Introduction
With advances in cancer treatment and prevention,
the survival of patients with gastric cancer (GC) has significantly
improved during the past decades (1). However, GC remains a common type of
malignancy worldwide and is a major cause of mortality among
patients due to its aggressive nature (2). In effect, GC has been the leading cause
of cancer-associated mortalities in China for the past 3 decades
(3). A major challenge in the
treatment of GC is the high prevalence of metastasis by the time of
initial diagnosis (4). It has been
reported that a considerable number of tumor suppressors and
oncogenes that are involved in the pathogenesis of GC, and these
genes may serve as potential therapeutic targets for GC (5). However, the tumor suppressors and
oncogenes identified so far may not be sufficient to explain the
complex mechanism underlying the pathogenesis of GC.
Long non-coding RNAs (lncRNAs) are transcripts
>200 nucleotides in length that have been identified to serve
important roles in cancer biology (6,7). lncRNAs
participate in the development and progression of cancer through
the regulation of gene expression at different levels (8,9).
Therefore, it is possible to target lncRNAs to indirectly regulate
the expression of specific genes involved in human diseases,
thereby assisting disease treatment (10). However, the function of the majority
of lncRNAs in cancer remains unknown. Therefore, future studies
investigating the roles of lncRNAs in different aspects of cancer
biology are required. A previous study reported that mitotically
associated long non coding RNA (MANCR) affects cell division and
genomic stability in breast cancer (11). In breast cancer, MANCR is upregulated
and expression levels of MANCR are closely associated with cell
cycle progression (11). LncRNA may
interact with different downstream protein-coding genes or other
non-coding RNAs, including microRNAs (miRNAs/miRs) (10). miR-101 is downregulated in GC and
inhibits cancer cell invasion and migration in vitro
(12). miR-101 and MANCR were
inversely associated in GC (revealed by deep sequencing data, data
not shown). The present study was therefore carried out to
investigate the interaction between miR-101 and MANCR in GC.
Materials and methods
Patients
A total of 102 pateints were diagnosed and treated
for GC in The Third Affiliated Hospital of Nanchang University
between July 2011 and July 2013. The present study selected 69
cases (39 males and 30 females; age range, 35–67 years; mean age,
49.9±7.1 years) based on the following inclusion criteria: i)
Patients who had not received any therapy prior to admission, ii)
patients who were diagnosed for the first time, iii) patients with
no previous history of maligancy and iv) patients willing to
participate in a five-year follow-up. The exclusion criteira were
as follows: i) Patients with other medical disorders and ii)
patients who were transferred from other hospitals. Accoring to the
American Joint Committee on Cancer staging criteria (13), there were 12, 18, 20 and 19 patients
with stage I, II, III and IV GC, respectively. The Ethics Committee
of The Third Affiliated Hospital of Nanchang University approved
the study prior to patient enrolment and written informed consent
was obtained from all patients.
Tissue specimens and cell lines
Patients underwent a gastric biopsy to collect GC
and adjacent (2 cm) non-cancerous tissues prior to receiving
therapy. All tissue specimens were analyzed by five experienced
histopathologists. Tissue specimens were stored in liquid nitrogen
prior to use.
The human GC cell lines HGC-27 (Sigma-Aldirch; Merck
KGaA), and SNU-1 (American Type Culture Collection) were used for
in vitro experiments. Cells were cultured in Eagle's Minimum
Essential Medium (EMEM) containing 2 mM glutamine, 1% non-essential
amino acids and 10% fetal bovine serum supplemented with 100 IU/ml
penicillin-streptomycin. All medium and cell culture reagents were
from Sigma-Aldrich (Merck KGaA). Cell culture conditions were at
37°C, 5% CO2 and 95% humidity, and were collected once
they reached ~80% confluency.
Follow-up
All patients were monitored for five years.
Follow-up was performed through either outpatient visits or
telephone calls. Patients who were lost to follow-up or succumbed
to accidents or unrelated clinical disorders were not included in
the present study.
Reverse-transcription quantitative
polymerase chain reaction (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from GC and
adjacent non-cancerous tissues and HGC-27 and SNU-1 cell lines. For
MANCR RT-qPCR, AMV Reverse Transcriptase (Promega Corporation) was
used to perform reverse transcription through following conditions:
50°C for 20 min and 80°C for 10 min. All operations were performed
according to the manufacturer's protocols. qPCR was subsequently
performed using SYBR® Green Master Mix (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocols. 18S
rRNA was used as an endogenous control. miRNA extraction was
performed using the mirPremier™ microRNA Isolation kit
(Sigma-Aldrich; Merck KGaA). TaqMan MicroRNA Reverse Transcription
kit (Thermo Fisher Scientific, Inc.) was used to perform reverse
transcription and qPCR was subsequently performed using TaqMan
Real-Time PCR Master Mix (Thermo Fisher Scientific, Inc.). miR-101
expression was normalized to the U6 endogenous control. Primer
sequences were as follows: 5′-CTAATCCTGTCAGGCAGCCT-3′ (forward) and
5′-ATTCTCAGGCTTTCTTTGCC-3′ (reverse) for MANCR;
5′-CTACCACATCCAAGGAAGC3′ (forward) and 5′-TTTCGTCACTACCTCCCCG-3′
(reverse) for human 18S rRNA. miR-101 forward primer was:
5′-CAGTTATCACAGTGCTGATG-3′. miR-101 reverse primer and U6 primers
were included in the kit. The thermocycling conditions were as
follows: 95°C for 50 sec, followed by 40 cycles of 95°C for 20 sec
and 60°C for 45 sec. All qPCR reactions were performed in
triplicate and data were analyzed using the 2−ΔΔCq
method (14).
Transient cell transfections
Full length MANCR cDNA was inserted into a pcDNA3.1
vector (Invitrogen; Thermo Fisher Scientific, Inc.) to establish a
MANCR overexpression vector. The vector was constructed by Sangon
Biotech Co., Ltd. Negative control (NC) miRNA
(5′-CAGGUTUACCUAUGCCCUUUGCU-3′) and an miR-101
(5′-CAGUUAUCACAGUGCUGAUGCU-3′) mimic were purchased from
Sigma-Aldrich, Merck KGaA. MANCR small interfering (si) RNA
(5′-GAGAACUGCCGGGACCUCCAG-3′) and NC siRNA
5′-(UUGGACCCUUUCGCUGUACUC-3′) were purchased from Shanghai
GenePharma Co., Ltd. HGC-27 and SNU-1 cells were cultured overnight
in aforementioned medium under aforementioned conditions in a
6-well cell culture medium (1×106 cells/well) overnight
to reach 70–80% confluency. All cell transfections were performed
using Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocols. The vector was transfected at a concentration of 10 nM
and the miRNAs and siRNAs were transfected at a concentration of 35
nM. A control (C) group consisting of untransfected cells and a NC
group consisting of cells transfected with NC miRNA, NC siRNA or an
empty vector were included. Subsequent experiments were performed
24 h following transfection.
Cell Counting Kit-8 (CCK-8) assay
HGC-27 and SNU-1 cells were harvested 24 h following
transfection and single cell suspensions using EMEM were prepared.
The cell density was adjusted to 4×104 cells/ml. Cells
were subsequently cultured under aforementioned conditions in a
96-well plate (0.1 ml per well). A total of 10 µl CCK-8 solution
(Sigma-Aldrich; Merck KGaA) was added to each well every 24 h over
a four-day period. Cells were subsequently incubated under
aforementioned conditions for an additional 4 h. The optical
density values were measured at a wavelength of 450 nM.
Statistical analysis
All experiments in the current study were performed
in triplicate. Data were expressed as mean ± standard deviation.
Differences among different clinical stages or among different cell
transfection groups were analyzed by the one-way ANOVA followed by
the Tukey post hoc test. Differences between GC and adjacent
non-cancerous tissues were analyzed using the paired t-test. Linear
regression was used to analyze the association between the
expression levels of MANCR and miR-101. Patients were divided into
high and low MANCR expression level groups according to Youden's
index, with 32 and 37 patients in each group, respectively.
Survival curves were plotted using the Kaplan-Meier method and
compared using the log-rank test. Univariate regression was
performed to analyze the effects of patient clinical data on MANCR
expression. P<0.05 was considered to indicate a statistically
significant difference.
Results
MANCR is upregulated in patients with
GC and is not affected by clinical stages
Expression of MANCR in GC and adjacent non-cancerous
tissues was analyzed by RT-qPCR. MANCR expression was significantly
upregulated in GC tissues compared with adjacent non-cancerous
tissues (Fig. 1A; P<0.05). No
significant differences in MANCR expression levels were observed
among patients with different clinical stages of GC (Fig. 1B). In addition, univariate regression
was performed to analyze the effects of patient clinical data on
MANCR expression. Patient age, sex, smoking and drinking habits BMI
and history of gastric diseases did not have significant effects on
MANCR expression (data not shown; P<0.05).
Patients with high MANCR expression
levels exhibit worse survival compared with patients with low
expression
Patients were divided into high and low MANCR
expression level groups based on Youden's index, with 32 and 37
cases in each group, respectively. Survival curves were plotted
using the Kaplan-Meier method and compared by the log-rank test.
Patients in the high MANCR expression level group had a
significantly worse overall survival rate compared with patients in
the low expression group (Fig. 2;
P<0.05).
miR-101 is inversely associated with
MANCR
Expression of miR-101 in tissues was analyzed by
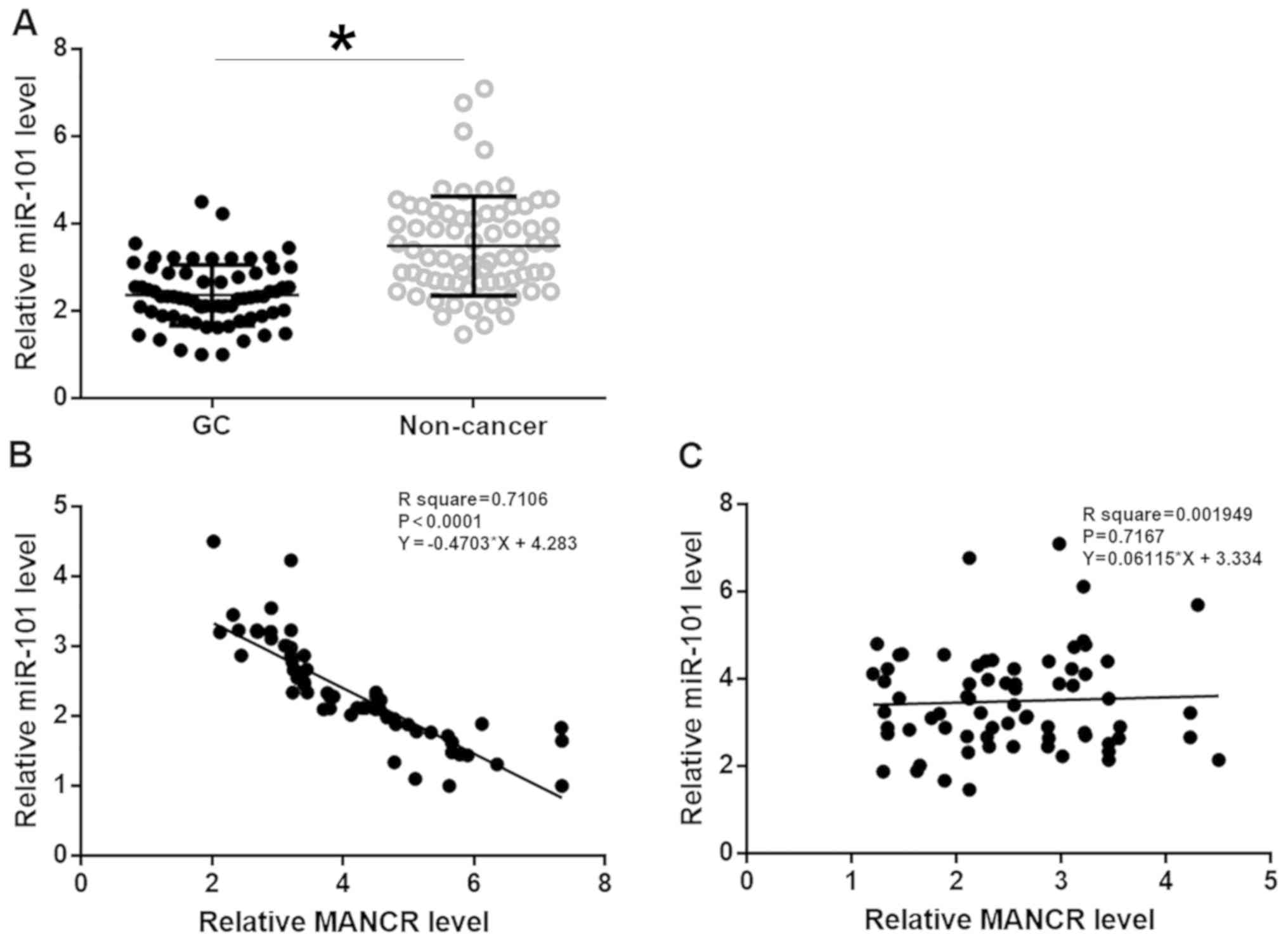
RT-qPCR. miR-101 was significantly downregulated in GC tissues
compared with adjacent non-cancerous tissues (Fig. 3A; P<0.05). Linear regression was
used to analyze the association between expression levels of MANCR
and miR-101. The expression level of miR-101 was significantly and
inversely associated with the MANCR expression level in GC tissues
(Fig. 3B; P<0.05), but not in
adjacent non-cancerous tissues (Fig.
3C).
miR-101 expression is regulated by
MANCR in GC cells
The MANCR expression vector, MANCR siRNA and miR-101
mimic were transfected into HGC-27 and SNU-1 cell lines. The
expression levels of MANCR (Fig. 4A)
and miR-101 (Fig. 4B) were
significantly increased 24 h following transfection compared with
the C and NC groups (P<0.05), indicating successful
transfection. MANCR overexpression resulted in miR-101
downregulation in the HGC-27 and SNU-1 cells compared with the C
and NC groups (Fig. 4C; P<0.05),
while miR-101 overexpression did not alter MANCR expression
compared with the C and NC groups (Fig.
4D. Furthermore, MANCR siRNA resulted in miR-101 upregulation
compared with the C and NC groups (Fig.
4E; P<0.05).
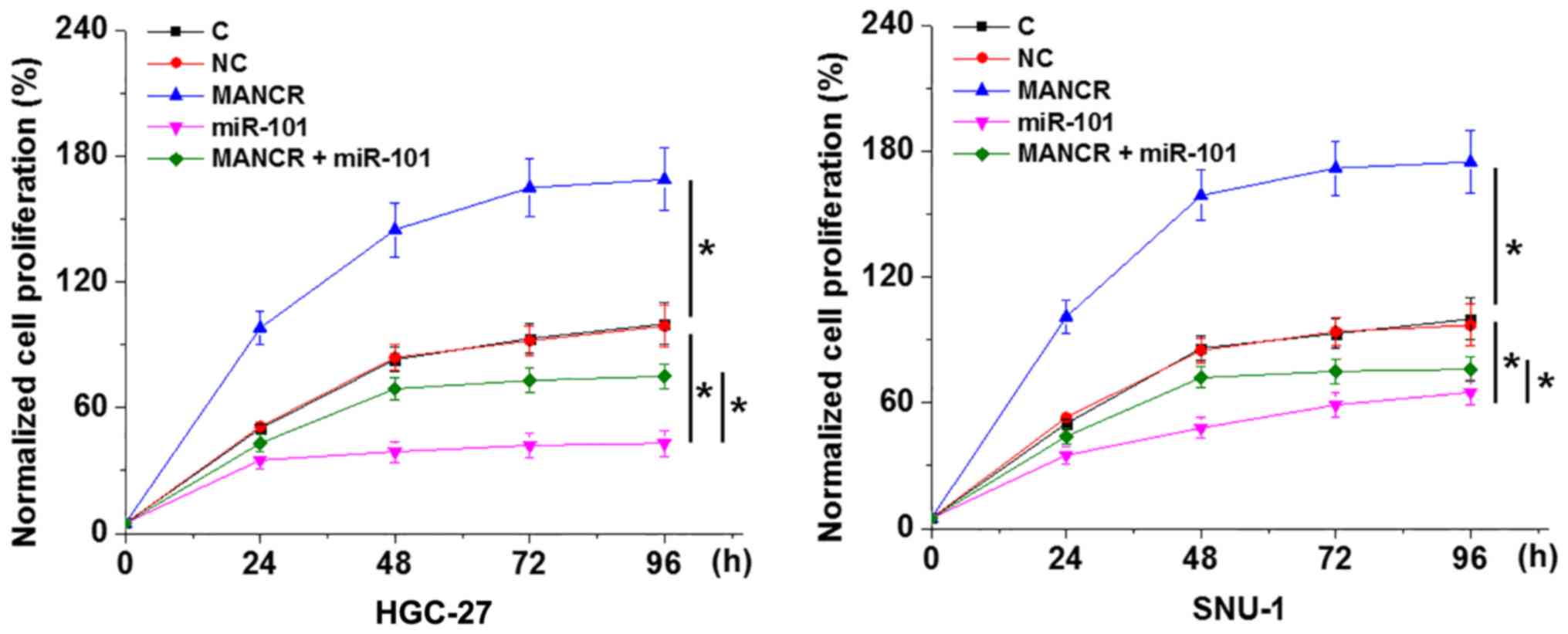
MANCR regulates GC cell proliferation
through miR-101
The CCK-8 assay revealed that, compared with the C
and NC groups, MANCR overexpression promoted, while miR-101
overexpression inhibited GC cell proliferation (Fig. 5; P<0.05). In addition, miR-101
overexpression decreased the effect of MANCR overexpression on cell
proliferation (Fig. 5;
P<0.05).
Discussion
To the best of our knowledge, the function of MANCR
has only been characterized in breast cancer (11). The present study analyzed the
expression pattern of MANCR in GC and explored its effects on
cancer cell behavior. MANCR may serve an oncogenic role in gastric
cancer by downregulating miR-101, which has been identified as a
tumor suppressive miRNA in GC (12).
Despite improvements in treatment, the overall
survival rate for patients with GC remains poor (15,16).
Furthermore, the mortality rate in developing countries, such as
China, is higher than that in Western countries, such as the United
States of America (16). The present
study focused on the prognostic value of MANCR in GC. MANCR was
upregulated in GC tissues compared with adjacent non-cancerous
tissues but was not affected by clinical stages, suggesting that
MANCR upregulation may be implicated in the development of GC.
Analysis of survival data revealed that patients with high MANCR
expression levels in GC tissues had a significantly decreased
overall survival rate compared with patients with low MANCR
expression. Therefore, MANCR may serve as a prognosis indicator in
GC.
lncRNAs participate in cancer biology by regulating
downstream regulatory molecules, including miRNAs (17,18). The
present study revealed that MANCR is a likely upstream inhibitor of
miR-101 in GC cells. A previous study demonstrated that
overexpression of miR-101 inhibits GC cell migration and invasion
in vitro (12). The present
study revealed that miR-101 overexpression inhibited the
proliferation of GC cells. The results obtained in the present
study are consistent with the role of MANCR in breast cancer, where
it mediates cancer cell division (11). lncRNAs may serve as an miRNA sponge
(19); however, a binding site for
miR-101 was not found on MACNR. Further investigation is required
to elucidate the interactions between MANCR and miR-101.
The present study investigated the role of MACNR in
regulating cancer cell proliferation. Further studies are required
to investigate the roles of this lncRNA in regulating other cancer
cell behaviors, including cell cycle progression and apoptosis.
In conclusion, the results obtained in the present
study revealed that MANCR was upregulated in GC tissues and
promoted GC cell proliferation in vitro by downregulating
miR-101.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81660408).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NF and LY designed the experiments. LY, JY and LG
performed experiments and data analysis. SH and FC analyzed data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from The Third
Affiliated Hospital of Nanchang University Medical Research Ethics
Committee, and written informed consent was obtained from all
patients and controls.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bertuccio P, Chatenoud L, Levi F, Praud D,
Ferlay J, Negri E, Malvezzi M and La Vecchia C: Recent patterns in
gastric cancer: A global overview. Int J Cancer. 125:666–673. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu S, Mao J, Shao Y, Chen F, Zhu X, Xu D,
Zhang X and Guo J: Reduced expression of the long non-coding RNA
AI364715 in gastric cancer and its clinical significance. Tumour
Biol. 36:8041–8045. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bernard D, Prasanth KV, Tripathi V,
Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L,
Coulpier F, et al: A long nuclear-retained non-coding RNA regulates
synaptogenesis by modulating gene expression. EMBO J. 29:3082–3093.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wahlestedt C: Targeting long non-coding
RNA to therapeutically upregulate gene expression. Nat Rev Drug
Discov. 12:433–446. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tracy KM, Tye CE, Ghule PN, Malaby HLH,
Stumpff J, Stein JL, Stein GS and Lian JB: Mitotically-associated
lncRNA (MANCR) affects genomic stability and cell division in
aggressive breast cancer. Mol Cancer Res. 16:587–598. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT,
Xia YJ, Ye ZY and Tao HQ: MicroRNA-101 is down-regulated in gastric
cancer and involved in cell migration and invasion. Eur J Cancer.
46:2295–2303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He X, Wu W, Lin Z, Ding Y, Si J and Sun
LM: Validation of the American Joint Committee on Cancer (AJCC)
stage system for gastric cancer patients: A population-based
analysis. Gastric Cancer. 21:391–400. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Strong VE, Song KY, Park CH, Jacks LM,
Gonen M, Shah M, Coit DG and Brennan MF: Comparison of gastric
cancer survival following R0 resection in the United States and
Korea using an internationally validated nomogram. Ann Surg.
251:640–646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Strong VE, Wu AW, Selby LV, Gonen M, Hsu
M, Song KY, Park CH, Coit DG, Ji JF and Brennan MF: Differences in
gastric cancer survival between the U.S. and China. J Surg Oncol.
112:31–37. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jalali S, Bhartiya D, Lalwani MK,
Sivasubbu S and Scaria V: Systematic transcriptome wide analysis of
lncRNA-miRNA interactions. PLoS One. 8:e538232013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|