
targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple
myeloma. Cell Death Dis. 8:e29752017. View Article : Google Scholar : PubMed/NCBI
|