Introduction
Melanoma is the most aggressive type of skin cancer,
the incidence of which has increased in recent decades (1–3). Despite
the improvement in diagnosis and clinical therapy (4–8), there
is still a high mortality rate among melanoma patients (9–11). In
addition, melanoma cells develop drug resistance to clinical
treatments and survival (12–14).
Hence, there is an urgent need to identify novel drugs and
strategies to improve melanoma treatment (15–18).
The function of aminoacyl-tRNA synthetases is to
catalyse the aminoacylation of tRNA through their cognate amino
acids (19). There are two forms of
isoleucine-tRNA synthetase: Cytoplasmic and mitochondrial. IARS2
encodes for mitochondrial form of isoleucyl-tRNA synthetase
(20). Recent studies have shown
that IARS2 is involved in several diseases (21,22).
IARS2 expression is higher in tumour tissues than surrounding
tissue and knockdown of IARS2 suppresses proliferation of the RKO
cells (23). IARS2 mutation was
found in a patient with neurotrophic keratitis and corneal
opacification (21). Approximately
59% of the colorectal cancers patients harbour a mutation at 5′
upstream region of the mitochondrial IARS2 (24). Thus, IARS2 may be considered as a
cancer-promoting gene (23,25–27).
To date, the association between IARS2 and melanoma
remains unclear. In the present study, the function of IARS2 was
elucidated in melanoma cell proliferation and apoptosis.
Materials and methods
Cell lines
The human melanoma cell lines A375, MUM-2B, and C918
were purchased from Cell Bank of the Chinese Academy of Sciences
(Shanghai, China). A375 and C918 cell lines were cultured in
Dulbecco's Modified Eagle Medium (GE Healthcare Life Sciences),
while MUM-2B cell line was cultured in Roswell Park Memorial
Institute (RPMI)-1640 medium (GE Healthcare Life Sciences) at 37°C
in a 5% CO2 incubator. Both media were supplemented with
10% fetal bovine serum (Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin, and 0.1 mg/ml streptomycin (Merck KGaA).
RT-qPCR
Total RNA was extracted from A375, MUM-2B and C918
cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) and was
quantified using NanoDrop 2000 (Thermo Fisher Scientific, Inc.). A
total of 2 µg of RNA was reverse-transcribed to cDNA using miRNA
1st strand cDNA synthesis kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instruction. A total of 1 µl of
cDNA was assessed by SYBR Green real time-quantitative PCR
(RT-qPCR). Primers were designed and synthesised by Guangzhou
RiboBio Co., Ltd. The sequences are as follows: IARS2 forward,
5′-GGCAACCCATGACAATCCCA-3′, and reverse,
5′-TGGACCTCCTTATGCAAACGG-3′; Glyceralde-hyde-3-phosphate
dehydrogenase (GAPDH) forward, 5′-TGACTTCAACAGCGACACCCA-3′ and
reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. RT-qPCR was performed at 95°C
for 4 min, then 40 cycles of 95°C for 15 sec and 60°C for 45 sec by
a LightCycler 480 (Roche Diagnostics). The results were analysed by
a GeneChip Scanner 3000 (Thermo Fisher Scientific, Inc.). All
reactions were performed in triplicate. GAPDH was used to normalise
expression. Relative expression level of target genes was
calculated using 2−ΔΔCq method (28).
Lentiviral packaging and cell
infection
The shIARS2 lentivirus and vector control were
constructed by GeneChem, Inc. IARS2 oligonucleotides were designed
to target the complementary DNA sequence (ACTTGCAGTCATCCATTAA). The
hairpin sequence of shIARS2 was
CCGGGTACTTGCAGTCATCCATTAATTCAAGAGATTAATGGATGACTGCAAGTACTTTTTG. The
shRNA was synthesized and inserted into the GV115 vector (GeneChem,
Inc.) at AgeI and EcoRI restriction sites. Lentivirus package was
performed as described (29). Then
shIARS2-lentivirus or negative control (shCtrl) lentivirus was
transfected into A375 cells using Lipofectamine 2000 (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
After 72 h of infection, the cells were observed under a
fluorescence microscope, and subsequently harvested to determine
knockdown efficiency by RT-qPCR and western blot analysis.
Western blot analysis
After 72 h of lentivirus infection, the cells were
washed with cold phosphate buffer saline (PBS) and lysed in
radioimmunoprecipitation assay (RIPA) lysis buffer supplemented
with protease and phosphatase inhibitor cocktails for 15 min. Cell
lysate was centrifuged at 12,000 × g for 10 min at 4°C and the
supernatants were collected. Protein concentration was measured by
bicinchoninic acid protein assay. Equal amounts of total protein
samples were analysed on sodium dodecyl sulphate polyacrylamide gel
electrophoresis and transferred onto polyvinylidene fluoride (PVDF)
membrane (EMD Millipore). PVDF membrane was blocked with 5% non-fat
dried milk for 1 h, and then incubated with the appropriate primary
antibodies overnight at 4°C. HRP-coupled secondary antibody was
added and protein bands were visualized by enhanced
chemiluminescence reagents (EMD Millipore). The antibodies for
IARS2 (cat. no. ab66012) was purchased from Abcam. The antibodies
for GAPDH (cat. no. sc-32233) and the secondary antibodies (cat.
no. sc-2004) were purchased from Santa Cruz Biotechnology, Inc.
Cell growth assay
After infection with either shCtrl or shIARS2
lentivirus, the infected A375 cells were seeded into 96-well plates
(1×103 cells per well). The plates were incubated at
37°C in 5% CO2 for 5 days. During this period, cell
number was imaged and counted by Celigo® Image Cytometer
(Nexcelom Bioscience) every day.
Moreover, cell viability was measured every day by
3-(4,5-dimethylthiazol-2-Yl)-2,5-diphenyltetrazolium bromide (MTT)
reagent. Melanoma cells transfected with shIARS2 or shCtrl were
cultured at 37°C with 5% CO2 for 24 h, then detached
with 0.25% trypsin solution, and placed in 96-well plates at a
final concentration of 2,000 cells/ml. MTT assay was performed at
24, 48, 72, 96 and 120 h following transfection. Briefly, the
culture medium was replaced and 10 µl MTT solution/well was added.
The cells with MTT were incubated for 4 h at 37°C in 5%
CO2. Post incubation, the cells were washed and 200 µl
dimethyl sulfoxide/well was added. Finally, the cells were
incubated at room temperature for 10 min. Absorbance of stained
supernatants was detected at 490 nm using a spectrophotometer
(Thermo Fisher Scientific, Inc.). Cell viability / proliferation is
directly proportional to the absorbance rate. The experiment was
performed in triplicate.
Colony formation assay
A375 shCtrl and IARS2 knockdown cells were plated
(1,000 cells/well) in a 6-well culture plate in triplicate. The
culture medium were refreshed every three days for 10 days or until
colonies were formed. Following colony formation, they were washed
twice with PBS, fixed with 4% paraformaldehyde for 30 min, and
stained with 0.4% crystal violet for 15 min. The number of colonies
containing ≥50 cells was counted under a microscope.
Flow cytometry analysis
Cell apoptosis was evaluated by flow cytometry
according to the manufacturer's instructions. After A375 cells
infected with shIARS2-lentivirus or shCtrl lentivirus were cultured
in 6-well plates for 5 days, they were collected and washed with
cold PBS. Then, a 100 μl cell suspension was prepared and stained
with 5 μl of Annexin V-APC and 5 μl of 7-AAD (BD Biosciences) for
15 min. The ratio of apoptotic cells was analysed by a flow
cytometer (EMD Millipore).
For analysis of caspase 3/7 activity, the
lentivirus-infected A375 cells (2×104 cells per well)
were cultured in 96-well plates for 5 days. Then 100 µl Caspase-Glo
3/7 reagent (Promega Corporation) was added and the cells were
incubated for 2 h. The luminescence of each sample was measured in
a luminometer plate reader according to the manufacturer's
instruction.
For cell cycle analysis, the lentiviral
vector-transduced A375 cells were labelled with propidium iodide
(Merck KGaA) and analysed using a flow cytometer. Annexin
V-APC-positive cells were considered apoptotic regardless of the
7-AAD status.
Clinical samples and
immunohistochemistry (IHC) staining
In this study, tumour samples of patients diagnosed
with malignant melanoma between January 2012 and December 2017 were
collected. A total of 30 melanoma tissues and 30 surrounding skin
tissue were examined for the expression of IARS2 protein using IHC.
All paraffin-embedded tissues were fixed in 4% paraformaldehyde.
Briefly, the slides were deparaffinized with xylene and rehydrated
in graded alcohol. Antigen was retrieved and endogenous peroxidase
activity was blocked by 3% hydrogen peroxide for 30 min. The slides
were then incubated with goat serum to block the non-specific
proteins (Bioz, Inc.). Subsequently, the slides were incubated
overnight at 4°C with primary antibody against IARS2 (Abcam), and
finally incubated with biotinylated secondary antibody (Bioz, Inc.)
for 1 h at room temperature. Tris buffered saline with Tween-20 was
used in the washing steps. All sections were scored blindly by two
investigators under a light microscope and recorded. The tissue
staining was scored by the percentage of positive staining cells.
The percentage of positive cells was scored as negative (score 0:
<1% of tumour cells stain positive, or 1: 1–5% of tumour cells
stain positive) or positive (score 2: 6–25%, score 3: 26–50%, score
4: >50% positive of the tumour area). Scores of each subgroup of
clinicopathological parameters are presented as mean ± standard
deviation (SD). Man-Whitney U test was applied for comparisons
between two groups.
Ethical approval was obtained from the clinical
research Ethics Committee of Shandong Provincial Hospital
affiliated to Shandong University (2016142) (Jinan, China). Written
informed consent for the acquisition and use of tissue samples was
obtained from all patients.
Statistical analysis
Statistical analysis were performed using SPSS 16.0
(SPSS, Inc.). The statistical data for each group are presented as
the mean ± SD. T-test was used for comparison between two groups.
Continuous data from multiple groups were analyzed by using one-way
ANOVA, with the Tukey's post hoc test. P<0.05 was accepted as
statistically significant.
Results
Expression of IARS2 in four melanoma
cell lines
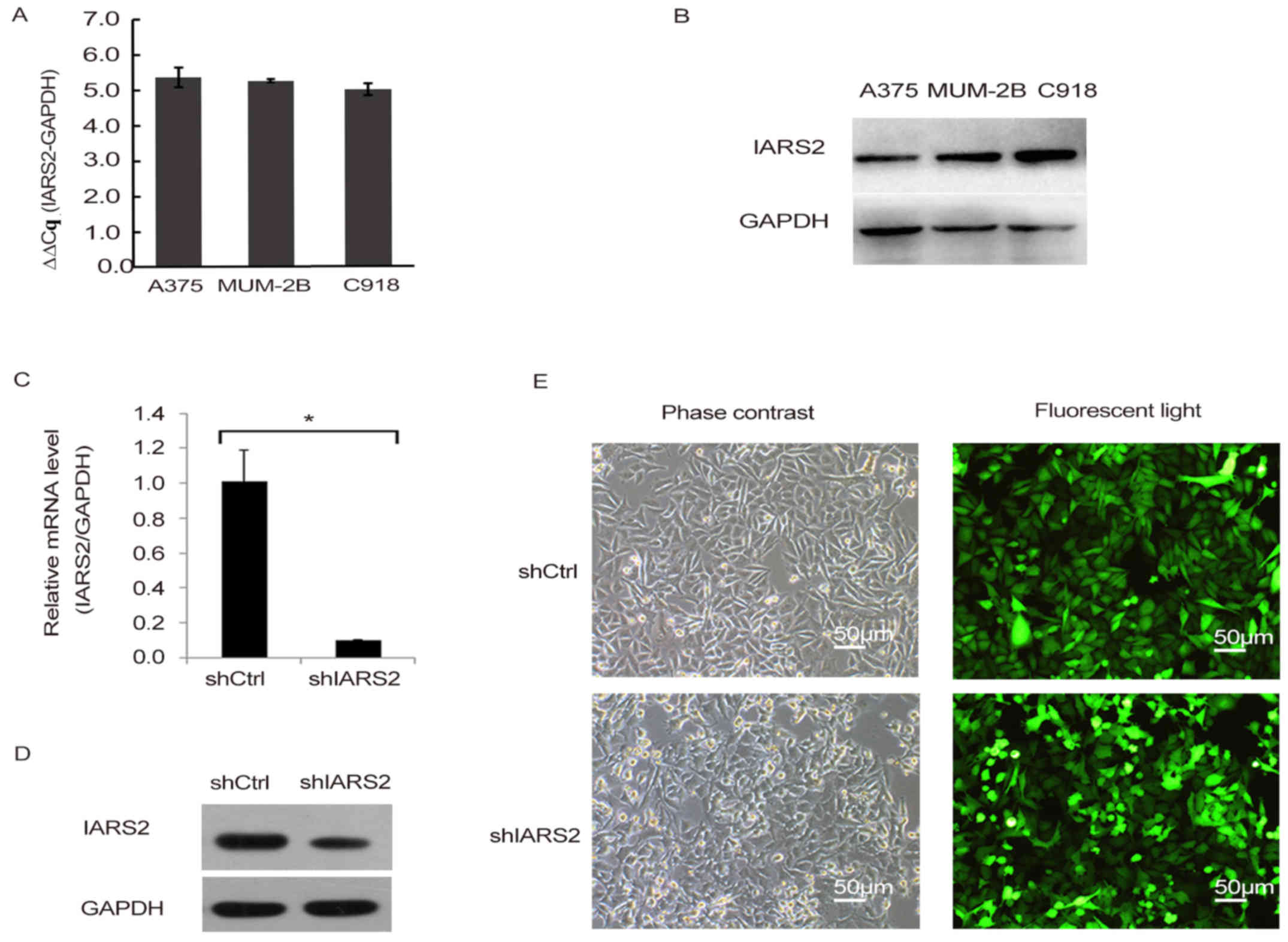
To explore the role of IARS2 in human melanoma
development, we first detected the expression of IARS2 in three
human melanoma cell lines, including A375, MUM-2B, and C918, using
RT-qPCR. We also detected the protein levels of IARS2 in the three
melanoma cell lines. The results showed that IARS2 mRNA and protein
were expressed in all three cell lines (Fig. 1A and B).
Knockdown of IARS2 in A375 cells
To further investigate the function of IARS2 in
human melanoma, we conducted lentivirus-mediated knockdown of IARS2
in the human melanoma cell line A375, and quantified its expression
by RT-qPCR (Fig. 1C-E). By day 3
post-infection, fluorescence microscopy was used to detect the GFP
expression. As shown in Fig. 1E, the
proportion of infected cells was >80%. As shown in Fig. 1C, IARS2 mRNA was significantly
downregulated in A375 cells infected with shIARS2 lentivirus
compared to cells treated with shCtrl lentivirus (P<0.05).
Knockdown efficiency was also examined by western blot analysis,
IARS2 protein expression was significantly reduced in shIARS2 A375
cells compared to shCtrl A375 cells (Fig. 1D). All the results demonstrated
effective knockdown of IARS2 in A375 cells.
Knockdown of IARS2 inhibits human
melanoma cell growth and colony formation
To determine the effect of IARS2 expression on human
melanoma cell growth, we infected A375 cells with shIARS2
lentivirus or shCtrl lentivirus, and analysed the cell growth every
day for 5 days by counting the number of cells. We found that the
cell growth rate of shIARS2 A375 cells was dramatically decreased
compared to shCtrl A375 cells (Fig.
2A-B).
To confirm the inhibitory effect of IARS2 on human
melanoma cell growth, we detected the proliferation of A375 cells
by MTT assay every day for 5 days. MTT assay showed that melanoma
cell proliferation was significantly inhibited in shIARS2 A375
cells compared with shCtrl A375 cells (Fig. 2C).
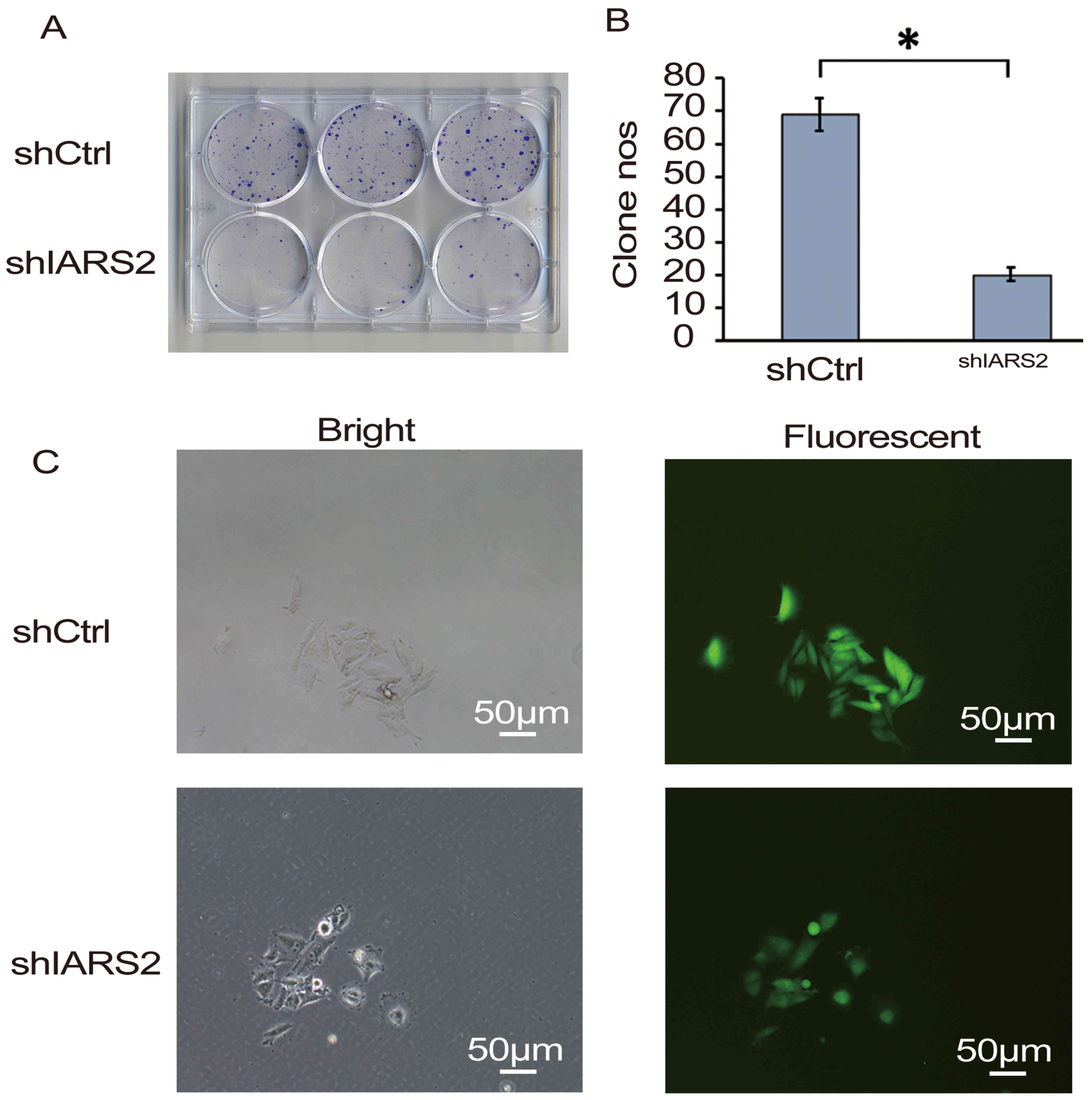
Furthermore, a colony formation assay was performed.
The results of the assay confirmed that IARS2 knockdown
significantly inhibited the colony formation of A375 cells
(Fig. 3; P<0.05).
Effect of IARS2 knockdown on apoptosis
and cell cycle
As is well known, apoptosis is associated with the
mechanism of tumour progression. We detected whether knockdown of
IARS2 affects cell apoptosis. Double staining with Annexin V-APC
and 7-AAD showed that the proportion of apoptotic A375 cells in the
shCtrl and shIARS2 groups was 2.62±0.35% and 8.77±1.32%,
respectively (P<0.05), suggesting that knockdown of IARS2
significantly enhanced cell apoptosis compared with the negative
control group (Fig. 4A). Moreover,
this result was further validated by caspase 3/7 activity analysis.
We observed that caspase 3 and 7 activities were significantly
upregulated in shIARS2 A375 cells compared to shCtrl A375 cells
(Fig. 4B). Taken together, these
data suggested that knockdown of IARS2 promoted human melanoma cell
apoptosis.
Cell cycle analysis showed that knockdown of IARS2
affected the distribution of cell cycle phases (Fig. 4C). Compared to control cells, the
proportion of G0/G1 cells increased, while that of S cells
decreased in IARS2-siRNA group (Fig.
4C, P<0.05).
Expression of IARS2 protein in
melanoma tissues
To further demonstrate the role of IARS2 in human
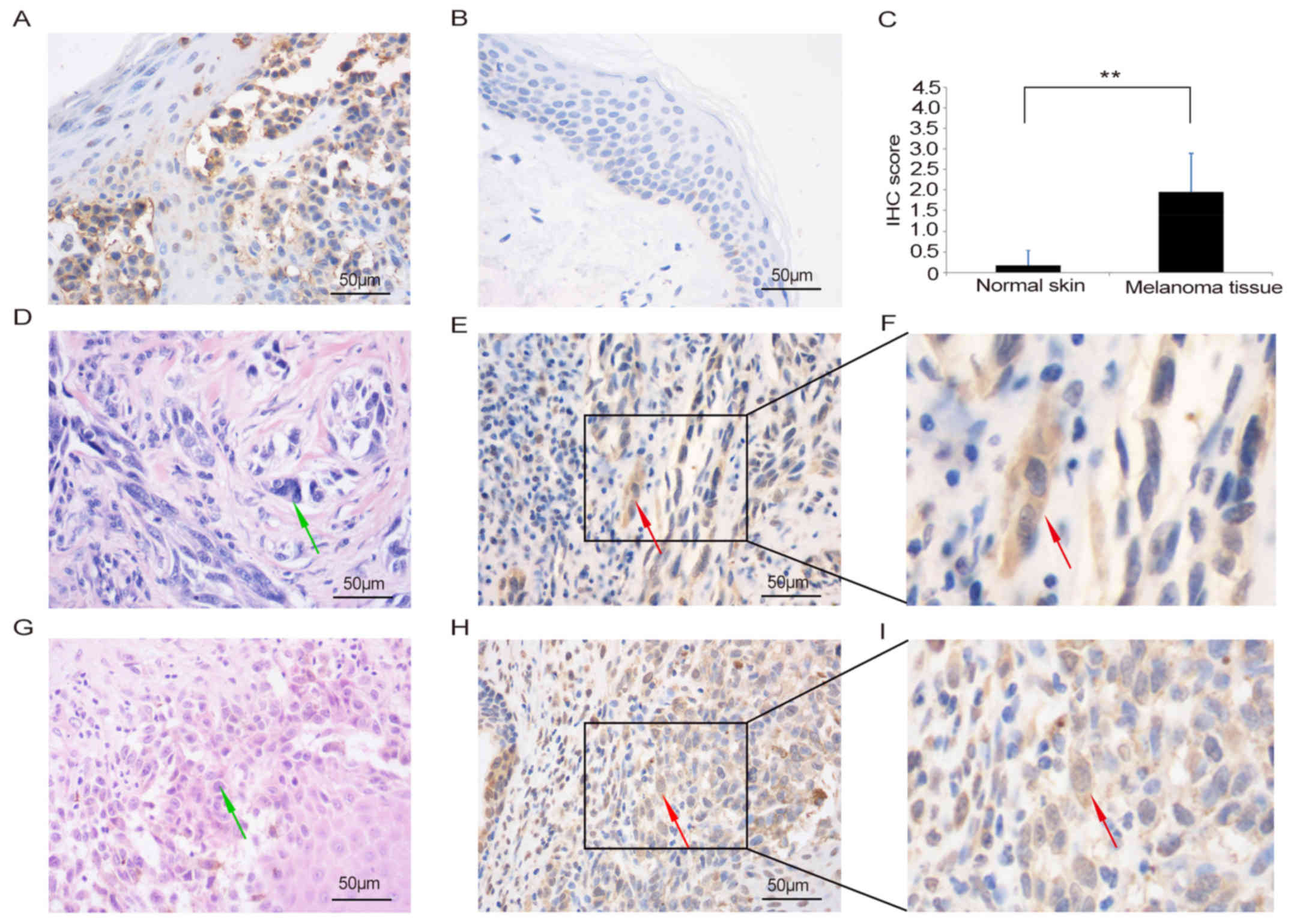
melanoma development, we detected the expression of IARS2 in
clinical melanoma patient tissues by IHC (Fig. 5). Tumour tissues and surrounding skin
tissues from 30 patients were examined for the expression of IARS2
protein in this study. The male-to-female ratio was 1.3:1. The age
of the patients ranged from 23–79 years, and a median age was 61
years. The results showed that expression of IARS2 was higher in
tumour tissues (Fig. 5A) than in
adjacent skin tissue (Fig. 5B and C;
P<0.05). The staining of the IARS2 revealed that it was mainly
localised in the cytoplasm with brown positive granules in tumour
tissues (Fig. 5A, E and H). Fig. 5E shows the IHC staining of IARS2 in
tumour tissue obtained from an IARS2-positive melanoma patient
(pathological no. S1302026). Fig. 5H
shows the same staining pattern of IARS2 in a tissue section
obtained from another IARS2-positive melanoma patient (pathological
no. S1408271).
Discussion
Melanoma is the most dangerous skin cancer and
accounts for the most skin cancer-related deaths worldwide
(30,31). The improvement in diagnosis and
clinical therapy has led to 5-year survival of melanoma patients.
However, doctors have been able to do very little to treat
metastatic melanoma. In addition, development and progression of
melanoma still remain poorly understood. In order to identify more
effective therapeutic strategy for treating melanoma, it is very
important to identify the novel factors that are related to
melanoma incidence and progression and to unravel the underlying
mechanisms (32,33). Hence, there is an urgent need to
identify novel factors associated with melanoma and strategies to
improve melanoma treatment.
Aminoacyl-tRNA synthetases (AARS) identify the
cognate amino acid to catalyse the aminoacylation of tRNA (34). Apart from protein synthesis, recent
studies have shown that AARS are also involved in several other
pathophysiological processes, including inflammation, angiogenesis,
and tumorigenesis. It has been reported that genetic mutation in
the glycyl-tRNA synthetase coding region is associated with
Charcot-Marie-Tooth disease type 2D and distal spinal muscular
atrophy type V (35). Human
tyrosyl-tRNA synthetase promotes angiogenesis but human
tryptophanyl-tRNA synthetase suppresses vascular growth (36). The human glutamyl-tRNA synthetase
regulates cell apoptosis via interaction with apoptosis
signal-regulating kinase 1 (37).
AARS are aberrantly expressed in several types of cancers,
suggesting their potential association with tumorigenesis (38).
IARS2 encodes for mitochondrial form of
isoleucyl-tRNA synthetase, which belongs to the class-I AARS
family. The mitochondrial isoleucine-tRNA synthetase was first
synthesized in the cytoplasm, and then, transported into the
mitochondrion. Recent studies have shown that IARS2 is involved in
several diseases. In 2014, the clinical features of patients with
IARS2 mutation were reported including growth hormone deficiency,
partial deafness, cataracts, and peripheral neuropathy (22). High expression level of IARS2
attenuated the clinical manifestations of patients with mtDNA
mutation (39). Importantly, IARS2
exhibited a vital role in the incidence and development of colon
cancer (23). IARS2 was highly
expressed in human colon cancer tissue compared to surrounding
tissues. Several studies have indicated that IARS2 may be a
cancer-promoting gene (23,25–28).
In our study, melanoma cancer cell lines, A375,
MUM-2B, and C918, were selected for the experiment. First it was
found that the mRNA expression level of IARS2 was high in all four
cell lines (Fig. 1A). IHC analysis
also confirmed a higher expression of IARS2 protein in clinical
melanoma tissues than in surrounding tissues (Fig. 5). Further cytological study showed
that knockdown of IARS2 inhibited A375 cell proliferation and
colony formation (Figs. 2 and
3). Flow cytometry revealed that
knockdown of IARS2 promoted cell apoptosis and influenced cell
cycle distribution (Fig. 4). Our
results suggested that IARS2 may be involved in the development and
progression of melanoma.
MicroRNAs (miRNAs) are a class of small non-coding
RNAs that regulate gene expression at the post-transcriptional
level. They play pivotal roles during several biological processes,
including tumorigenesis in melanoma (40,41).
MiRNA expression levels are regulated in response to various
factors, including toxicant stresses in the environment (42). Based on their genomic location,
miRNAs are generally classified as ‘intergenic’ or ‘intronic.’ The
‘intronic’ miRNAs share the promoter with their host gene and they
are usually expressed along with their host gene indicating the
functional association with their host gene (43). IARS2 is the host gene of miR-215, a
member of the miR-192/215 family. In colon tissues, miR-215
expression was shown to be positively correlated with expression of
its host gene, IARS2, which indicated that miR215 was transcribed
together with IARS2 (44). The
expression of miR-215 might be abnormal after IARS2 was knocked
down in A375 melanoma cells. Previous studies showed that miR-215
plays an important role in tumour development and metastasis
(45–47). It may promote cell proliferation,
repress apoptosis, and alter the cell cycle (45,46).
Thus, the inhibition of proliferation of A375 cells with IARS2
knockdown might be also associated with the alteration of miR-215
expression in shIARS2 cells. However, this speculation still needs
to be proved.
In conclusion, our study showed that downregulation
of IARS2 expression by lentivirus-delivered small interfering RNA
in A375 cells inhibited cell proliferation and colony formation,
induced cell cycle arrest, and upregulated caspase 3/7 activity,
which then promoted cell death through apoptosis. Therefore,
knockdown of IARS2 might be a potential therapeutic approach for
melanoma in which IARS2 is highly expressed.
Acknowledgments
We thank Dr Lei Zhao (Institute of Basic Medical
Sciences, Qilu Hospital of Shandong University (Jinan, China) for
providing technical assistance and experimental materials.
Funding
This project was sponsored by a grant from the Key
Research and Development Program of Shandong Province (grant nso.
2015GSF118171 and 2016GSF201212).
Availability of data and materials
The datasets used and/or analysed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
DM conceived the study and drafted the manuscript.
DM was responsible for RNA extraction and quantitative RT-PCR
assay. XN and LC performed the western blot assay and cell growth
assay. SL was responsible for lentiviral packaging and cell
infection. NC and DH conducted colony formation assay and cell
apoptosis assay. XL and BG were responsible for clinical samples
and IHC staining. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from the clinical
research Ethics Committee of Shandong Provincial Hospital
affiliated to Shandong University (approval no. 2016142; Jinan,
China). Written informed consent for the acquisition and use of
tissue samples was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rastrelli M, Tropea S, Rossi CR and
Alaibac M: Melanoma: epidemiology, risk factors, pathogenesis,
diagnosis and classification. In Vivo. 28:1005–1011.
2014.PubMed/NCBI
|
|
2
|
Minini R, Rohrmann S, Braun R, Korol D and
Dehler S: Incidence trends and clinical-pathological
characteristics of invasive cutaneous melanoma from 1980 to 2010 in
the Canton of Zurich, Switzerland. Melanoma Res. 27:145–151. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin AY, Wang PF, Li H and Kolker JA:
Multicohort model for prevalence estimation of advanced malignant
melanoma in the USA: An increasing public health concern. Melanoma
Res. 22:454–459. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Read T, Webber S, Thomas J, Wagels M,
Schaider H, Soyer HP and Smithers BM: Protocol for the TIDAL
melanoma study: Topical imiquimod or diphenylcyclopropenone for the
management of cutaneous in-transit melanoma metastases-a phase II,
single centre, randomised, pilot study. BMJ Open. 7:e0168162017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pike E, Hamidi V, Saeterdal I,
Odgaard-Jensen J and Klemp M: Multiple treatment comparison of
seven new drugs for patients with advanced malignant melanoma: A
systematic review and health economic decision model in a Norwegian
setting. BMJ Open. 7:e0148802017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen J, Lei QQ, Chen X, Cao C and Cen Y:
Diagnostic performance of micropthalmia transcription factor for
melanoma: A systematic review and meta-analysis. Eur Rev Med
Pharmacol Sci. 18:798–805. 2014.PubMed/NCBI
|
|
7
|
Sun J, Zager JS and Eroglu Z:
Encorafenib/binimetinib for the treatment of BRAF-mutant advanced,
unresectable, or metastatic melanoma: Design, development, and
potential place in therapy. OncoTargets Ther. 11:9081–9089. 2018.
View Article : Google Scholar
|
|
8
|
Cosgarea I, Ritter C, Becker JC,
Schadendorf D and Ugurel S: Update on the clinical use of kinase
inhibitors in melanoma. J Dtsch Dermatol Ges. 15:887–893. 2017.
View Article : Google Scholar
|
|
9
|
Robsahm TE, Helsing P, Nilssen Y, Vos L,
Rizvi SMH, Akslen LA and Veierød MB: High mortality due to
cutaneous melanoma in Norway: A study of prognostic factors in a
nationwide cancer registry. Clin Epidemiol. 10:537–548. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boniol M, Autier P and Gandini S: Melanoma
mortality following skin cancer screening in Germany. BMJ Open.
5:e0081582015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bottoni U, Paolino G, Didona D, Corsetti
P, Clerico R, Cantisani C, Richetta AG, Arcidiacono V, Scali E and
Pranteda G: Improvement of survival in patients with melanoma and
non-melanoma skin cancers compared to patients without double
cutaneous malignancies. Eur Rev Med Pharmacol Sci. 19:1640–1644.
2015.PubMed/NCBI
|
|
12
|
Filitis DC, Rauh J and Mahalingam M: The
HGF-cMET signaling pathway in conferring stromal-induced
BRAF-inhibitor resistance in melanoma. Melanoma Res. 25:470–478.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mak G, Arkenau HT and Chin M: Resistance
surveillance in a BRAF mutant melanoma patient on long-term
BRAF-inhibitor treatment. Melanoma Res. 24:408–412. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cohen-Solal KA, Kaufman HL and Lasfar A:
Transcription factors as critical players in melanoma invasiveness,
drug resistance, and opportunities for therapeutic drug
development. Pigment Cell Melanoma Res. 31:241–252. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gladfelter P, Darwish NHE and Mousa SA:
Current status and future direction in the management of malignant
melanoma. Melanoma Res. 27:403–410. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Najem A, Krayem M, Perdrix A, Kerger J,
Awada A, Journe F and Ghanem G: New drug combination strategies in
melanoma: Current status and future directions. Anticancer Res.
37:5941–5953. 2017.PubMed/NCBI
|
|
17
|
Zhu Z, Liu W and Gotlieb V: The rapidly
evolving therapies for advanced melanoma - Towards immunotherapy,
molecular targeted therapy, and beyond. Crit Rev Oncol Hematol.
99:91–99. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bogusławska J and Małecki M: siRNA
preparations in gene therapy of melanoma. Med Wieku Rozwoj.
17:196–201. 2013.PubMed/NCBI
|
|
19
|
Rajendran V, Kalita P, Shukla H, Kumar A
and Tripathi T: Aminoacyl-tRNA synthetases: Structure, function,
and drug discovery. Int J Biol Macromol. 111:400–414. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee SW, Cho BH, Park SG and Kim S:
Aminoacyl-tRNA synthetase complexes: Beyond translation. J Cell
Sci. 117:3725–3734. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jabbour S and Harissi-Dagher M: Recessive
mutation in a nuclear-encoded mitochondrial tRNA synthetase
associated with infantile cataract, congenital neurotrophic
keratitis, and orbital myopathy. Cornea. 35:894–896. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schwartzentruber J, Buhas D, Majewski J,
Sasarman F, Papillon-Cavanagh S, Thiffault I, Sheldon KM,
Massicotte C, Patry L, Simon M, et al FORGE Canada Consortium, :
Mutation in the nuclear-encoded mitochondrial isoleucyl-tRNA
synthetase IARS2 in patients with cataracts, growth hormone
deficiency with short stature, partial sensorineural deafness, and
peripheral neuropathy or with Leigh syndrome. Hum Mutat.
35:1285–1289. 2014.PubMed/NCBI
|
|
23
|
Zhong L, Zhang Y, Yang JY, Xiong LF, Shen
T, Sa YL, O'Yang YM, Zhao SH and Chen JY: Expression of IARS2 gene
in colon cancer and effect of its knockdown on biological behavior
of RKO cells. Int J Clin Exp Pathol. 8:12151–12159. 2015.PubMed/NCBI
|
|
24
|
Miyaki M, Iijima T, Shiba K, Aki T, Kita
Y, Yasuno M, Mori T, Kuroki T and Iwama T: Alterations of repeated
sequences in 5′ upstream and coding regions in colorectal tumors
from patients with hereditary nonpolyposis colorectal cancer and
Turcot syndrome. Oncogene. 20:5215–5218. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yin J, Liu W, Li R, Liu J, Zhang Y, Tang W
and Wang K: IARS2 silencing induces non-small cell lung cancer
cells proliferation inhibition, cell cycle arrest and promotes cell
apoptosis. Neoplasma. 63:64–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fang Z, Wang X, Yan Q, Zhang S and Li Y:
Knockdown of IARS2 suppressed growth of gastric cancer cells by
regulating the phosphorylation of cell cycle-related proteins. Mol
Cell Biochem. 443:93–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li H, Tian Y, Li X, Wang B, Zhai D, Bai Y,
Dong C and Chao X: Knockdown of IARS2 inhibited proliferation of
acute myeloid leukemia cells by regulating p53/p21/PCNA/eIF4E
pathway. Oncol Res. 27:673–680. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lois C, Hong EJ, Pease S, Brown EJ and
Baltimore D: Germline transmission and tissue-specific expression
of transgenes delivered by lentiviral vectors. Science.
295:868–872. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Garbe C, Peris K, Hauschild A, Saiag P,
Middleton M, Bastholt L, Grob JJ, Malvehy J, Newton-Bishop J,
Stratigos AJ, et al European Dermatology Forum (EDF); European
Association of Dermato-Oncology (EADO); European Organisation for
Research and Treatment of Cancer (EORTC), : Diagnosis and treatment
of melanoma. European consensus-based interdisciplinary guideline -
Update 2016. Eur J Cancer. 63:201–217. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tas F: Metastatic behavior in melanoma:
Timing, pattern, survival, and influencing factors. J Oncol.
2012:6476842012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sarkar D, Leung EY, Baguley BC, Finlay GJ
and Askarian-Amiri ME: Epigenetic regulation in human melanoma:
Past and future. Epigenetics. 10:103–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Diodato D, Ghezzi D and Tiranti V: The
mitochondrial aminoacyl tRNA synthetases: Genes and syndromes. Int
J Cell Biol. 2014:7879562014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Antonellis A, Ellsworth RE, Sambuughin N,
Puls I, Abel A, Lee-Lin SQ, Jordanova A, Kremensky I, Christodoulou
K, Middleton LT, et al: Glycyl tRNA synthetase mutations in
Charcot-Marie-tooth disease type 2D and distal spinal muscular
atrophy type V. Am J Hum Genet. 72:1293–1299. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yao P and Fox PL: Aminoacyl-tRNA
synthetases in medicine and disease. EMBO Mol Med. 5:332–343. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ko YG, Kim EY, Kim T, Park H, Park HS,
Choi EJ and Kim S: Glutamine-dependent antiapoptotic interaction of
human glutaminyl-tRNA synthetase with apoptosis signal-regulating
kinase 1. J Biol Chem. 276:6030–6036. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim S, You S and Hwang D: Aminoacyl-tRNA
synthetases and tumorigenesis: More than housekeeping. Nat Rev
Cancer. 11:708–718. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Perli E, Giordano C, Tuppen HA, Montopoli
M, Montanari A, Orlandi M, Pisano A, Catanzaro D, Caparrotta L,
Musumeci B, et al: Isoleucyl-tRNA synthetase levels modulate the
penetrance of a homoplasmic m.4277T>C mitochondrial tRNA(Ile)
mutation causing hypertrophic cardiomyopathy. Hum Mol Genet.
21:85–100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fattore L, Costantini S, Malpicci D,
Ruggiero CF, Ascierto PA, Croce CM, Mancini R and Ciliberto G:
MicroRNAs in melanoma development and resistance to target therapy.
Oncotarget. 8:22262–22278. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bennett PE, Bemis L, Norris DA and
Shellman YG: miR in melanoma development: miRNAs and acquired
hallmarks of cancer in melanoma. Physiol Genomics. 45:1049–1059.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gonzalez H, Lema C, Kirken RA, Maldonado
RA, Varela-Ramirez A and Aguilera RJ: Arsenic-exposed keratinocytes
exhibit differential microRNAs expression profile; Potential
implication of miR-21, miR-200a and miR-141 in melanoma pathway.
Clin Cancer Drugs. 2:138–147. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim YK and Kim VN: Processing of intronic
microRNAs. EMBO J. 26:775–783. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lei H, Li H, Xie H, Du C, Xia Y and Tang
W: Role of MiR-215 in Hirschsprung's Disease pathogenesis by
targeting SIGLEC-8. Cell Physiol Biochem. 40:1646–1655. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li N, Zhang QY, Zou JL, Li ZW, Tian TT,
Dong B, Liu XJ, Ge S, Zhu Y, Gao J, et al: miR-215 promotes
malignant progression of gastric cancer by targeting RUNX1.
Oncotarget. 7:4817–4828. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gao X, Cai Y and An R: miR 215 promotes
epithelial to mesenchymal transition and proliferation by
regulating LEFTY2 in endometrial cancer. Int J Mol Med.
42:1229–1236. 2018.PubMed/NCBI
|
|
47
|
Wei Y, Sun J and Li X: MicroRNA-215
enhances invasion and migration by targeting retinoblastoma tumor
suppressor gene 1 in high-grade glioma. Biotechnol Lett.
39:197–205. 2017. View Article : Google Scholar : PubMed/NCBI
|