Introduction
In 2018, thyroid cancer was the most common thyroid
malignancy, accounting for ~1% of all malignant tumors, and
includes papillary, follicular, undifferentiated and medullary
carcinomas (1). Thyroid cancer
occurs mostly in young adults aged ~40 years, and the ratio men to
women is 1.0:2.5 to 1.0:3.0 (2).
Papillary thyroid carcinoma is typically exhibits lower malignancy,
and better prognosis and is the most common type of thyroid cancer,
accounting for 85–90% of cases (3).
In recent years, the incidence of thyroid cancer has steadily
increased (4). Most patients (~90%)
with PTC can be effectively treated via surgical removal and have a
good 5-year survival rate (5).
However, a small proportion of patients with PTC have poor
prognosis and survival rates (6).
Therefore, it is necessary to illustrate the molecular mechanism
underlying proliferation and invasion of thyroid cancer, in order
to identify potential therapeutic targets (7).
Long non-coding RNA (lncRNAs) is a type of RNA
>200 nucleotides in length, with little or no coding potential
(8). Recently, lncRNAs were found to
serve a pivotal role in several biological processes (9–11), such
as embryo development (12,13), immunology (14,15) and
cancer (16–18). Dysregulation of lncRNAs have been
shown to be involved in tumorigenesis and tumor progression in PTC
(19–21), suggesting the potential of lncRNAs as
diagnostic markers or therapeutic agents for PTC (22,23). For
example, lncRNA LINC00460 promotes carcinogenesis by sponging
microRNA (miR)-613 in PTC and upregulating the expression of
sphingosine kinase 2 (24).
Additionally, lncRNA FOXD2-AS1 was found to be upregulated in PTC
tissues and function as a competing endogenous RNA (ceRNA) to
enhance the expression of KLK7, by sponging miR-485-5p (25).
lncBRM was first reported to be highly expressed in
liver stem cells and was shown to be associated with the
progression of liver cancer, by interacting with BRM and regulating
the YAP signaling pathway (26).
Moreover, lncBRM was proven to exhibit a pivotal role in ovarian
cancer, through the upregulation of SOX4 and thereby facilitating
proliferation, migration and invasion of ovarian cancer cells
(27). Furthermore, lncBRM was shown
to regulate the proliferation and invasion of colorectal cancer
cells, by sponging miR-204-3p and upregulating
translationally-controlled tumor protein 1 (TPT1) (28). However, the role of lncBRM in PTC
remains unclear. In the present study, the expression patterns,
biological functions and mechanisms of action of lncBRM in PTC
progression were elucidated.
Materials and methods
Samples and cell lines
PTC samples and matched normal tissues were obtained
from 90 patients (mean age, 63.5±5.1 years; age range, 37.5–72.4
years; 32 men and 58 women) with PTC undergoing surgery at The
People's Hospital of Tong Liang District (Chongqing, China). The
pathological diagnosis of all specimens was graded according to the
classification of thyroid malignancy of the World Health
Organization (2004) by two experienced pathologists (29). Among these patients, 41 cases were
high-grade and 41 cases were low-grade, and 46 cases were
non-metastasis and 44 cases were metastasis. All tissues were
frozen in liquid nitrogen until further use. The study was approved
by the Ethics Committee of The People's Hospital of Tong Liang
District (permission no. 2015KT57). All patients provided written
informed consent.
The normal human thyroid follicular epithelial cell
line Nthy-ori 3-1, and human thyroid cancer cell lines TPC-1 and
SW1736, were obtained from the American Type Culture Collection.
All cells were cultured in RPMI-1640 medium (Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (HyClone; GE Healthcare
Life Sciences), 100 U/ml penicillin and 100 µg/ml streptomycin
(both from Sigma-Aldrich; Merck KGaA) at 37°C in 5%
CO2.
Plasmids and transfections
TPC-1 and SW1736 cells were seeded into six-well
plates at a density of 1×105 cells/well, and cultured
overnight until they reach 70–80% confluence. miR-331-3p mimic,
empty vector control, miR-331-3p inhibitor were used at 50 nM. The
short hairpin (sh)RNA specifically targeting lncBRM, SLC25A1 and
scrambled negative control shRNA were provided by GenePharma Co.
Ltd and used at 1 mg/ml. The coding sequence of SLC25A1 was
constructed into pMy vectors (Addgene, Inc.) to overexpress SLC25A1
that was used at 1 mg/ml. The empty control, which served as a
negative control, was also used at 1 mg/ml. Plasmids were
transfected into the cells using Lipofectamine™ 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. After 6 h, successfully transfected cells were
confirmed by reverse transcription-quantitative PCR (RT-qPCR)
analysis and cultured in six-well plates using RPMI-1640 medium
containing 10% FBS for two days at 37°C and 5% CO2 to
expand for subsequent experiments. The sequences of the primers
used were as follows: miR-331-3p mimics,
5′-GCCCCUGGGCCUAUCCUAGAA-3′; and miR-331-3p inhibitor,
5′-TTCTUGGUTUGGCCCUGGGGC-3′. pMIR-SLC25A1-3′UTR (wild type or
mutant) or pMIR-lncBRM (wild type or mutant) and miR-331-3p mimics
were transfected into TPC-1 or SW1736 cell lines along with pRL-TK
vectors (Promega Corporation). Following culture for 24 h,
luciferase activity was measured using a dual Glo™ Luciferase assay
system (Promega Corporation) according to the manufacturer's
protocols and normalized to Renilla luciferase activity.
RNA preparation and RT-qPCR
Total RNA was isolated from tissues or cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Total RNA was
subsequently reverse-transcribed into cDNA using the PrimeScript RT
reagent kit (Promega Corporation) according to the manufacturer's
protocol. The reaction was performed at 42°C for 1 h, and the
enzyme was subsequently inactivated at 85°C for 5 min. qPCR was
performed using SYBR Green PCR Master mix reagents (Takara Bio,
Inc.) in a 7300 Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). miR-331-3p expression was determined
using SYBR Premix Ex Taq II (GeneCopoeia, Inc.) according to the
following conditions: 10 min of pre-denaturation at 95°C, followed
by 40 cycles of 10 sec denaturation at 95°C, 20 sec annealing at
60°C and 30 sec extension at 72°C. SLC25A1 mRNA expression was
measured using a SYBR Green PCR Master Mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with the following procedures: 10
min pre-denaturation at 95°C, followed by 36 cycles of 10 sec
denaturation at 95°C, 20 sec annealing at 60°C and 34 sec extension
at 72°C. 18S was used as internal reference for miR-331-3p and
SLC25A1 mRNA expression. Relative expression of miR-6852 and LEF1
mRNA was calculated using 2−ΔΔCq method (30). The sequences of the primers used were
as follows: SLC25A1 forward, 5′-CCGTCAGGTTTGGAATGTTCG-3′ and
reverse, 5′-TAACCCCGTGGAAGAATCCTC-3′; lncBRM forward,
5′-GGTCAAGAGGCCAGGAAGAG-3′ and reverse,
5′-TTCTCACTTCAGCCCAATGCT-3′; and 18S forward,
5′-CAGCCACCCGAGATTGAGCA-3′ and reverse,
5′-TAGTAGCGACGGGCGGTGTG-3′.
Cell counting Kit-8 (CCK-8)
assays
TPC-1 and SW1736 cells (1×104) were
seeded in a 96-well plate and cultured in RPMI-1640 medium
supplemented with 10% FBS at 37°C and 5% CO2. CCK-8
solution (10 µl; Beyotime Institute of Biotechnology) was added
into each well for 4 h at 37°C. Cell proliferation was assessed 24,
48 and 72 h post-transfection. The absorbance was measured at 450
nm using a microplate reader (BioTek Instruments, Inc.).
Colony-formation assays
Cells in the logarithmic growth phase were treated
with 0.25% trypsin to give single-cell suspensions. Each group was
inoculated at a density of ~1,000 cells/well in the culture medium.
The culture was terminated when colonies were visible, after 2–3
weeks. The supernatant was discarded and the colonies were fixed at
room temperature with 4% paraformaldehyde for 15 min and stained
with 0.5% Giemsa for 30 min at room temperature. The number of
colonies were examined under a light microscope (magnification,
×100).
Bioinformatics analysis
miR-331-3p was predicted as a potential lncBRM
target by using miRDB tool (http://mirdb.org/miRDB/index.html). miRDB is an online
database for miRNA target prediction and functional annotations.
All the targets in miRDB were predicted by a bioinformatics tool,
MirTarget, which was developed by analyzing thousands of
miRNA-target interactions from high-throughput sequencing
experiments. SLC25A1 was predicted as a potential miR-331-3p target
by using TargetScan7 tool (http://www.targetscan.org/vert_72/). TargetScan
predicts biological targets of miRNAs by searching for the presence
of conserved 8mer, 7mer, and 6mer sites that match the seed region
of each miRNA.
Transwell experiments
Two days after transfection, TPC-1 and SW1736 cells
were prepared as single cell suspensions (1×105
cells/ml) in serum-free RPMI-1640. Transwell chambers (8 mm pore;
EMD Millipore) were inserted into 24-well plates containing 600 µl
RPMI-1640 supplemented with 10% FBS in the lower chamber. A 100 µl
cell suspension containing 1×104 cell was added into the
upper chamber. Cells were cultured for 48 h at 37°C. Non-migrated
cells were scraped off and migrated cells were fixed using 4%
formaldehyde for 30 min at room temperature, stained using 0.5%
crystal violet for 30 min at room temperature and counted under a
light microscope (magnification, ×200). The aforementioned
procedure was also used to detect cell invasion; however, for the
cell invasion assay, Transwell were pre-coated with 100 ml Matrigel
(1 mg/ml; BD Biosciences) for 30 min at 37°C, prior to cell
seeding.
Western blot analysis
Total proteins in each cell sample were extracted
using RIPA lysis buffer (Beyotime Institute of Biotechnology).
Proteins were quantified using a bicinchoninic acid kit (Pierce;
Thermo Fisher Scientific, Inc.), and 40 mg protein/lane were
separated by 10% SDS-PAGE. Proteins were then transferred onto
polyvinylidene difluoride membranes. Membranes were subsequently
incubated with 5% skimmed milk for 1 h at room temperature. TBS
supplemented with 0.1% Tween 20 (TBST) was used to wash the
membrane. Membranes were incubated with primary antibodies against
SLC25A1 (1:2,000; cat. no. 15235-1-AP; ProteinTech Group, Inc.) and
β-actin (1:5,000; cat. no. 60008-1-Ig; ProteinTech Group, Inc.) for
12 h at 4°C. Membranes were then incubated with goat anti-mouse
(cat. no. 70-GAM007) and goat anti-rabbit (cat. no. 70-GAR007)
secondary antibodies [1:5,000; Multisciences (Lianke) Biotech Co.,
Ltd.] for 2 h at room temperature. Membranes were washed with three
times TBST for 15 min. Protein expression was monitored using the
Pierce™ ECL Western Blotting Substrate (Beijing Solarbio Science
& Technology Co., Ltd.) and quantified using Quantity One
software v4.62 (Bio-Rad Laboratories Inc.).
Statistical analysis
GraphPad Prism version 6 (GraphPad Software, Inc.)
was used to analyze the data. All results are expressed as the mean
± standard deviation. A Student's t-test was used to analyze the
differences between two groups. A one-way ANOVA with a pos-hoc
Tukey's test was used for multiple comparisons. Kaplan-Meier
analysis followed by a log-rank test was used to analyze survival
rate. The association between lncBRM expression and clinical
features listed in Table I was
analyzed using a χ2 test. P<0.05 was considered to
indicate a statistically significant difference.
 | Table I.Associations between lncBRM
expression and clinicopathological features in 90 patients with
papillary thyroid carcinoma. |
Table I.
Associations between lncBRM
expression and clinicopathological features in 90 patients with
papillary thyroid carcinoma.
| Variables | Low (n=45) | High (n=45) | P-value |
|---|
| Age, years |
|
| 0.397 |
|
>60 | 23 | 18 |
|
|
≤60 | 22 | 27 |
|
| Sex |
|
| 0.833 |
|
Male | 21 | 23 |
|
|
Female | 24 | 22 |
|
| TNM stage |
|
| 0.033 |
|
I–II | 30 | 19 |
|
|
III–IV | 15 | 26 |
|
| Lymph node
metastasis |
|
| 0.020 |
|
Yes | 16 | 28 |
|
| No | 29 | 17 |
|
Results
lncBRM is upregulated in PTC tissues
and cell lines
In order to determine the role of lncBRM in PTC
progression, RT-qPCR was performed, which revealed increased
expression of lncBRM in PTC tissues compared with the adjacent
normal tissue (P<0.05; Fig. 1A).
Consistently, lncBRM was also upregulated in the PTC cell lines
TPC-1 and SW1736, compared with the normal thyroid Nthy-ori 3-1
cell line (both P<0.05; Fig. 1B).
To further examine the prognostic significance of lncBRM, overall
survival time was analyzed. Based on the median level of lncBRM
expression, the 90 tissues were divided into two groups, lncBRM
high and lncBRM low expression groups. Higher expression of lncBRM
was associated with poor overall survival time in patients with PTC
(P<0.05; Fig. 1C). Moreover, high
expression of lncBRM was associated with metastasis and advanced
stages of PTC (both P<0.05; Fig. 1D
and E). The potential association between lncBRM expression and
the clinicopathological features of patients with PTC was also
explored. High lncBRM expression was associated with late
Tumor-Node-Metastasis (P=0.033) stages and lymph node metastasis
(P=0.02; Table I). Overall, lncBRM
was upregulated in PTC and was associated with a poor
prognosis.
lncBRM knockout inhibits the
progression of PTC cells
In order to elucidate the mechanism by which lncBRM
regulates PTC progression, TPC-1 and SW1736 cells were transfected
with shRNA targeting lncBRM and expression of lncBRM was
significantly decreased in the transfected cells (both P<0.05;
Fig. 2A). A CCK-8 assay was
performed to examine the proliferation of shcontrol or shlncBRM
TPC-1 and SW1736 cells. lncBRM knockout resulted in significantly
decreased proliferation of TPC-1 and SW1736 cells after 48 and 72 h
(both P<0.05; Fig. 2B). Transwell
assays revealed significantly decreased invasion and migration of
TPC-1 cells and SW1736 cells following lncBRM knockout (all
P<0.05; Fig. 2C). Furthermore,
colony-formation assays revealed that lncBRM promoted the
proliferation ability of TPC-1 and SW1736 cells (P<0.05;
Fig. 2D).
lncBRM negatively regulates miR-331-3p
expression in PTC
Bioinformatics analysis was conducted in order to
elucidate the mechanism by which lncBRM promotes PTC progression.
lncBRM formed complementary base pairing with miR-331-3p (Fig. 3A). To verify whether lncBRM binds to
miR-331-3p directly, a luciferase reporter assay was performed. The
luciferase activity was significantly decreased following
co-transfection with WT-lncBRM and miR-331-3p mimics (P<0.05;
Fig. 3B). However, luciferase
activity remained unchanged in cells co-transfected with WT-lncBRM
and miR-331-3p mimics (Fig. 3B).
Moreover, the expression of miR-331-3p was increased following
lncBRM knockout compared with the control cells (P<0.05;
Fig. 3C). In order to investigate
the association between miR-331-3p and lncBRM, cells were
transfected with miR-331-3p mimics or inhibitor plasmids and
transfection efficiency was confirmed (Fig. 3D). Transfection of miR-331-3p mimic
significantly inhibited the expression of lncBRM in TPC-1 cells,
whereas transfection with miR-331-3p inhibitor increased lncBRM
expression compared with control (both P<0.05; Fig. 3E). Consistently, CCK-8 assays showed
proliferation was decreased after lncBRM knockout in TPC-1 and
SW1736 cells compared with control after 48 and 72 h (both
P<0.05). However, the decreased proliferation caused by lncBRM
knockout was reversed by inhibition of miR-331-3p in TPC-1 and
SW1736 cells (Fig. 3F). Overall,
these data suggest that lncBRM can bind to miR-331-3p and regulate
TPC-1 cell proliferation.
 | Figure 3.lncBRM interacts with miR-331-3p. (A)
Schematic representation of the predicted target site for
miR-331-3p and lncBRM. (B) Luciferase reporter assay was performed
in TPC-1 cells. Cells were co-transfected with the reporter plasmid
(or the corresponding mutant reporter) and the indicated miRs.
RT-qPCR was performed to examine the relative expression of
miR-331-3p, following transfection with (C) negative control or
shlncBRM plasmids and (D) negative control, miR-331-3p mimic or
miR-331-3p inhibitor. (E) RT-qPCR was performed to examine the
relative expression of lncBRM in cells transfected with negative
control or ectopic miR-331-3p expression or inhibition of
miR-331-3p. Fold changes were normalized to 18S for all RT-qPCR
experiments. (F) Proliferative ability of TPC-1 and SW1736 cells,
following transfection with the negative control, shlncBRM alone or
with miR inhibitor, were determined through CCK-8 assays.
*P<0.05. RT-qPCR, reverse transcription-quantitative PCR; lnc,
long non-coding; sh, short hairpin; CCK-8, Cell Counting Kit-8; WT,
wild type; MUT, mutant; miR, microRNA. |
lncBRM regulates SLC25A1 expression
through miR-331-3p
A TargetScan7 analysis was performed to identify
potential genes that are regulated by miR-331-3p, and SLC25A1 was
identified as a potential candidate gene. There was a potential
binding site at SLC25A1-3′UTR with miR-331-3p (Fig. 4A). The role of SLC25A1 in PTC
progression was explored in the present study. Firstly, ectopic
expression of miR-331-3p or lncBRM knockout resulted in
significantly decreased luciferase activity of wild type
(P<0.05; Fig. 4B) but not mutant
SLC25A1-3′UTR. Inhibition of miR-331-3p restored the decreased
luciferase activity caused by lncBRM knockout (Fig. 4B). Ectopic expression of miR-331-3p
resulted in decreased SLC25A1 expression (P<0.05), whereas
miR-331-3p inhibitor promoted SLC25A1 expression (P<0.05;
Fig. 4C). Furthermore,
overexpression of miR-331-3p or knockout of lncBRM resulted in
decreased SLC25A1 expression, whereas inhibiting miR-331-3p
restored this phenotype (all P<0.05; Fig. 4D and E). These data suggest that
lncBRM regulates SLC25A1 expression by targeting miR-331-3p.
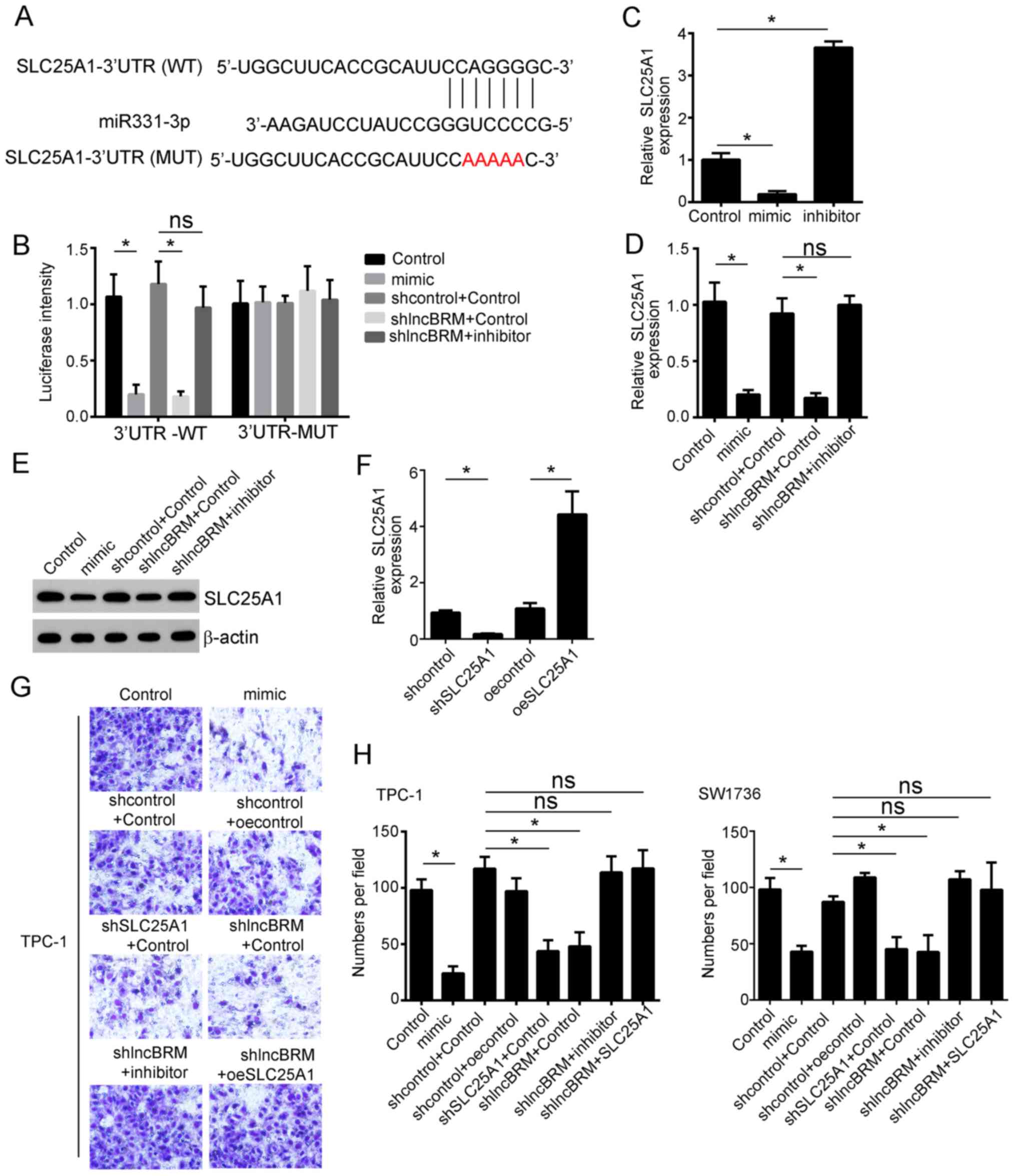 | Figure 4.lncBRM regulates PTC progression
through sponging miR-331-3p and targeting SLC25A1. (A) Schematic
representation of the predicted target site for miR-331-3p and
SLC25A1. (B) Luciferase activity assay in the WT or MUT reporter
containing SLC25A1 3′UTR when transfected with different vectors in
TPC-1 cells. Cells were co-transfected with the reporter plasmid
(or the corresponding mutant reporter) and the indicated miRs. The
relative expression of SLC25A1 following (C) ectopic miR-331-3p
expression or inhibition of miR-331-3p in TPC-1 cells and (D)
ectopic miR-331-3p expression or knockout of lncBRM and inhibition
of miR-331-3p in TPC-1 cells, examined by RT-qPCR. Samples were
normalized to 18S. (E) The protein expression of SLC25A1 was
examined by western blot analysis. Samples were normalized to
β-actin. (F) The relative expression of SLC25A1 following knockout
or overexpression of SLC25A1. Samples were normalized to 18S. (G
and H) Migratory abilities of TPC-1 cells and SW1736 cells
transfected with control and different plasmids, examined using
transwell assays. *P<0.05. RT-qPCR, revere
transcription-quantitative PCR; lnc, long non-coding; sh, short
hairpin; WT, wild type; MUT, mutant; miR, microRNA; UTR,
untranslated region, NS, not significant. |
To determine whether lncBRM exerts its biological
effects through miR-331-3p and SLC25A1, rescue assays were
performed by overexpressing of SLC25A1 or inhibiting SLC25A1 in
lncBRM knockout TPC-1 and SW1736 cells (Fig. 4F). Transwell assays demonstrated that
overexpression of miR-331-3p and knockout of lncBRM or SLC25A1
resulted in significantly decreased migration of cells (all
P<0.05). Inhibition of miR-331-3p or overexpression of SLC25A1
in lncBRM-depleted TPC-1 and SW1736 cells significantly restored
cell migration abilities (Fig. 4G and
H).
Discussion
Thyroid cancer is one of the most prevalent
endocrine tumors and the incidence rate is increasing steadily.
Therefore, it is important to illustrate the mechanisms of thyroid
cancer (31). In recent years,
lncRNAs have been shown to serve vital roles in many biological
processes (32). Moreover, several
studies revealed the roles of lncRNAs in the regulation of PTC
progression. For example, Xia et al (33) reported that lncRNA HOXA-AS2 was
upregulated in PTC tissues and promoted migration and invasion of
PTC cells by regulating the miR-520c-3p/S100A4 pathway. Wang et
al (34) demonstrated that
LINC01186 suppressed proliferation and invasion of PTC cells by
decreasing the expression of YAP1 and increasing LATS1
expression.
lncBRM was found to play vital roles in some cancer
types. Xi et al (27) found
that lncBRM facilitated ovarian cancer cell proliferation,
migration and invasion via SOX4 upregulation. Li et al
(28) reported that lncBRM can
sponge miR-204-3p and target TPT1 to promote colorectal cancer cell
migration, invasion and proliferation. However, the role of lncBRM
in thyroid cancer remains unclear. In the present study, lncBRM was
determined to be upregulated in PTC tissues and cell lines. lncBRM
knockout decreased TPC-1 cell proliferation, migration and
invasion. Furthermore, bioinformatics analysis identified
miR-331-3p as a potential target of lncBRM.
Solute carrier (SLC) members are a type of membrane
transport proteins (35). SLC25A1 is
a mitochondrial carrier, which promotes the flux of
citrate/isocitrate across the mitochondria (35). Previously, SLC25A1 was reported to
play a vital role in promoting the mitochondrial pool of citrate
and redox balance and therefore promoting self-renewal abilities of
cancer stem cells in non-small cell lung cancer (36). In addition, SLC25A1 is required for
the conversion of glucose into acetyl-CoA for fatty acid synthesis
and is increased by PGC1α to promote tumor growth in liver and
colon cancer (37). However, the
role of SLC25A1 in thyroid cancer remains unclear. In the present
study, lncBRM was demonstrated to sponge miR-331-3p and target
SLC25A1. The knockout of SLC25A1 resulted in decreased TPC-1 cell
migration and invasion, which was rescued by the overexpression of
SLC25A1. Thus, SLC25A1 is involved in PTC progression. However, the
role of SLC25A1 in other cancer types remains to be explored.
Despite several studies demonstrating the role of
lncRNAs in promoting or inhibiting cancer progression (38–40),
there are still unanswered questions. For example, most lncRNAs
have no coding potential, however, some may regulate cancer
progression by coding for small peptides. The ceRNA hypothesis
suggests that most lncRNAs are implicated in the pathogenesis of
cancer, by serving as miRNA sponges to modulate the expression of
miRNA target genes (41). However,
whether other mechanisms exist, requires further exploration.
In conclusion, the present study demonstrated
increased expression of lncBRM in PTC tissues and cell lines.
Bioinformatics analysis predicts lncBRM as a sponge for miR-331-3p
that target SLC25A1. Overexpression of lncBRM promotes PTC cells
proliferation, migration and invasion, which were rescued by
ectopic expression of miR-331-3p or SLC25A1 knockout.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in the published article.
Authors' contributions
SL and XH designed the study, analyzed and
interpreted the data, and wrote the manuscript. DZ, LC and SG
performed some experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
For the use of human samples, the protocol for this
study was approved by the Institutional Ethics Committee of The
People's Hospital of Tong Liang District and all enrolled patients
signed a written informed consent document.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen C, Huang S, Huang A, Jia Y, Wang J,
Mao M, Zhou J and Wang L: Total endoscopic thyroidectomy versus
conventional open thyroidectomy in thyroid cancer: A systematic
review and meta-analysis. Ther Clin Risk Manag. 14:2349–2361. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang F, Zhang Q, Chen W, Zhang H, Lu G,
Chen J and Qiu C: Long noncoding RNA cancer susceptibility
candidate 2 suppresses papillary thyroid carcinoma growth by
inactivating the AKT/ERK1/2 signaling pathway. J Cell Biochem.
120:10380–10390. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu J, Wang Y and Zhang B: Advances in
diagnosis and treatment of thyroid cancer in children and
adolescents. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 40:838–842.
2018.(In Chinese). PubMed/NCBI
|
|
4
|
Davis PJ, Tang HY, Hercbergs A, Lin HY,
Keating KA and Mousa SA: Bioactivity of thyroid hormone analogs at
cancer cells. Front Endocrinol (Lausanne). 9:7392018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Albano D, Durmo R, Bertagna F and Giubbini
R: 18F-choline PET/CT incidental thyroid uptake in patients studied
for prostate cancer. Endocrine. 63:531–536. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bolf EL, Sprague BL and Carr FE: A linkage
between thyroid and breast cancer: A common etiology? Cancer
Epidemiol Biomarkers Prev. 28:643–649. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Enokida T and Tahara M: I. genomic
medicine in thyroid cancer. Gan To Kagaku Ryoho. 45:1711–1715.
2018.(In Japanese). PubMed/NCBI
|
|
8
|
Fu XM, Guo W, Li N, Liu HZ, Liu J, Qiu SQ,
Zhang Q, Wang LC, Li F and Li CL: The expression and function of
long noncoding RNA lncRNA-ATB in papillary thyroid cancer. Eur Rev
Med Pharmacol Sci. 21:3239–3246. 2017.PubMed/NCBI
|
|
9
|
Guo Y, Zhang P, Sheng Q, Zhao S and
Hackett TA: lncRNA expression in the auditory forebrain during
postnatal development. Gene. 593:201–216. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang F, Chen W, Peng J, Li Y, Zhuang Y,
Zhu Z, Shao C, Yang W, Yao H and Zhang S: lncRNA PVT1 triggers
Cyto-protective autophagy and promotes pancreatic ductal
adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol
Cancer. 17:982018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun C, Luan S, Zhang G, Wang N, Shao H and
Luan C: CEBPA-mediated upregulation of the lncRNA PLIN2 promotes
the development of chronic myelogenous leukemia via the GSK3 and
Wnt/β-catenin signaling pathways. Am J Cancer Res. 7:1054–1067.
2017.PubMed/NCBI
|
|
12
|
Ye B, Liu B, Yang L, Zhu X, Zhang D, Wu W,
Zhu P, Wang Y, Wang S, Xia P, et al: lncKdm2b controls self-renewal
of embryonic stem cells via activating expression of transcription
factor Zbtb3. EMBO J. 37(pii): e971742018.PubMed/NCBI
|
|
13
|
Zheng JL, Sun J, Zhang H and Zhang Y: Role
of microRNA and lncRNA in lens development and cataract formation.
Zhonghua Yan Ke Za Zhi. 54:390–395. 2018.(In Chinese). PubMed/NCBI
|
|
14
|
Guo CJ, Zhang W and Gershwin ME: Long
noncoding RNA lncKdm2b: A critical player in the maintenance of
group 3 innate lymphoid cells. Cell Mol Immunol. 15:5–7. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu
P, Du Y, Wu J, Qin X, et al: Long noncoding RNA lncKdm2b is
required for ILC3 maintenance by initiation of Zfp292 expression.
Nat Immunol. 18:499–508. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiang Y, Huang Y, Sun H, Pan Y, Wu M and
Zhang J: Deregulation of miR-520d-3p promotes hepatocellular
carcinoma development via lncRNA MIAT regulation and EPHA2
signaling activation. Biomed Pharmacother. 109:1630–1639. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song Y, Zou L, Li J, Shen ZP, Cai YL and
Wu XD: lncRNA SNHG8 promotes the development and chemo-resistance
of pancreatic adenocarcinoma. Eur Rev Med Pharmacol Sci.
22:8161–8168. 2018.PubMed/NCBI
|
|
18
|
Yu Y, Shen HM, Fang DM, Meng QJ and Xin
YH: lncRNA HCP5 promotes the development of cervical cancer by
regulating MACC1 via suppression of microRNA-15a. Eur Rev Med
Pharmacol Sci. 22:4812–4819. 2018.PubMed/NCBI
|
|
19
|
Jiang L, Wu Z, Meng X, Chu X, Huang H and
Xu C: lncRNA HOXA-AS2 facilitates tumorigenesis and progression of
papillary thyroid cancer through modulating miR-15a-5p/HOXA3 axis.
Hum Gene Ther. 30:618–631. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang W, Zhan H, Jiao Y, Li S and Gao W: A
novel lncRNA-miRNA-mRNA network analysis identified the hub lncRNA
RP11-159F24.1 in the pathogenesis of papillary thyroid cancer.
Cancer Med. 7:6290–6298. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang XM, Liu Y, Fan YX, Liu Z, Yuan QL,
Jia M, Geng ZS, Gu L and Lu XB: lncRNA PTCSC3 affects drug
resistance of anaplastic thyroid cancer through STAT3/INO80
pathway. Cancer Biol Ther. 19:590–597. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu X, Yan Y, Li H, Ji N, Yu T, Huang Y,
Shi W, Gao L, Ma L and Hu Y: DNA copy number gain-mediated lncRNA
LINC01061 upregulation predicts poor prognosis and promotes
papillary thyroid cancer progression. Biochem Biophys Res Commun.
503:1247–1253. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gu Y, Feng C, Liu T, Zhang B and Yang L:
The downregulation of lncRNA EMX2OS might independently predict
shorter recurrence-free survival of classical papillary thyroid
cancer. PLoS One. 13:e02093382018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feng L, Yang B and Tang XD: Long noncoding
RNA LINC00460 promotes carcinogenesis via sponging miR-613 in
papillary thyroid carcinoma. J Cell Physiol. 234:11431–11439. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Hu J, Zhou W and Gao H: lncRNA
FOXD2-AS1 accelerates the papillary thyroid cancer progression
through regulating the miR-485-5p/KLK7 axis. J Cell Biochem. Nov
19–2018.(Epub ahead of print). doi: 10.1002/jcb.28072.
|
|
26
|
Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B,
Du Y, Gao G, Tian Y, He L and Fan Z: lncBRM initiates YAP1
signalling activation to drive self-renewal of liver cancer stem
cells. Nat Commun. 7:136082016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xi J, Feng J and Zeng S: Long noncoding
RNA lncBRM facilitates the proliferation, migration and invasion of
ovarian cancer cells via upregulation of Sox4. Am J Cancer Res.
7:2180–2189. 2017.PubMed/NCBI
|
|
28
|
Li R, Zhu H, Yang D, Xia J and Zheng Z:
Long noncoding RNA lncBRM promotes proliferation and invasion of
colorectal cancer by sponging miR-204-3p and upregulating TPT1.
Biochem Biophys Res Commun. 508:1259–1263. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
DeLellis RA and Williams ED: Tumours of
the thyroid and parathyroid. World Health Organization
Classification of Tumours. Pathology and Genetics of Endocrine
Organs. IARC Press; Lyon: pp. 58–70. 2004
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Saini S, Maker AV, Burman KD and Prabhakar
BS: Molecular aberrations and alterations in signaling cascades
implicated in the pathogenesis of anaplastic thyroid cancer.
Biochim Biophys Acta Rev Cancer. Dec 31–2018.(Epub ahead of print).
doi: 10.1016/j.bbcan.2018.12.003. PubMed/NCBI
|
|
32
|
Brajic A, Franckaert D, Burton O,
Bornschein S, Calvanese AL, Demeyer S, Cools J, Dooley J, Schlenner
S and Liston A: The Long Non-coding RNA Flatr Anticipates Foxp3
expression in regulatory T cells. Front Immunol. 9:19892018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xia F, Chen Y, Jiang B, Du X, Peng Y, Wang
W, Huang W, Feng T and Li X: Long noncoding RNA HOXA-AS2 promotes
papillary thyroid cancer progression by regulating
miR-520c-3p/S100A4 pathway. Cell Physiol Biochem. 50:1659–1672.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang N, Duan H, Zhang C, Zhou Y and Gao R:
The LINC01186 suppresses cell proliferation and invasion ability in
papillary thyroid carcinoma. Oncol Lett. 16:5639–5644.
2018.PubMed/NCBI
|
|
35
|
Palmieri F: The mitochondrial transporter
family SLC25: Identification, properties and physiopathology. Mol
Aspects Med. 34:465–484. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fernandez HR, Gadre SM, Tan M, Graham GT,
Mosaoa R, Ongkeko MS, Kim KA, Riggins RB, Parasido E, Petrini I, et
al: The mitochondrial citrate carrier, SLC25A1, drives stemness and
therapy resistance in non-small cell lung cancer. Cell Death
Differ. 25:1239–1258. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bhalla K, Hwang BJ, Dewi RE, Ou L, Twaddel
W, Fang HB, Vafai SB, Vazquez F, Puigserver P, Boros L and Girnun
GD: PGC1α promotes tumor growth by inducing gene expression
programs supporting lipogenesis. Cancer Res. 71:6888–6898. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chi H, Yang R, Zheng X, Zhang L, Jiang R
and Chen J: lncRNA RP11-79H23.3 functions as a competing endogenous
RNA to regulate PTEN expression through sponging hsa-miR-107 in the
development of bladder cancer. Int J Mol Sci. 19:25312018.
View Article : Google Scholar
|
|
39
|
Cui S, Yang X, Zhang L, Zhao Y and Yan W:
lncRNA MAFG-AS1 promotes the progression of colorectal cancer by
sponging miR-147b and activation of NDUFA4. Biochem Biophys Res
Commun. 506:251–258. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hao Y, Yang X, Zhang D, Luo J and Chen R:
Long noncoding RNA LINC01186, regulated by TGF-β/SMAD3, inhibits
migration and invasion through Epithelial-Mesenchymal-Transition in
lung cancer. Gene. 608:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Long J, Xiong J, Bai Y, Mao J, Lin J, Xu
W, Zhang H, Chen S and Zhao H: Construction and Investigation of a
lncRNA-associated ceRNA regulatory network in cholangiocarcinoma.
Front Oncol. 9:6492019. View Article : Google Scholar : PubMed/NCBI
|


















