Introduction
The most common type of endocrine tumors is thyroid
cancer (TC) (1). In 2012, TC caused
more than 40,000 deaths and affected about 300,000 new cases
worldwide (2). Moreover, the
incidence rate of TC is continuously increasing in recent years in
several countries, such as China (3). Therefore, it is important to develop
effective approaches to prevent the occurrence of TC and improve
treatment outcomes. Papillary TC (PTC) is the major histological
subtype of TC and accounts for about 80% of all cases (4). With appropriate treatment, such as
thyroid hormone suppression, surgical resection and radioactive
iodine therapy, most PTC cases can be cured. However, tumor
metastasis may occur in a small portion of PTC patients and the
prognosis is generally poor (5).
Compared with normal cells, glucose metabolism is
accelerated in cancer cells, and the increased glucose metabolism
rate in cancer cell supports the continuous growth of tumors
(6). In cancer treatment, the
inhibition of glucose uptake has been considered as a promising
target (7). Glucose transporter 1
(GLUT1) is one of the major players in glucose uptake, which
mediates glucose across plasma membranes (8). GLUT1 is usually upregulated in
different types of cancer types, including in PTC (9). Some microRNAs (miRNAs/miRs), such as
miR-1291 may regulate the expression of GLUT1 to inhibit cancer
progression (10). In a recent
study, Huang et al (11)
showed that miR-125b is involved in glucose-induced reactive oxygen
species generation, indicating the involvement of miR-125b in
glucose metabolism. The present study was conducted to investigate
the role of miR-125b in PTC and explore its possible involvement in
glucose metabolism.
Materials and methods
Patients and specimens
A total of 109 patients with PTC were admitted to
Yongchuan Hospital of Chongqing Medical University between December
2016 and November 2018. From these 109 patients, 56 PTC patients
(30 males and 26 females; age range, 34 to 62 years; mean age,
48.1±6.2 years) were selected to serve as the research subjects of
the present study. The inclusion criteria were as follows: i)
First-time diagnosed cases; and ii) patients who received no
therapies within 100 days before admission. The exclusion criteria
were as follows: i) Recurrence cases (n=2); ii) patients with other
clinical disorders (n=30); iii) and patients who were treated for
any other clinical disorders within 100 days before admission
(n=21). Based on clinical findings, there were 16, 20, 10 and 10
patients at clinical stages (AJCC) I–IV (12), respectively. All 56 patients signed
informed consent. This study was approved by the Ethics Committee
of Yongchuan Hospital of Chongqing Medical University.
Adjacent normal tissues (within 2 cm around tumors)
and PTC tissues were obtained from each patient during the
diagnosis, through histopathological biopsy. Weights of tissue
samples ranged from 0.015 to 0.022 g. At least three pathologists
checked the tissues and confirmed that all tissue samples were
correctly classified (nomal tissues contained no cancer cells and
PTC tissues contained >80% cancer cells).
PTC cells and cell transfections
HTH83 and IHH-4 human PTC cell lines (ATCC) were
used. Cells were cultivated under conditions of 37°C and 5%
CO2. Cell culture medium was DMEM (10% FBS, Invitrogen;
Thermo Fisher Scientific, Inc.).
Negative non-targeting control miRNA
(5′-CCGGUGUACGUAGUGGGCAGUG-3′) and miR-125b mimic
(5′-UCCCUGAGACCCUAACUUGUGA-3′) were obtained from Sigma-Aldrich
(Merck KGaA). GLUT1 expression pcDNA3 vector and empty pcDNA3
vector were purchased from Sangon Biotech Co., Ltd. HTH83 and IHH-4
cells were harvested at the confluence of 70–80%, and 40 nM
negative control (NC) miRNA, 40 nM miR-125b mimic, 15 nM empty
pcDNA3 vector (NC), or 10 nM GLUT1 expression pcDNA3 vector was
transfected into the cells using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Cells without transfections were
control (C) cells. The interval between transfection and the
subsequent experiments was 24 h.
Reverse transcription-quantitative PCR
(RT-qPCR)
HTH83 and IHH-4 cells (1×105 cells) or
0.01 g tissue of each sample (ground in liquid nitrogen) was mixed
with 1 ml Ribozol reagent (VWR Life Science) to extract total RNAs.
Following digestion with DNase I, reverse transcriptions (55°C for
15 min and 80°C for 10 min) were performed using Quantitect Reverse
Transcription kit (Qiagen China Co., Ltd.) and qPCR reaction
mixtures were prepared using the Universal One-Step RT-qPCR kit
(SYBR, New England BioLabs, Inc.). The expression of GLUT1 mRNA was
detected using GAPDH as endogenous control. The PCR thermocycling
conditions were: 95°C for 1 min, followed by 40 cycles of 95°C for
10 sec and 60°C for 40 sec.
miRNAs were extracted from the aforementioned cells
and tissues using mirVana miRNA Isolation kit (Thermo Fisher
Scientific, Inc.), following miRNA reverse transcriptions (55°C for
10 min and 80°C for 10 min) performed using Taqman®
MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.;
cat. no. 4366596). The expression of miR-125b was detected using
the TaqMan™ microRNA assay (Applied
Biosystems™; Thermo Fisher Scientific, Inc.; cat. no.
4427975) with U6 as the endogenous control. The PCR thermal cycling
conditions were: 95°C for 1 min, followed by 40 cycles of 95°C for
10 sec and 58°C for 20 sec.
The primer sequences were: GLUT-1 forward,
5′-CTGCTCATCAACCGCAA-3′; GLUT-1 reverse, 5′-CTTCTTCTCCCGCATCATC-3′;
GAPDH forward, 5′-ACCCAGAAGACTGTGGATG-3′; and GAPDH reverse,
5′-GTAGAGGCAGGGATGATGTT-3′. The forward primer of miR-125b was
5′-UCCCUGAGACCCUAACUUG-3′. The reverse primer of miR-125b and U6
primers were from the kit.
All data were processed using the 2−∆∆Cq
method (13). Each experiment was
replicated three times.
Glucose uptake analysis
The Krebs-Ringer-HEPES (KRH) buffer was prepared
before glucose uptake assay, using the following reagents: 1.3 mM
CaCl2, 1.2 mM MgSO4, 120 mM NaCl, 5 mM KCl,
1.3 mM KH2PO4 and 25 mM HEPES (pH 7.4). HTH83
and IHH-4 cells were harvested at 24 h post-transfections.
Following washing with KRH buffer, 4×105 cells were
dissolved in fresh KRH buffer. Subsequently,
[3H]-2-deoxyglucose (1µCi; Perkin Elmer Life Sciences)
was added and cells were cultivated at 37°C for 2 min. Glucose
uptake was terminated by adding ice-cold KRH (3 volumes). Cells
were separated from the buffer by centrifugation at 1,200 × g for
10 min at room temperature. A scintillation spectrometry was used
to measure radioactivity and disintegrations per minute, which
represents the amount of glucose in cells. DMP was normalized to
cellular protein mass.
Cell proliferation analysis
HTH83 and IHH-4 cells were resuspended in Eagle's
Minimum Essential Medium (10% FBS, Invitrogen; Thermo Fisher
Scientific, Inc.) with a ratio of 4×104 cells per 1 ml
medium to make single-cell suspensions. The cells were cultivated
under the aforementioned conditions, and Cell Counting Kit-8
solution (10 µl; Sigma-Aldrich; Merck KGaA) was added to each well
at 4 h before the end of cell culture. Cell culture was terminated
every 24 h until 96 h, followed by the addition of 10 µl DMSO.
Subsequently, the OD values (450 nm) were measured.
Western blotting
HTH83 and IHH-4 cells (1×105 cells) were
mixed with 1 ml pre-cold (4°C) RIPA buffer (Invitrogen; Thermo
Fisher Scientific, Inc.) to extract total protein. BCA assay was
performed to quantify all protein samples. Protein samples were
boiled for 5 min, followed by SDS-PAGE using gel (10%)
electrophoresis with 30 µg per lane. Proteins were then transferred
onto PVDF membranes. Subsequently, the membranes were blocked in 5%
non-fat milk for 2 h at 22°C, and treated with primary antibodies
of rabbit polyclonal GAPDH (1:800; cat. no. ab8245; Abcam) and
GLUT1 (1:800; cat. no. ab15309; Abcam) overnight at 4°C. IgG-HRP
goat anti-rabbit (1:800; cat. no. MBS435036; MyBioSource, Inc.) was
used as the secondary antibody and incubation was performed for 2 h
at room temperature. Signals were developed using ECL
(Sigma-Aldrich; Merck KGaA) and analyzed by Image J version 1.46
(National Institutes of Health).
Statistical analysis
Each experiment was repeated three times and the
data from three biological replicates were used to calculate the
mean ± standard deviation values. The differences between normal
and PTC tissues were analyzed using the paired t-test. The
differences among different cell and patient groups were analyzed
using ANOVA (one-way) in combination with Tukey's test.
Correlations were analyzed using linear regression. P<0.05 was
considered to indicate a statistically significant difference.
Results
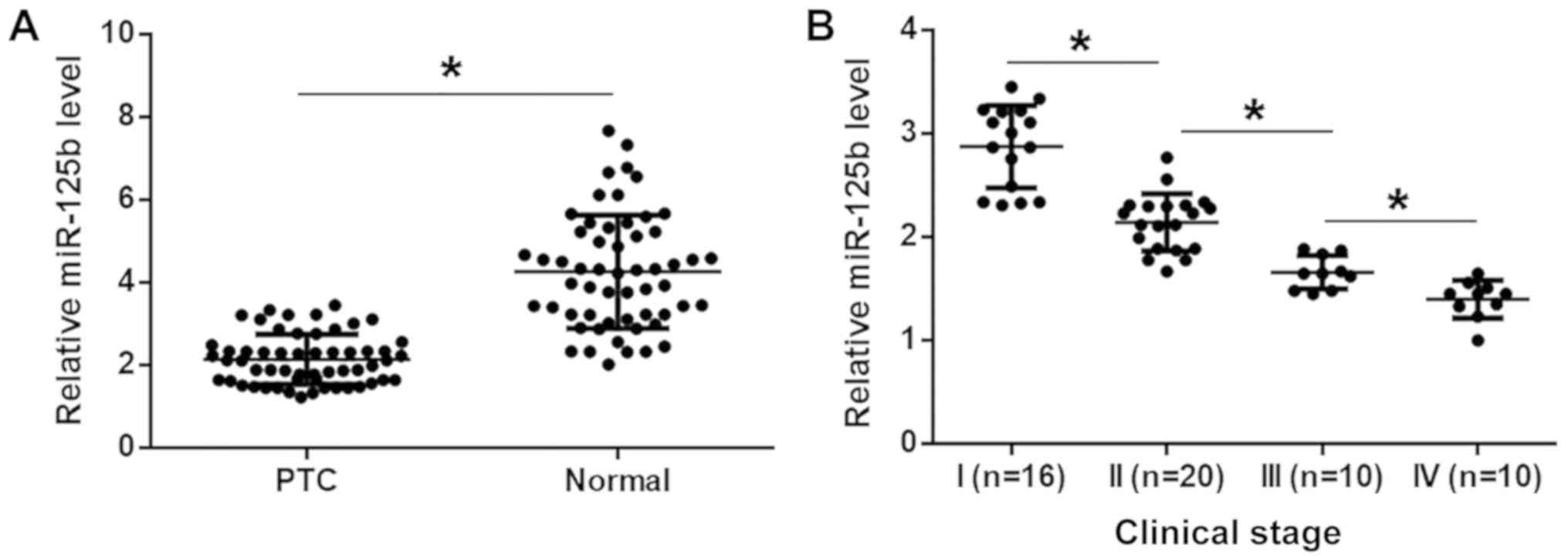
miR-125b is downregulated in PTC
The levels of miR-125b expression were measured in
the two types of tissues from 56 patients with PTC, by RT-qPCR.
Compared with normal tissues, the expression levels of miR-125b
were significantly lower in PTC tissues (Fig. 1A; P<0.05). The 56 patients with
PTC included 16, 20, 10 and 10 patients at clinical stages (AJCC)
I–IV, respectively. It was observed that the levels of miR-125b
expression decreased with increasing clinical stage (Fig. 1B; P<0.05).
GLUT1 is negatively correlated with
miR-125b in PTC
GLUT1 mRNA level was detected by RT-qPCR. Paired
t-test analysis showed that GLUT1 mRNA levels were significantly
higher in PTC tissues compared with normal tissues (Fig. 2A; P<0.05). Linear regression
analysis showed that GLUT1 mRNA and miR-125b were significantly and
inversely correlated in PTC tissues, (Fig. 2B). In normal tissues, GLUT1 mRNA and
miR-125b were not significantly correlated (Fig. 2C).
miR-125b suppresses glucose uptake in
PTC cell by downregulating GLUT1
HTH83 and IHH-4 cells were transfected with miR-125b
mimic and GLUT1 expression vectors. Compared with NC and C groups,
the expression levels of miR-125b (Fig.
3A-a) as well as GLUT1 protein (Fig.
3A-b) and GLUT1 mRNA (Fig. 3A-c)
were significantly increased at 24 h post-transfection (Fig. 3A; P<0.05). Moreover, compared with
the two controls, miR-125b overexpression resulted in downregulated
GLUT1 (Fig. 3B-a) and decreased
glucose uptake (Fig. 3B-b). GLUT1
overexpression decreased the effects of miR-125b overexpression on
glucose uptake (Fig. 3B;
P<0.05).
 | Figure 3.miR-125b suppresses glucose uptake in
PTC cell by downregulating GLUT1. (A) Compared with NC and C
groups, the expression levels of miR-125b (A-a) and GLUT1 (A-b,
protein levels and A-c, mRNA levels) were significantly increased
at 24 h post-transfection, following the transfections with
miR-125b mimic and GLUT1 expression vectors. Moreover, compared
with the two controls, miR-125b overexpression resulted in
downregulated GLUT1 (B-a) and decreased glucose uptake (B-b). (B-b)
GLUT1 overexpression reduced the effects of miR-125b overexpression
on glucose uptake. *P<0.05. GLUT1, glucose transporter 1; miR,
microRNA; PTC, papillary thyroid carcinoma; NC, negative control
miRNA transfection (in cases of miR-125b overexpression) or empty
pcDNA3 vector transfection (in cases of GLUT1 overexpression); C,
control (cells without transfections); DPM, disintegrations per
minute. |
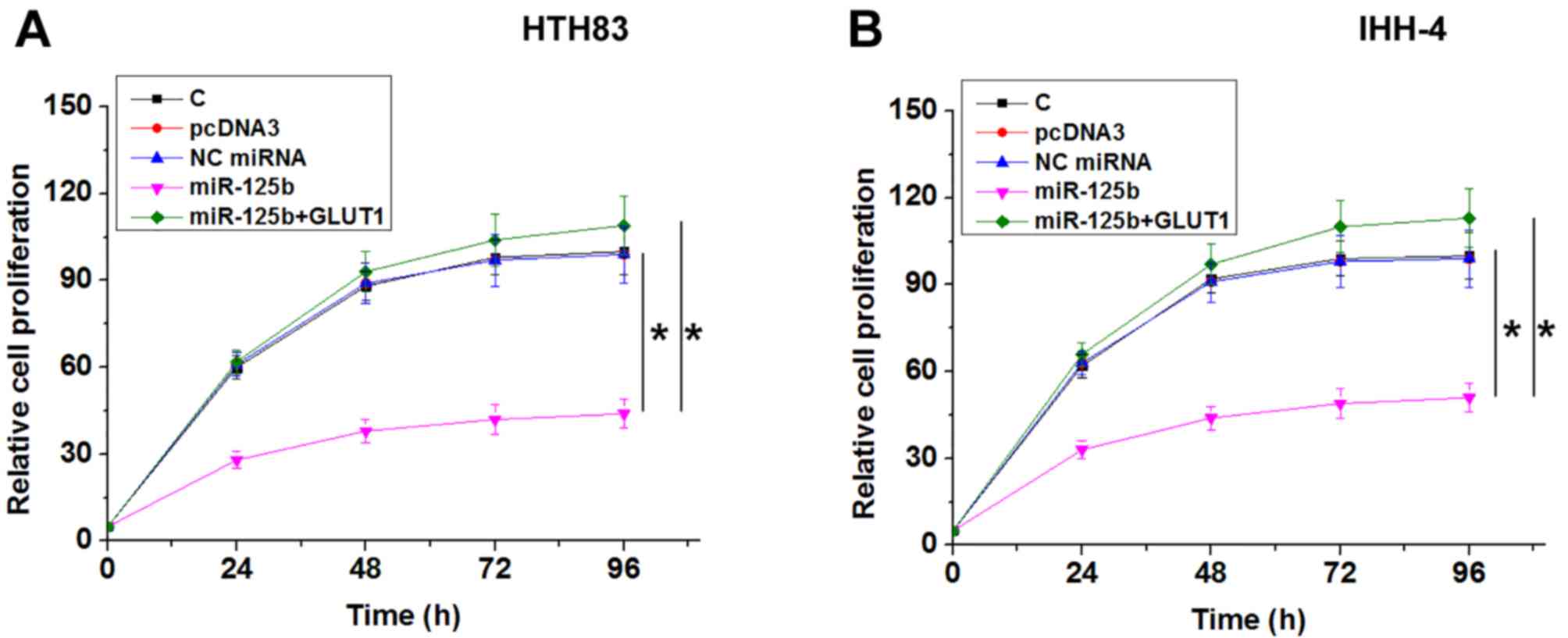
miR-125b suppresses PTC cell
proliferation by downregulating GLUT1
Compared with NC and C groups, miR-125b
overexpression resulted in decreased, while GLUT1 overexpression
resulted in increased proliferation rate of PTC cells. GLUT1
overexpression reduced the effects of miR-125b overexpression on
cancer cell proliferation (Fig. 4;
P<0.05).
Discussion
In the present study, the role of miR-125b was
investigated in PTC. It was found that miR-125b was downregulated
in PTC and may inhibit the glucose uptake and proliferation of PTC
cells, by downregulating GLUT1.
miR-125b has been characterized as a
tumor-suppressive miRNA or oncogenic miRNA in different types of
cancer. In ovarian cancer, miR-125b is downregulated and the
overexpression of miR-125b suppresses the proliferation of cancer
cells by directly targeting BCL3 (14). In invasive breast cancer, miR-125b is
methylated, while the transcription of miR-125b gene downregulates
the expression of oncogenic ETS1, thereby inhibiting cancer
progression (15). In contrast, Shi
et al (16) established a
xenograft prostate tumor model to study the roles of miR-125b in
prostate cancer, and found that miR-125b inhibited the expression
of pro-apoptotic genes in prostate cancer cell and promoted the
growth of tumor, indicating that miR-125b played an oncogenic role
in prostate cancer. To the best of our knowledge, the involvement
of miR-125b in PTC and other types of TC is still unknown. The
present study found that miR-125b was downregulated in PTC and that
the upregulation of miR-125b led to decreased glucose uptake in PTC
cells and inhibited PTC cell proliferation. Therefore, miR-125b has
tumor-suppressive roles in PTC.
The inhibition of glucose metabolism is an effective
approach to inhibiting cancer progression. It is known that certain
tumor-associated miRNAs can regulate glucose metabolism to
participate in cancer biology (17–19). In
the present study, it was shown that miR-125b can downregulate
GLUT1 to suppress glucose uptake in PTC cells and inhibit cell
proliferation. Therefore, overexpression of miR-125b may serve as a
potential therapeutic target to inhibit the growth of PTC tumors;
however, clinical trials are needed to test this conclusion. In
addition, the mechanism that mediates the downregulation of GLUT1
by miR-125b is unclear. In the present study, a significant
correlation between GLUT-1 and miR-125b was observed across PTC
tissues, however not across normal tissues. Therefore, the
interaction between GLUT-1 and miR-125b is indirect and may be
mediated by certain PTC-associated pathological factors; however,
the factors involved in this interaction have not been identified.
Thus, further studies are still required. The present study also
showed that the altered expression of miR-125b can be used to
assist the diagnosis of HCC. However, future studies with larger
sample sizes are needed to further confirm these conclusions.
In conclusion, miR-125b is downregulated in PTC and
miR-125b overexpression may inhibit the growth of PTC by
suppressing glucose uptake, by downregulating GLUT1.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GZ: experimental work, data analysis, and manuscript
writing. SHZ and QY: experiment work, literature research, and data
analysis. FL: study design, research concept and manuscript
editing. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from the Ethics
Committee of Yongchuan Hospital of Chongqing Medical University
(approval no. YCH201611063565CQMU). All the patients signed
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rosenbaum MA and McHenry CR: Contemporary
management of papillary carcinoma of the thyroid gland. Expert Rev
Anticancer Ther. 9:317–329. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brito JP, Hay ID and Morris JC: Low risk
papillary thyroid cancer. BMJ. 348:g30452014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Annibaldi A and Widmann C: Glucose
metabolism in cancer cells. Curr Opin Clin Nutr Metab Care.
13:466–470. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cairns RA, Harris IS and Mak TW:
Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kraegen EW, Sowden JA, Halstead MB, Clark
PW, Rodnick KJ, Chisholm DJ and James DE: Glucose transporters and
in vivo glucose uptake in skeletal and cardiac muscle: Fasting,
insulin stimulation and immunoisolation studies of GLUT1 and GLUT4.
Biochem J. 295:287–293. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ciavardelli D, Bellomo M, Consalvo A,
Crescimanno C and Vella V: Metabolic alterations of thyroid cancer
as potential therapeutic targets. Biomed Res Int. 2017:25450312017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamasaki T, Seki N, Yoshino H, Itesako T,
Yamada Y, Tatarano S, Hidaka H, Yonezawa T, Nakagawa M and Enokida
H: Tumor-suppressive microRNA-1291 directly regulates glucose
transporter 1 in renal cell carcinoma. Cancer Sci. 104:1411–1419.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang YF, Zhang Y, Liu CX, Huang J and
Ding GH: microRNA-125b contributes to high glucose-induced reactive
oxygen species generation and apoptosis in HK-2 renal tubular
epithelial cells by targeting angiotensin-converting enzyme 2. Eur
Rev Med Pharmacol Sci. 20:4055–4062. 2016.PubMed/NCBI
|
|
12
|
Lang BHH, Lo CY, Chan WF, Lam KY and Wan
KY: Staging systems for papillary thyroid carcinoma: A review and
comparison. Ann Surg. 245:366–378. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guan Y, Yao H, Zheng Z, Qiu G and Sun K:
miR-125b targets BCL3 and suppresses ovarian cancer proliferation.
Int J Cancer. 128:2274–2283. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Yan LX, Wu QN, Du ZM, Chen J,
Liao DZ, Huang MY, Hou JH, Wu QL, Zeng MS, et al: miR-125b is
methylated and functions as a tumor suppressor by regulating the
ETS1 proto-oncogene in human invasive breast cancer. Cancer Res.
71:3552–3562. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi XB, Xue L, Ma AH, Tepper CG, Kung HJ
and White RW: miR-125b promotes growth of prostate cancer xenograft
tumor through targeting pro-apoptotic genes. Prostate. 71:538–549.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang R, Xiao T, Fang Z, Sun Y, Li F, Gao
Y, Feng Y, Li L, Wang Y, Liu X, et al: MicroRNA-143 (miR-143)
regulates cancer glycolysis via targeting hexokinase 2 gene. J Biol
Chem. 287:23227–23235. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiao X, Huang X, Ye F, Chen B, Song C, Wen
J, Zhang Z, Zheng G, Tang H and Xie X: The miR-34a-LDHA axis
regulates glucose metabolism and tumor growth in breast cancer. Sci
Rep. 6:217352016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen B, Liu Y, Jin X, Lu W, Liu J, Xia Z,
Yuan Q, Zhao X, Xu N and Liang S: MicroRNA-26a regulates glucose
metabolism by direct targeting PDHX in colorectal cancer cells. BMC
Cancer. 14:4432014. View Article : Google Scholar : PubMed/NCBI
|