Introduction
Pancreatic cancer (PC) is one of the most highly
malignant cancers, with a mean 5-year survival rate of <9% in
the United States (1,2). According to a previous report, PC is
responsible for ~227,000 deaths annually worldwide due to its high
recurrence rate, asymptomatic onset and <5% resectable localized
tumors (3). Unfortunately, early
diagnostic and effective treatment strategies are lacking for PC
and there is an urgent need to explore diagnostic tools with high
sensitivity, specificity and repeatability (4). Thus, there is an urgent need to
investigate the underlying molecular mechanisms of PC and identify
novel diagnostic biomarkers for the disease. Identifying a
minimally invasive/non-invasive and sensitive diagnostic method has
recently become a popular topic of research and there has been some
progression in the study of serum markers (5,6). Further
elucidation of the associated biological processes involved is
crucial as it may uncover novel potential biomarkers for early
diagnosis of PC.
MicroRNAs (miRNAs) are 18–22 nucleotides in length,
endogenous non-coding single-stranded RNAs that regulate gene
expression by binding to the 5′ or 3′-untranslated regions
post-transcriptionally or to the open reading frames (7,8). mRNA or
protein is degraded or translation suppressed by miRNAs through
this interference process (9).
Numerous studies have demonstrated that miRNAs are involved in a
variety of malignancies, such as PC, lung, breast and prostate
cancer (10–12). In particular, the clinical
applications of miRNAs have been investigated as biomarkers for
early diagnosis and treatment of tumors, clinical response and
histological classification (13).
Due to the repeatability and ease of sample collection, circulating
miRNAs are currently being investigated as non-invasive biomarkers
in the early diagnosis of different types of cancer (14). Valadi et al (15) first extracted and verified miRNAs in
exosomes, and demonstrated that exosomes are secreted by different
types of cells and may be used for transporting proteins, mRNAs and
miRNAs to target cells. The study of miRNAs in exosomes is becoming
a new research focus. Furthermore, it was revealed that exosomal
(ex)-miRNAs may play crucial roles in biological processes such as
tumor progression, cell proliferation, invasion, metastasis,
apoptosis and differentiation (16–19), and
may be of value as novel serum biomarkers for the early diagnosis
of cancer, such as PC.
Exosomes originate from internal multivesicular
bodies and are membranous vesicles with a diameter of 30–120 nm
(20,21). Exosomes have been identified in
serum, plasma, breast milk and other human bodily fluids (22). Recent findings have reported that
ex-miRNAs contain miRNA, mRNA and cell-specific proteins, and have
been used as diagnostic biomarkers in tumor cells as they may
reflect genetic information and molecular signatures of the various
cells of origin (23–25). Tumor cell derived exosomes contain
tumor-specific miRNAs and their roles are emerging in malignant
progression (26). However, to the
best of our knowledge, the role of circulating ex-miRNAs in PC has
not yet been extensively investigated.
Previous studies have demonstrated that miRNA-21 and
miRNA-210 are associated with a poor prognosis in PC; however, the
findings of these studies were based on tumor tissues and a small
sample size limited their clinical significance (27,28).
Furthermore, there are currently few reports on the significance of
circulating ex-miRNAs as diagnostic markers of PC (29–31). The
aim of the present study was to investigate the diagnostic
relevance of two circulating serum ex-miRNAs, miRNA-21 and
miRNA-210 as novel serological biomarkers for PC.
Materials and methods
Patients
A total of 40 patients at the Renmin Hospital of
Wuhan University (Wuhan, China) were enrolled between March 2018
and August 2019 in the present study and their clinicopathological
characteristics were evaluated. Among the patients, 30 had PC and
10 had CP. A diagnosis of PC was made by pathology or cytology,
while a diagnosis of chronic pancreatitis (CP) was made by
pathology or clinical criteria based on the World Health
Organization Classification of Tumours of the Digestive System
(32). The enrollment criteria
included subjects who had not received neoadjuvant
chemoradiotherapy. Patients were excluded if the diagnosis of PC
was not pathologically confirmed. Cases were staged according to
the modified European Neuroendocrine Tumor Society
Tumor-Node-Metastasis (TNM) staging system (33). The serum ex-microRNAs in patients
with PC were detected prior to surgical resection. The study
protocol was approved by the Ethics Committee of Renmin Hospital of
Wuhan University (approval no. 20180212; Wuhan, China). Written
informed consent was provided by all patients.
Isolation and identification of
exosomes from the serum of patients with PC
Blood samples (5–6 ml) were collected from all
patients and centrifuged at 500 × g for 5 min at 4°C. The
supernatants were preserved at −80°C until further use. Exosomes
were isolated from serum samples using the Exosome extraction kit
(System Biosciences). Briefly, 500 µl of serum was mixed with 120
µl ExoQuick solution and incubated at 4°C for 30 min. The mixed
solution (ExoQuick/serum) was centrifuged at 12,000 × g for 2 min.
for obtaining exosome pellets. Transmission electron microscopy
(TEM; cat. no. HT7800; Hitachi High-Technologies Corporation; HC
mode, ×200-200,000; HR mode, ×4,000-600,000; LowMag mode,
×50-1,000) was used to visualize and verify the exosomes that were
extracted from the serum. The nanoparticle size distribution and
concentration of vesicles were analyzed on the NanoSight LM10-HS
instrument (NanoSight, Ltd.) using NTA 3.0 software (34).
Western blotting
The exosomes marker proteins were detected by
western blotting. Total protein of exosomes was extracted with a
cell lysis buffer (cat. no. 9803; Cell Signaling Technology, Inc.)
supplemented with 1 mM protease inhibitor cocktail (cat. no. 5871;
Cell Signaling Technology, Inc.). Equal amounts of extracted
protein (0.5 µg/lane) were separated by 10% SDS-PAGE and
transferred to polyvinylidene difluoride (PVDF) membranes (Thermo
Fisher Scientific, Inc.). The blots were blocked with 5% non-fat
milk at 37°C for 2 h prior to incubation overnight at 4°C with
primary antibodies anti-cluster of differentiation (CD)63 (1:500;
cat. no. ARG57952; Arigo Biolaboratories, Corp.), antitumor
susceptibility gene (TSG)101 (1:1,000; cat. no. ab125011; Abcam)
and anti-CD81 (1:500; cat. no. MA1-10290; Thermo Fisher Scientific,
Inc.). GAPDH (1:5,000; cat. no. AF7021; Affinity Biosciences) was
used as the internal loading control. The blots were incubated with
horseradish peroxidase-conjugated anti-IgG secondary antibodies
(1:5,000; cat. no. ANT022; Wuhan AntGene Biotechnology Co., Ltd.)
at room temperature for 2 h, and subsequently developed using an
enhanced chemiluminescence kit (cat. no. 34095; Thermo Fisher
Scientific, Inc.) and semi-quantified using the Quantity One
software version 4.62 (Bio-Rad Laboratories Inc.).
miRNA extraction
miRNA extraction from the isolated exosomes was
performed using a miRNeasy Mini kit (Qiagen GmbH) according to the
manufacturer's instructions. The final volumes of RNA were
standardized by dilution with 30 µl nuclease-free water (cat. no.
ST876; Beyotime Institute of Biotechnology). The RNA concentration
in 2 µl was quantified using a Microvolume UV–Vis Spectrophotometer
(cat. no. ND-ONE-W; Thermo Fisher Scientific, Inc.).
Measurement of miRNA levels using
reverse transcription-quantitative (RT-q) PCR
In order to investigate ex-miRNAs, miRNA-21 and
miRNA-210 were selected (34,35).
Total RNA extraction from the isolated exosomes was performed as
aforementioned. The RNA was reverse-transcribed using the TaqMan
MicroRNA Reverse Transcription kit (Qiagen GmbH) at 37°C for 60 min
and 94°C for 5 min. qPCR was performed using the TaqMan miRNA kit
according to the manufacturer's protocol (Qiagen GmbH) and Cq
values were calculated. The primers used were as follows: miR-21
forward, 5′-CGCTAGCTTATCAGACTG-3′ and reverse,
5′-GAGCAGGCTGGAGAA-3′; miR-210 forward,
5′-ACACTCCAGCTGGGCTGTGCGTGTG-3′ and reverse,
5′-CAACTGGTGTCGTGGAGTCG-3′. PCR amplifications were performed in
triplicate for each sample. The thermocycling conditions used were
as follows: 95°C for 15 min, followed by 45 cycles at 94°C for 15
sec and finally 55°C for 30 sec. The relative miRNA expression
values were normalized to cel-miR-39 (36,37) and
calculated using the 2−ΔΔCq method (38).
Statistical analysis
Data were statistically analyzed using SPSS version
20.0 (IBM Corp.). Data are presented as the median (interquartile
range) or n (%) values as appropriate. The association between
miRNA expression levels and clinicopathological features was
examined by χ2 or Fisher's exact test. Comparisons of
continuous data were made using the unpaired Student's t-test. The
optimal cut-off was established as the mean threshold or critical
value that maximized the sum of sensitivity and specificity using
time-dependent receiver operating characteristic (ROC) curves and
the areas under curves (AUC). The diagnostic value of the candidate
miRNAs was evaluated by calculating specificity and sensitivity.
The cut-off points were determined using the Youden index and were
0.09 and 0.0012 for miRNA-21 and miRNA-210, respectively. AUCs for
miRNAs and CA19-9 were compared using Z tests. P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient characteristics
Clinicopathological characteristics of the patients
are presented in Table I. The mean
age was 62 years for patients with PC and 50.5 years for patients
with CP. The median diameter of the tumor was 4.2 cm in patients
with PC (data not shown). The mean level of serum carbohydrate
antigen (CA) 19-9 was significantly higher in patients with PC
compared with patients with CP (P<0.05), whereas no
statistically significant differences were observed in sex or age
(Table I).
 | Table I.Comparison of mean values of age,
sex, location, stage, T factor and CA19-9 levels between patients
with PC and CP. |
Table I.
Comparison of mean values of age,
sex, location, stage, T factor and CA19-9 levels between patients
with PC and CP.
|
Characteristics | PC | CP | P-value |
|---|
| Mean age (range),
years | 62.0 (30–80) | 50.5 (34–80) | 0.17 |
| Sex, male/female,
n | 18/12 | 8/2 | 0.64 |
| Location,
head/body/tail, n | 12/8/10 | – | NA |
| Stagea, 0/IA/IB/IIA/IIB/III/IV | 1/0/1/4/14/2/8 | – | NA |
| T factor,
cis/1/2/3/4 | 1/1/4/20/4 | – | NA |
| Mean CA19-9
(range), U/ml | 104.3
(0.5–15,00.0) | 15.0
(4.0–100.5) | 0.01b |
Identification of circulating serum
exosomes
Exosomes were collected and observed using TEM
(Fig. 1A and B) in accordance with
the characteristics of exosomes previously described (39). Numerous small vesicles were observed
by a NanoSight in the range of 50–100 nm and the size and overall
concentration were ~93 nm and 1.3×109 particles/ml,
respectively (Fig. 1C). Known
exosome-specific markers, such as CD63, CD81 and TSG101 were
identified in the vesicles by western blotting, and the exosome
vesicles obtained from patients with PC were compared with those
from patients with CP; it was observed by western blot analysis
that the expression of these markers in patients with PC was
stronger than that in patients with CP (Fig. 1D).
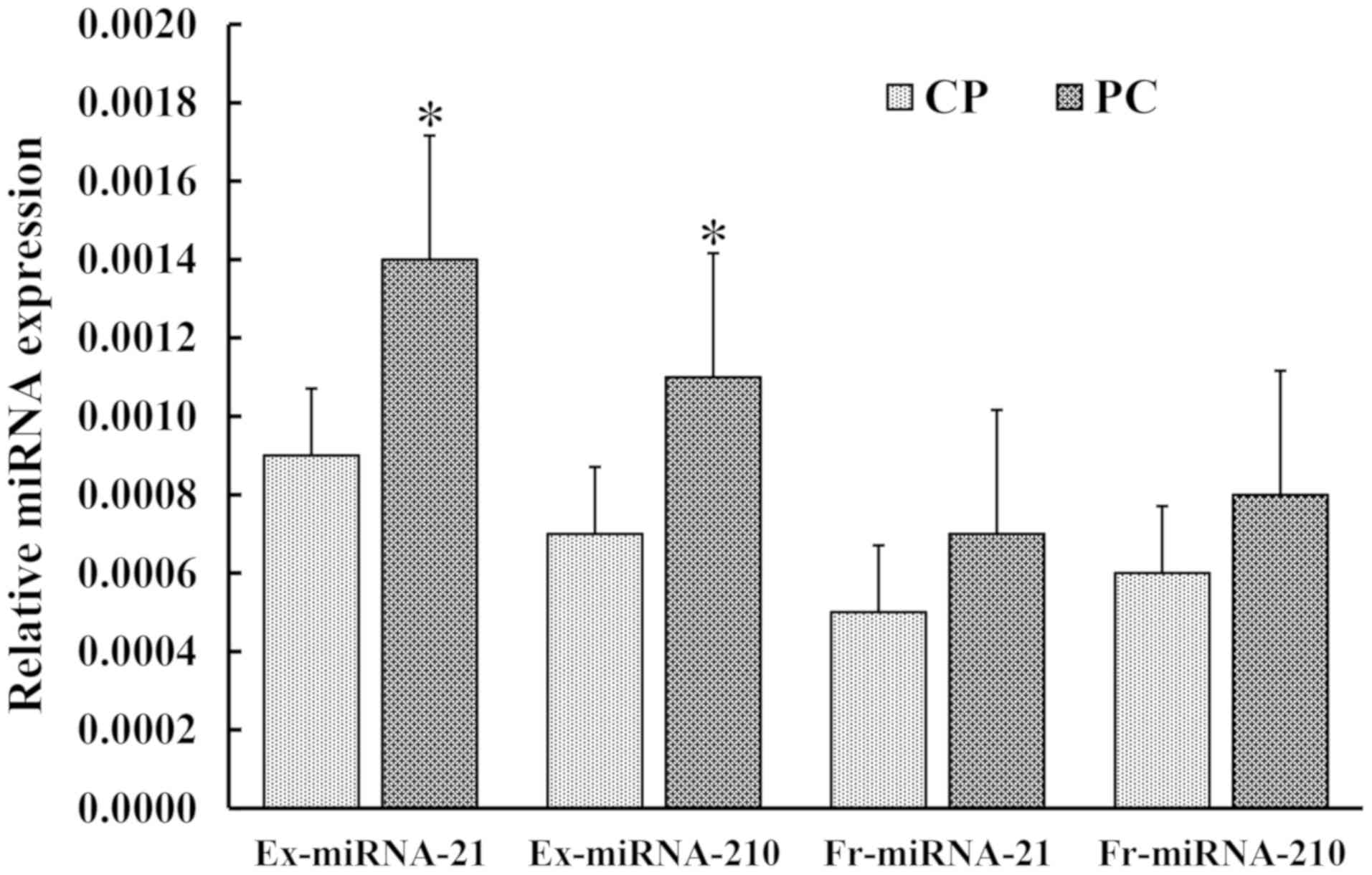
Expression level of serum ex- and
serum fr-circulating miRNAs in patients with PC and CP by
RT-qPCR
The expression levels of serum ex-miRNA-21 and
miRNA-210 were significantly higher in patients with PC (P<0.01;
Fig. 2). There was a significant
difference between the AUC values for ex-miRNA-21 and fr-miRNA-21
(P=0.003; Fig. 3A). Similarly, there
was a significant difference between the AUC values for
ex-miRNA-210 and fr-miRNA-210 (P=0.009; Fig. 3B).
 | Figure 3.Receiver operating characteristic
curve analysis was performed to distinguish the diagnostic value of
miRNA and serum CA19-9. (A) Diagnostic value of Ex-21 levels
compared with levels of Fr-21, with AUC values of 0.869 and 0.696,
respectively. (B) Diagnostic value of Ex-210 level compared with
levels of Fr-210, with AUC values of 0.823 and 0.650, respectively.
(C) Diagnostic value of Ex-21 level and CA19-9 level, with AUC
values of 0.869 and 0.758, respectively. (D) Diagnostic value of
Ex-210 level and CA19-9 level, with AUC values of 0.823 and 0.758,
respectively. AUC, area under the curve; Ex-21, exosomal miRNA-21;
Ex-210, exosomal miRNA-210; Fr-21, free-miRNA-21; Fr-210
free-miRNA-210; CA19-9, carbohydrate antigen 19-9. |
Comparison of the diagnostic values of
ex-miRNAs, serum circulating miRNAs and CA19-9 quantitation
Diagnostic values of serum circulating free or
exosomal miRNA-21 and miRNA-210 were investigated. The AUC values
for ex-miRNA-21 and ex-miRNA-210 levels were significantly higher
compared with those for serum CA19-9 levels (both P=0.038; Fig. 3C and D). The cut-off values for
ex-miRNA-21 and ex-miRNA-210 levels were determined and used to
stratify patients into two groups (positive or negative) for each
ex-miRNA. The accuracy of ex-miRNA-21 (83%) and ex-miRNA-210 (85%)
were superior compared with serum fr-miRNA-21 (73%) and
fr-miRNA-210 (78%) (Table II). When
combining the results of ex-miRNA-21 and ex-miR-210, the accuracy,
specificity and sensitivity were 90, 80 and 93%, respectively
(Table II). Taking the positive
results of either ex-miRNA-21/ex-miRNA-210 levels or serum cytology
as the criterion for diagnosis of PC, the specificity of the
combined test (with CA19-9) and sensitivity was 90% (Table II).
 | Table II.Diagnostic value of Ex-miR or serum
Fr -miRNA21 and -miRNA210 level for patients with PC calculated via
receiver operating characteristic curve analysis. |
Table II.
Diagnostic value of Ex-miR or serum
Fr -miRNA21 and -miRNA210 level for patients with PC calculated via
receiver operating characteristic curve analysis.
| Ex-miR | TP, n | FN, n | FP, n | TN, n | Sensitivity, % | Specificity, % | Accuracy, % | PPV, % | NPV, % |
|---|
| Ex-miR-21 | 24 | 6 | 1 | 9 | 80 | 90 | 83 | 96 | 60 |
| Ex-miR-210 | 25 | 5 | 1 | 9 | 83 | 90 | 85 | 96 | 64 |
| Ex-miR-21/210 | 28 | 2 | 2 | 8 | 93 | 80 | 90 | 93 | 80 |
| Fr-miR21 | 22 | 8 | 3 | 7 | 73 | 70 | 73 | 88 | 47 |
| Fr-miR210 | 23 | 7 | 2 | 8 | 76 | 80 | 78 | 92 | 53 |
|
Ex-miR-21/CA19-9 | 27 | 3 | 1 | 9 | 90 | 90 | 90 | 96 | 75 |
|
Ex-miR-210/CA19-9 | 27 | 3 | 1 | 9 | 90 | 90 | 90 | 96 | 75 |
Serum ex-miRNA-21 and miRNA-210 levels
are associated with multiple prognostic factors of PC
Associations between the relative levels of these
miRNA-21 and miRNA-210 and clinical characteristics were evaluated.
High and low expression levels of miRNA-21 and miRNA-210 were based
on the cut-off values of 0.09 and 0.0012, respectively. The
expression levels of miRNA-21 and miRNA-210 were associated with
the Tumor-Node (TN) stage (31), and
the levels of these miRNAs were closely associated with advanced
disease (P<0.05) and other prognostic factors, including TN
stage, age, treatment history, location, CA19-9, size of tumor and
CRP, while they were not associated with sex and M stage (Table III).
 | Table III.Associations between expression
levels of both serum ex-miRNA-21 and ex-miRNA-210 level and
clinical characteristics in patients with pancreatic cancer
(n=30). |
Table III.
Associations between expression
levels of both serum ex-miRNA-21 and ex-miRNA-210 level and
clinical characteristics in patients with pancreatic cancer
(n=30).
|
| Serum ex-miRNA-21
expression level |
| Serum ex-miRNA-210
expression level |
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | Low expression
group, n=12 | High expression
group, n=18 | P- value | Low expression
group, n=16 | High expression
group, (n=14) | P-value |
|---|
| Age, <60/≥60
years, n | 5/7 | 7/11 | 0.060 | 5/11 | 6/8 | 0.007a |
| Male/female, n | 8/4 | 12/6 | >0.999 | 12/4 | 11/3 | 0.311 |
| Treatment history,
No/Yes, n | 10/2 | 15/3 | 0.046a | 9/7 | 8/6 | 0.026a |
| Location,
head/body/tail, n | 5/3/4 | 5/7/6 | 0.010a | 4/6/6 | 3/5/6 | 0.004a |
| Size of tumor,
<5/≥5 cm, n | 7/5 | 10/8 | 0.038a | 10/6 | 8/6 | 0.016a |
| bT stage, 1–2/3-4, n | 4/8 | 7/11 | 0.011a | 16/0 | 12/2 | 0.006a |
| bN stage, 0/1, n | 9/3 | 17/1 | 0.030a | 8/8 | 7/7 | 0.020a |
| bM stage, 0/1, n | 10/2 | 18/0 | 0.250 | 14/2 | 11/3 | 0.201 |
| CA19-9, <37/≥37,
U/ml | 7/5 | 10/8 | 0.016a | 7/9 | 10/4 | 0.018a |
| CRP, <10/≥10,
mg/l | 4/8 | 10/8 | 0.020a | 9/5 | 10/4 | 0.013a |
Discussion
Recently, the abnormal expression of serum ex-miRNAs
has emerged as a potential novel biomarker of tumor diagnosis and
progression due to their stability in plasma/serum and their
specific expression profiles which reflect the gene information and
biological properties of tumor cells (40,41).
These serum ex-miRNAs may be used as non-invasive, specific and
sensitive biomarkers for the diagnosis and prognosis of tumors
(42). Alterations of circulating
miRNAs have been investigated in patients with PC (43). However, studies on serum ex-miRNAs in
PC are lacking. In the present study, the levels of fr- and
ex-miRNA-21 and miRNA-210 were investigated in patients with
PC.
Numerous studies have indicated that serum ex-miRNAs
are associated with tumor-derived microRNAs and that these miRNAs
may be of value as diagnostic biomarkers for cancer (44,45). In
the present study, compared with patients with CP, the expression
levels of serum ex-miRNA-21 and miRNA-210 were significantly higher
in patients with PC. In contrast, the expression levels of serum
fr-miRNA-21 and miRNA-210 did not differ significantly between
patients with CP and PC. Whole circulating peripheral blood
contains other miRNAs in addition to ex-miRNAs, which are easily
degraded probably because these miRNAs are not enclosed in exosomes
(46,47). In the present study, it was
hypothesized that ex-miRNAs rather than serum fr- miRNAs were
preferable and may serve as new biomarkers for PC. In fact, serum
ex-miRNAs originating from tumor-derived miRNAs may be used as
novel diagnostic biomarkers (48).
In addition, ex-miRNAs in the serum or plasma and other bodily
fluids may be more stable and valuable as biomarkers for PC
detection compared with serum fr-miRNAs.
miRNA-21 is upregulated in different types of
tumors, such as esophageal squamous cell carcinoma and breast, lung
and liver cancer, and targets tumor-suppressive mRNAs (49,50). It
was demonstrated that overexpression of serum ex-miRNA-21 was
diagnostic of pancreatic ductal adenocarcinoma (51). In contrast, miRNA-210 acts as an
oncogene, and has been demonstrated to be upregulated in various
types of tumors compared with adjacent normal tissues contributing
to the progression and development of various cancers, including
lung cancer and PC via different signaling pathways, including the
NF-κB, hypoxia and Akt- and p53-dependent signaling pathways
(52,53). The possible clinical application of
miRNA-210 in diagnosing and detecting different types of cancer,
such as PC, breast, lung and colorectal cancer, was previously
investigated (54). In addition, the
serum levels of miRNA-210 were previously found to be induced under
hypoxic conditions and linked to adverse prognosis in some types of
cancer, including colon and breast carcinoma, esophageal
adenocarcinoma and liver cancer (55), as a hypoxic environment is common in
PC (56,57), it may be worth investigating the
potential diagnostic value of serum ex-miRNAs for patients with PC.
The analysis of ex-miRNA-21 and miRNA-210 levels among circulating
serum miRNAs may represent a feasible strategy for PC diagnosis. It
may be inferred that ex-miRNAs in the serum directly reflect the
properties of tumor cells, as well as various diseases as the
quantity and content of exosomes may be reflective of the
pathophysiological state of the cells (58).
In the present study, the potential diagnostic value
of serum ex-miRNAs was compared with the serum levels of CA19-9.
Ex-miRNA-21 and miRNA-210 levels may be a new diagnostic or
therapeutic target for early PC, while CA19-9 levels remained
within the normal range. The sensitivity of the serum test was
increased to 90% when combining positive results for ex-miRNA-21
and miRNA-210 levels with serum CA19-9 levels, while the
specificity remained 90%. The findings of the present study
revealed the favorable sensitivity and specificity of circulating
fr-miRNA-21 and miRNA-210 for the diagnosis of PC, which was
consistent with other findings that the high levels of serum/plasma
miR-21 and miR-210 could be used as biomarkers for PC (59,60).
However, in the present study PC-associated secreted ex-miR-21 and
miR-210 may have clinical relevance in early detection; higher
sensitivity, specificity and accuracy compared with serum fr-miRNA.
Exosomes are highly heterogeneous and likely reflect the phenotype
of the cell that generate them (61). Similar to cells, exosomes are
composed of a lipid bilayer, this structure offers high stability
in body fluids, making exosomes highly attractive targets for
diagnostic markers (62). Serum
exosomes are a valuable tool and different methods may be used to
investigate their components, which may lead to more accurate,
sensitive, cost-effective, and high-throughput diagnoses (63). The present study demonstrated that
the serum ex-miRNA-21 and miRNA-210 levels were closely associated
with disease state and other prognostic factors, and that they may
be used for early diagnosis and prediction of prognosis
non-invasively. These data collectively revealed that these serum
exosomal miRNAs are potential valuable biomarkers for the
diagnosis, screening and prognosis of PC.
The limitations of the present study included the
small sample size, which may affect the diagnostic value of
exosomal miRNAs. In addition, the expression level of serum
ex-miRNAs was not compared between normal controls and PC tissues
i.e., the lack of healthy controls to compare with patients with PC
and CP, and the western blotting data was only qualitative rather
than quantitative. The obvious advantage of ex-miRNAs compared with
circulating fr-miRNA for the diagnosis of PC will be investigated
in depth in future studies. The mechanisms regulating transfer of
ex-miRNAs from tumor cells into the blood is rarely reported, while
the association between serum and tissue levels of ex-miRNAs should
be a focus of future research. In addition, exosomal miRNAs in the
plasma, pancreatic juice, bile, urine, saliva, breast milk and
other bodily fluids have not yet been investigated as diagnostic
biomarkers and will be examined in future studies. Another
limitation of the present study was that the level of change of
circulating ex-miRNAs was not examined before and after treatment
in patients with PC. Finally, the effect of the degree of
circulating ex-miRNA expression on disease progression, tumor cell
metastasis and overall survival cannot be concluded from the
results of the present study due to the small number of patients
with PC and the short follow-up period; this limitation must be
addressed by future studies.
The present study demonstrated that serum
ex-miRNA-21 and miRNA-210 levels distinguished effectively between
patients with PC and those with CP. Furthermore, the combination of
ex-miRNAs with CA19-9 was more specific and sensitive compared with
serum tests alone for the diagnosis of PC. These findings indicate
that circulating ex-miRNAs, miRNA-21 and miRNA-210, may be
potential novel biomarkers and therapeutic targets for PC. In
addition, the quantitation of ex-miRNAs may be used as a clinical
examination for further confirmation of diagnosis. Further
investigation of a large number of cases is needed to determine the
potential diagnostic, prognostic and clinical value of miRNA-21 and
miRNA-210 in patients with PC.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Scientific Foundation of China (grant no. 81872509), the Free
Exploration Project of Hubei University of Medicine (grant no.
FDFR201804), the Hubei Province Health and Family Planning
Scientific Research Project (grant no. WJ2019M054), the Natural
Science Foundation of the Bureau of Science and Technology of
Shiyan City (grant no. 18Y76, 17Y47 and 18Y77) and the Natural
Science Foundation of Hubei Provincial Department of Education
(grant nos. 2019CFB429 and Q20162113).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LW, WBZ, QHC and ZGT contributed to experimental
design, procedure and data acquisition. LW, JZ, WBZ, ZGT, WW and YW
contributed to data analysis, as well as drafting or revising the
manuscript. HMW, XDL, XCC, LY and LCY analyzed the data and
assisted in the experiments. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of Renmin Hospital of Wuhan University (approval no.
20180212; Wuhan, China). Written informed consent was provided by
all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qian L, Li Q, Kwaku B, Qiu W, Li K, Zhang
J, Yu Q, Xu D, Liu W, Brand RE, et al: Biosensors for early
diagnosis of pancreatic cancer: A review. Transl Res. 213:68–89.
2019. View Article : Google Scholar
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park J, Choi Y, Namkung J, Yi SG, Kim H,
Yu J, Kim Y, Kwon MS, Kwon W, Oh DY, et al: Diagnostic performance
enhancement of pancreatic cancer using proteomic multimarker panel.
Oncotarget. 8:93117–93130. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Akamatsu M, Makino N, Ikeda Y, Matsuda A,
Ito M, Kakizaki Y, Saito Y, Ishizawa T, Kobayashi T, Furukawa T and
Ueno Y: Specific MAPK-Associated MicroRNAs in serum differentiate
pancreatic cancer from autoimmune pancreatitis. PLoS One.
11:e01586692016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu H, Li B, Li T, Zhang S and Lin X:
Combination of noninvasive methods in diagnosis of infertile women
with minimal or mild endometriosis, a retrospective study in China.
Medicine (Baltimore). 98:e166952019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng H and Bai L: Hypoxia induced
microRNA-301b-3p overexpression promotes proliferation, migration
and invasion of prostate cancer cells by targeting LRP1B. Exp Mol
Pathol. 20:1043012019. View Article : Google Scholar
|
|
8
|
Fadaka AO, Pretorius A and Klein A:
Biomarkers for Stratification in Colorectal Cancer: MicroRNAs.
Cancer Control. 26:10732748198627842019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takagawa Y, Gen Y, Muramatsu T, Tanimoto
K, Inoue J, Harada H and Inazawa J: miR-1293, a Candidate for
miRNA-based cancer therapeutics, simultaneously targets BRD4 and
the DNA repair pathway. Mol Ther. 11:pii: S1525-0016. 30182–30189.
2020.
|
|
10
|
Örs Kumoğlu G, Döşkaya M and Gulce Iz S:
The biomarker features of miR-145-3p determined via meta-analysis
validated by qRT-PCR in metastatic cancer cell lines. Gene.
710:341–353. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Z, Meng Q, Pan A, Wu X, Cui J, Wang Y
and Li L: MicroRNA-455-3p promotes invasion and migration in triple
negative breast cancer by targeting tumor suppressor EI24.
Oncotarget. 8:19455–19466. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bai X, Lu D, Lin Y, Lv Y and He L: A
seven-miRNA expression-based prognostic signature and its
corresponding potential competing endogenous RNA network in early
pancreatic cancer. Exp Ther Med. 18:1601–1608. 2019.PubMed/NCBI
|
|
13
|
Kooshkaki O, Rezaei Z, Rahmati M, Vahedi
P, Derakhshani A, Brunetti O, Baghbanzadeh A, Mansoori B,
Silvestris N and Baradaran B: MiR-144: A new possible therapeutic
target and diagnostic/prognostic tool in cancers. Int J Mol Sci.
21(pii): E25782020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aggarwal V, Priyanka K and Tuli HS:
Emergence of circulating MicroRNAs in breast cancer as diagnostic
and therapeutic efficacy biomarkers. Mol Diagn Ther. 24:153–173.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang Y, Zhao Y, Song X, Song X, Niu L and
Xie L: Tumor-derived exosomal miRNA-320d as a biomarker for
metastatic colorectal cancer. J Clin Lab Anal. 16:e230042019.
|
|
17
|
Munson PB, Hall EM, Farina NH, Pass HI and
Shukla A: Exosomal miR-16-5p as a target for malignant
mesothelioma. Sci Rep. 9:116882019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang
G, Song J, Li Z, Zhang Z and Yuan W: Effect of exosomal miRNA on
cancer biology and clinical applications. Mol Cancer. 17:1472018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu H, Chen X, Ji J, Zhou R, Liu J, Ni W,
Qu L, Ni H, Ni R, Bao B and Xiao M: Progress of exosomes in the
diagnosis and treatment of pancreatic cancer. Genet Test Mol
Biomarkers. 23:215–222. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bortoluzzi S, Lovisa F, Gaffo E and
Mussolin L: Small RNAs in circulating exosomes of cancer patients:
A minireview. High Throughput. 6(pii): E132017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fanale D, Taverna S, Russo A and Bazan V:
Circular RNA in exosomes. Adv Exp Med Biol. 1087:109–117. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jayaseelan VP: Emerging role of exosomes
as promising diagnostic tool for cancer. Cancer Gene Ther. Sep
3–2019.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang W, Yin Y, Shan X, Zhou X, Liu P, Cao
Q, Zhu D, Zhang J and Zhu W: The value of plasma-based MicroRNAs as
diagnostic biomarkers for ovarian cancer. Am J Med Sci.
358:256–267. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He D, Wang H, Ho SL, Chan HN, Hai L, He X,
Wang K and Li HW: Total internal reflection-based single-vesicle in
situ quantitative and stoichiometric analysis of tumor-derived
exosomal microRNAs for diagnosis and treatment monitoring.
Theranostics. 9:4494–4507. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bai H, Lei K, Huang F, Jiang Z and Zhou X:
Exo-circRNAs: A new paradigm for anticancer therapy. Mol Cancer.
18:562019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yi M, Xu L, Jiao Y, Luo S, Li A and Wu K:
The role of cancer-derived microRNAs in cancer immune escape. J
Hematol Oncol. 13:252020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nguyen HV, Gore J, Zhong X, Savant SS,
Deitz-McElyea S, Schmidt CM, House MG and Korc M: MicroRNA
expression in a readily accessible common hepatic artery lymph node
predicts time to pancreatic cancer recurrence postresection. J
Gastrointest Surg. 20:1699–706. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xie Y, Hang Y, Wang Y, Sleightholm R,
Prajapati DR, Bader J, Yu A, Tang W, Jaramillo L, Li J, et al:
Stromal modulation and treatment of metastatic pancreatic cancer
with local intraperitoneal triple miRNA/siRNA nanotherapy. ACS
Nano. 14:255–271. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang Z, LaRiviere MJ, Ko J, Till JE,
Christensen T, Yee SS, Black TA, Tien K, Lin A, Shen H, et al: A
multi-analyte panel consisting of extracellular vesicle miRNAs and
mRNAs, cfDNA, and CA19-9 shows utility for diagnosis and staging of
pancreatic adenocarcinoma. Clin Cancer Res. 16:pii: clincanres.
3313:20192020.
|
|
30
|
Yoshizawa N, Sugimoto K, Tameda M, Inagaki
Y, Ikejiri M, Inoue H, Usui M, Ito M and Takei Y:
miR-3940-5p/miR-8069 ratio in urine exosomes is a novel diagnostic
biomarker for pancreatic ductal adenocarcinoma. Oncol Lett.
19:2677–2684. 2020.PubMed/NCBI
|
|
31
|
Ko J, Bhagwat N, Black T, Yee SS, Na YJ,
Fisher S, Kim J, Carpenter EL, Stanger BZ and Issadore D: miRNA
profiling of magnetic nanopore-isolated extracellular vesicles for
the diagnosis of pancreatic cancer. Cancer Res. 78:3688–3697.
2018.PubMed/NCBI
|
|
32
|
Matsuda Y, Fujii Y, Matsukawa M, Ishiwata
T, Nishimura M and Arai T: Overexpression of carbohydrate
sulfotransferase 15 in pancreatic cancer stroma is associated with
worse prognosis. Oncol Lett. 18:4100–4105. 2019.PubMed/NCBI
|
|
33
|
Luo G, Javed A, Strosberg JR, Jin K, Zhang
Y, Liu C, Xu J, Soares K, Weiss MJ, Zheng L, et al: Modified
staging classification for pancreatic neuroendocrine tumors on the
basis of the American joint committee on cancer and european
neuroendocrine tumor society systems. J Clin Oncol. 35:274–280.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee YR, Kim G, Tak WY, Jang SY, Kweon YO,
Park JG, Lee HW, Han YS, Chun JM, Park SY and Hur K: Circulating
exosomal noncoding RNAs as prognostic biomarkers in
humanhepatocellular carcinoma. Int J Cancer. 144:1444–1452. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shi M, Jiang Y, Yang L, Yan S, Wang YG and
Lu XJ: Decreased levels of serum exosomal miR-638 predict poor
prognosis in hepatocellular carcinoma. J Cell Biochem.
119:4711–4716. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Suehiro T, Miyaaki H, Kanda Y, Shibata H,
Honda T, Ozawa E, Miuma S, Taura N and Nakao K: Serum exosomal
microRNA-122 and microRNA-21 as predictive biomarkers in
transarterial chemoembolization-treated hepatocellular carcinoma
patients. Oncol Lett. 16:3267–3273. 2018.PubMed/NCBI
|
|
37
|
Shiotsu H, Okada K, Shibuta T, Kobayashi
Y, Shirahama S, Kuroki C, Ueda S, Ohkuma M, Ikeda K, Ando Y, et al:
The influence of pre-analytical factors on the analysis of
circulating MicroRNA. Microrna. 7:195–203. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Silverman JM and Reiner NE: Exosomes and
other microvesicles in infection biology: Organelles with
unanticipated phenotypes. Cell Microbiol. 13:1–9. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rahbarghazi R, Jabbari N, Sani NA, Asghari
R, Salimi L, Kalashani SA, Feghhi M, Etemadi T, Akbariazar E,
Mahmoudi M and Rezaie J: Tumor-derived extracellular vesicles:
Reliable tools for Cancer diagnosis and clinical applications. Cell
Commun Signal. 17:732019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Salehi M and Sharifi M: Exosomal miRNAs as
novel cancer biomarkers: Challenges and opportunities. J Cell
Physiol. 233:6370–6380. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang M, Yu F, Ding H, Wang Y, Li P and
Wang K: Emerging function and clinical values of exosomal MicroRNAs
in cancer. Mol Ther Nucleic Acids. 16:791–804. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Capula M, Mantini G, Funel N and
Giovannetti E: New avenues in pancreatic cancer: Exploiting
microRNAs as predictive biomarkers and new approaches to target
aberrant metabolism. Expert Rev Clin Pharmacol. 12:1081–1090. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rabinowits G, Gerçel-Taylor C, Day JM,
Taylor DD and Kloecker GH: Exosomal microRNA: A diagnostic marker
for lung cancer. Clin Lung Cancer. 10:42–46. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sohn W, Kim J, Kang SH, Yang SR, Cho JY,
Cho HC, Shim SG and Paik YH: Serum exosomal microRNAs as novel
biomarkers for hepatocellular carcinoma. Exp Mol Med. 47:e1842015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
de Carvalho IN, de Freitas RM and Vargas
FR: Translating microRNAs into biomarkers: What is new for
pediatric cancer? Med Oncol. 33:492016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Su J, Wu F, Xia H, Wu Y and Liu S:
Accurate cancer cell identification and microRNA silencing induced
therapy using tailored DNA tetrahedron nanostructures. Chem Sci.
11:80–86. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Luo D, Huang Z, Lv H, Wang Y, Sun W and
Sun X: Up-regulation of MicroRNA-21 indicates poor prognosis and
promotes cell proliferation in esophageal squamous cell carcinoma
via upregulation of lncRNA SNHG1. Cancer Manag Res. 12:1–14. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Goto T, Fujiya M, Konishi H, Sasajima J,
Fujibayashi S, Hayashi A, Utsumi T, Sato H, Iwama T, Ijiri M, et
al: An elevated expression of serum exosomal microRNA-191, - 21,
−451a of pancreatic neoplasm is considered to be efficient
diagnostic marker. BMC Cancer. 18:1162018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Feng S, He A, Wang D and Kang B:
Diagnostic significance of miR-210 as a potential tumor biomarker
of human cancer detection: An updated pooled analysis of 30
articles. Onco Targets Ther. 12:479–493. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hong L, Han Y, Zhang H, Zhao Q and Qiao Y:
miR-210: A therapeutic target in cancer. Expert Opin Ther Targets.
17:21–28. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ren CX, Leng RX, Fan YG, Pan HF, Wu CH and
Ye DQ: MicroRNA-210 and its theranostic potential. Expert Opin Ther
Targets. 20:1325–1338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bavelloni A, Ramazzotti G, Poli A, Piazzi
M, Focaccia E, Blalock W and Faenza I: MiRNA-210: A current
overview. Anticancer Res. 37:6511–6521. 2017.PubMed/NCBI
|
|
56
|
Li W, Liu H, Qian W, Cheng L, Yan B, Han
L, Xu Q, Ma Q and Ma J: Hyperglycemia aggravates microenvironment
hypoxia and promotes the metastatic ability of pancreatic cancer.
Comput Struct Biotechnol J. 16:479–487. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen S, Zhang J, Chen J, Wang Y, Zhou S,
Huang L, Bai Y, Peng C, Shen B, Chen H and Tian Y: RER1 enhances
carcinogenesis and stemness of pancreatic cancer under hypoxic
environment. J Exp Clin Cancer Res. 38:152019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Severino V, Dumonceau JM, Delhaye M, Moll
S, Annessi-Ramseyer I, Robin X, Frossard JL and Farina A:
Extracellular vesicles in bile as markers of malignant biliary
stenoses. Gastroenterology. 153:495–504.e8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang J, Chen J, Chang P, LeBlanc A, Li D,
Abbruzzesse JL, Frazier ML, Killary AM and Sen S: MicroRNAs in
plasma of pancreatic ductal adenocarcinoma patients as novel
blood-based biomarkers of disease. Cancer Prev Res. 2:807–813.
2009. View Article : Google Scholar
|
|
60
|
Qu K, Zhang X, Lin T, Liu T, Wang Z, Liu
S, Zhou L, Wei J, Chang H, Li K, et al: Circulating miRNA-21-5p as
a diagnostic biomarker for pancreatic cancer: Evidence from
comprehensive miRNA expression profiling analysis and clinical
validation. Sci Rep. 7:16922017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kowal J, Arras G, Colombo M, Jouve M,
Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M and Thery
C: Proteomic comparison defines novel markers to characterize
heterogeneous populations of extracellular vesicle subtypes. Proc
Natl Acad Sci USA. 113:E968–E977. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hu Q, Su H, Li J, Lyon C, Tang W, Wan M
and Hu TY: Clinical applications of exosome membrane proteins.
Precis Clin Med. 3:54–66. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Makler A and Asghar W: Exosomal biomarkers
for cancer diagnosis and patient monitoring. Expert Rev Mol Diagn.
20:387–400. 2020. View Article : Google Scholar : PubMed/NCBI
|