Introduction
Osteosarcoma (OS) is a common type of primary bone
cancer that affects ~3.4/1,000,000 people per year in the past
decades worldwide (1). Despite the
low incidence rate, osteosarcoma is a heavy burden on public
health; as it mainly affects teenagers and young adults, early
diagnosis is rare and curative therapies are lacking (2,3). With
the advances in OS treatment, the overall 5-year survival rate has
increased from 20 to 50% during the 20th century (4). However, there is still a significant
population of patients with OS diagnosed with metastatic tumors for
which there are no radical treatment regimens available (5). Thus, novel therapeutic approaches are
still needed.
OS occurrence is associated with a number of
physical factors, such as age and height (6). However, genetic factors are the most
critical causative factors of OS (7). Rho-associated coiled-coil containing
protein kinase 1 (ROCK1) functions downstream of GTPase RhoA and
regulates the generation of contractile force, motility and
metastasis of OS cells (8). ROCK1 is
upregulated in a number of cancers including OS and can promote
tumor metastasis by enhancing the mobility of cancer cell (9). A previous study has demonstrated that
ROCK1 expression can be downregulated by certain microRNAs
(miRNAs), such as microRNA (miR)-144 (10). Therefore, regulating the expression
of certain tumor-suppressive miRNAs may directly suppress cancer
metastasis through the downregulation of ROCK1.
Zinc finger protein 281 (ZNF281) is a recently
identified tumor-suppressive long non-coding (lncRNA) in glioma
(11). Our preliminary RNA-seq data
showed that ZNF281 was inversely associated with ROCK1 expression
in OS cells (data not shown). However, the mechanisms of
interaction among ZNF281, miR-144 and ROCK1 have not been explored.
The present study aimed to explore the role of ZNF281 in OS and
possible interactions with ROCK1 and miR-144.
Materials and methods
Collection of tissue specimens from
patients with OS
A total of 60 patients with OS [36 male and 24
female; aged 12–31 years; mean ± standard deviation (SD): 21.1±3.4
years] were enrolled in the present study from a total of 108
patients with OS admitted to Honghui Hospital affiliated to Xi'an
Jiaotong University between April 2011 and April 2014. This study
was approved by the Ethics Committee of Honghui Hospital Affiliated
to Xi'an Jiaotong University (Xi'an, China). All patients or their
guardians if the patient was <18 years old signed an informed
consent form. Inclusion criteria for the patients with OS were: i)
No prior treatment; ii) newly diagnosed; and iii) completion of
treatment and a 5-year follow-up at Honghui Hospital. Exclusion
criteria included: i) Prior treatment; ii) recurrent OS; iii)
patient transfer from another institute; and iv) the presence of
other severe diseases. The American Joint Committee on Cancer
staging system (12) was used to
stage the 60 patients with OS. A total of 12, 19, 16 and 13 cases
were clinical stage I–IV, respectively.
Patients with OS were subjected to MRI-guided fine
needle biopsy. During biopsy, OS and adjacent (within 2 cm of the
tumor) non-tumor tissue samples were collected from each patient.
All OS samples contained >98% cancer cells and all non-tumor
samples contained <1% cancer cells.
OS cells and transient
transfections
A human OS cell line U2OS (ATCC) was obtained and
used for subsequent experiments. Cells were cultured in a mixture
of 90% Eagle's Minimum Essential Medium (EMEM, Sigma-Aldrich; Merck
KGaG) and 10% FBS. Cell culture conditions were 95% humidity, 37°C
and 5% CO2. Cells were harvested when they were 70–80%
confluent for subsequent transfections. ZNF281 and ROCK1
overexpression vectors were constructed using the pcDNA3.1 vector
(Sangon Biotech Co., Ltd). Negative control (NC) miRNA
(5′-CACGUACGGUAGUACCCGUAUU-3′) and the miR-144 mimic
(5′-GGAUAUCAUCAUAUACUGUAAG-3′) were obtained from Guangzhou RiboBio
Co., Ltd. Cells were counted, and 3×106 cells were
transfected with 45 nM miRNA (NC miRNA as NC group) or 10 nM vector
(empty vector as NC group) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Cells were harvested
at 24 h post-transfection to perform subsequent experiments.
Control cells were untransfected cells.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
RNAzol (Merck KGaA) was used to extract the RNA from
tissue specimens from patients with OS (0.1 g tissue ground in
liquid nitrogen) and U2OS cells (6×105) according to the
manufacturer's instructions. RNA precipitation for harvesting
miRNAs was performed using 85% ethanol. RNA samples were first
digested with DNase I (2 h at 37°C, Sigma-Aldrich; Merck KGaG) and
then reverse transcribed into cDNA using AMV Reverse Transcriptase
kit (Promega Corporation) by incubating at 55°C for 10 min,
followed by 53°C for 20 min and 80°C for 10 min. Subsequently,
QuantiTect SYBR® Green PCR kit (Qiagen China Co., Ltd.)
was used to prepare the qPCR to assay the expression of ZNF281 and
ROCK1 mRNA. GAPDH was used as the internal reference gene.
Expression levels of miR-144 were measured using the All-in-One™
miRNA qRT-PCR Detection kit (GeneCopoeia, Inc.), which was used to
perform 3′-polyadenylation, reverse transcriptions and preparation
of qPCR mixtures. U6 was used as the endogenous control. PCR
conditions were: 95°C for 1 min, followed by 40 cycles of 95°C for
12 sec and 58.5°C for 45 sec. Primer sequences were: ZNF281,
forward 5′-CAGGGTATACAAATATGATG-3′ and reverse
5′-GCATTGAAAGGGCATCACATTA-3′; ROCK1, forward
5′-AGTATTTCTCCCATATGGATA-3′ and reverse
5′-ACCAATGGATTGTTCACCTGAA-3′; GAPDH, forward
5′-GTCTCCTCTGACTTCAACAGCG-3′ and reverse
5′-ACCACCCTGTTGCTGTAGCCAA-3′. Forward primer of miR-144 was
5′-GGATATCATCATATACTG-3′. Universal reverse primers and U6 primers
were from the kit (sequences unavailable). Each experiment included
three replicates and data were normalized using the
2−ΔΔCq method (13).
Western blotting
RIPA buffer (Sangon Biotech Co., Ltd) was used to
extract total proteins from U2OS cells. Protein concentrations were
measured using a bicinchoninic acid assay kit. Protein samples were
boiled for 5 min for denaturation. Subsequently, 10% SDS-PAGE was
used to separate the proteins with 30 µg protein per lane. The
proteins were transferred to PVDF membranes, followed by blocking
for 2 h in PBS containing 5% FBS (Sigma-Aldrich; Merck KGaA) at
room temperature. The membranes were probed with rabbit anti-ROCK1
(1:800; cat. no. ab97592; Abcam) and anti-GAPDH (1:800; cat. no.
ab37168; Abcam) for 12 h at 4°C, followed by incubation with
horseradish peroxidase-conjugated goat secondary antibody (IgG;
1:1,000; cat. no. ab6721; Abcam) for 2 h at 24°C. Signal
development was performed using the ECL Western Blotting Substrate
kit (cat. no. ab65623; Abcam), and the data were processed using
Image J v.1.48 software (National Institute of Health).
Transwell invasion and migration
assays
The effects of the various transfections on the
invasion and migration of U2OS cells were determined by Transwell
invasion and migration assays. Transwell membranes were precoated
with Matrigel at 37°C for 6 h for the invasion assay. Cells were
harvested, counted and mixed with serum-free EMEM at
3×104 cells/ml to prepare single-cell suspensions. Cells
were added into the upper chamber (0.1 ml/well), and the lower
chamber was filled with EMEM containing 20% FBS. Transwell chambers
were incubated at 37°C for 12 h. Subsequently, non-invasive and
non-migrated cells were removed using cotton swabs and the lower
surface of membranes was stained with 0.5% crystal violet (Merck
KGaA) for 20 min at room temperature and the cells were observed
under an optical microscope in five random visual fields
(magnification, ×40). The number of cells in the control group was
set to 100% and all other groups were normalized to this group.
Statistical analyses
All data are expressed as mean ± SD values of three
biological replicates. Differences between OS and non-tumor tissues
were analyzed by paired Student's t-test. Differences among
multiple cell transfection groups were analyzed by one-way ANOVA
followed by the Tukey's post-hoc test. Correlations were analyzed
by linear regression. To perform survival analysis, 60 patients
with OS were grouped into high and low ZNF281 groups according to
its median expression level in OS tissues. Survival curves were
plotted and compared by GraphPad Prism v6 software (GraphPad
Software, Inc.) using the log-rank test. P<0.05 was considered
to indicate a statistically significant difference.
Results
ZNF281 is downregulated in tissue
specimens from patients with OS and affected by the clinical
stages
Expression levels of ZNF281 in OS and non-tumor
tissues from the patients were measured by qPCR and compared by the
paired Student's t-test. The resulted revealed significantly higher
expression levels of ZNF281 in OS tissues compared with non-tumor
tissues (Fig. 1A). Expression levels
of ZNF281 in OS tissues were compared among different clinical
stages. Significantly decreased expression levels of ZNF281 were
observed with the increase of clinical stages (Fig. 1B). Of note, expression levels of
ZNF281 significantly decreased with the increase in clinical
stages.
Low levels of ZNF281 mRNA expression
predict the poor survival of patients with OS
Survival curves of the high and low ZNF281
expression groups were plotted. Compared with the low ZNF281
expression group, the 5-year overall survival rate of the high
ZNF281 expression group was significantly higher (Fig. 2).
ZNF281 expression levels are
significantly associated with ROCK1 mRNA and miR-144 expression in
OS tissues
Expression levels of ROCK1 mRNA and miR-144 in OS
tissues were also measured by qPCR (data not shown). Associations
between ZNF281 and ROCK1 mRNA/miR-144 were analyzed by linear
regression. The mRNA expression levels of ZNF281 and miR-144 were
significantly positively associated (Fig. 3A). However, the mRNA expression level
of ZNF281 was significantly inversely associated with that of ROCK1
(Fig. 3B).
ZNF281 upregulates miR-144, which
downregulates ROCK1 in U2OS cells
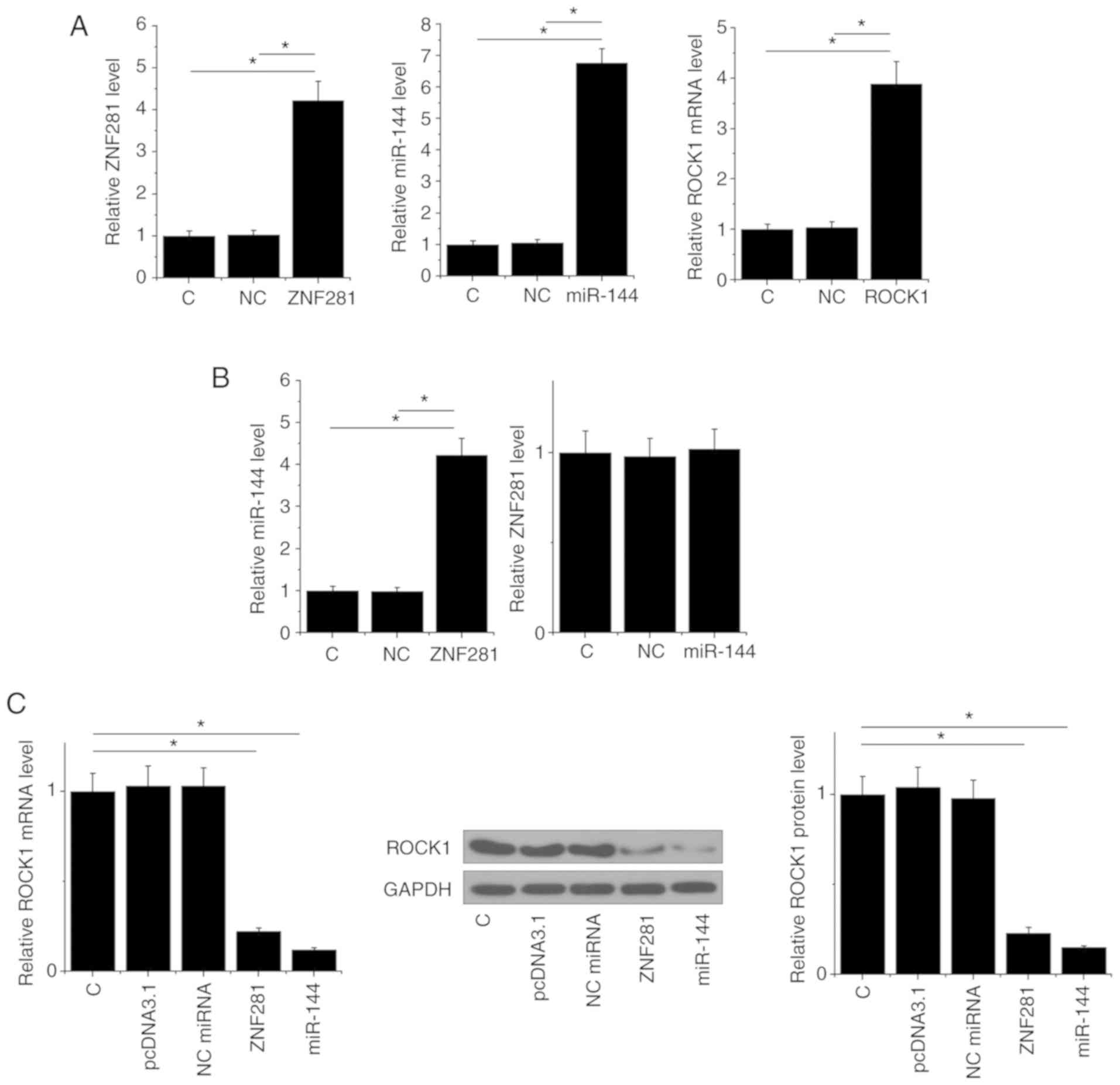
Transfections of ZNF281 and ROCK1 overexpression
vector, as well as a miR-144 mimic, were performed to analyze the
interactions among ZNF281, miR-144 and ROCK1. Compared to C
(untransfected cells) and NC groups, expression levels of ZNF281,
miR-144 and ROCK1 were increased at 24 h post-transfection,
indicating that the transfections were successful (Fig. 4A). Compared with the two control
groups, ZNF281 overexpression mediated the upregulation of miR-144,
whereas miR-144 over-expression failed to affect ZNF281 (Fig. 4B). Moreover, ZNF281 and miR-144
overexpression mediated the downregulation of ROCK1 (Fig. 4C).
ZNF281 suppresses U2OS cell invasion
and migration through miR-144 and ROCK1
Compared with the untransfected and NC groups, cell
invasion and migration analysis demonstrated that ZNF281 and
miR-144 resulted in decreased U2OS cell invasion (Fig. 5A). ROCK1 overexpression resulted in
increased invasion (Fig. 5A) and
migration (Fig. 5B) of OS cells. In
addition, ROCK1 overexpression attenuated the effects of ZNF281
overexpression in U2OS cells (Fig.
5B).
Discussion
The present study, investigated the functions of
ZNF281 in OS. In U2OS cells, ZNF281 overexpression upregulated
miR-144 which downregulated ROCK1, thus inhibiting the invasion and
migration of OS cells.
In a recent study, Li et al (11) identified a novel lncRNA termed ZNF281
in glioma with a tumor-suppressive role in regulating cancer cell
stemness, proliferation and invasion. ZNF281 was downregulated in
glioma, and the multiple functions of ZNF281 were associated with
numerous cancer-related molecular markers, such as tCD133, Nestin,
OCT4, Nanog and the NF-κB1 signaling pathways (11). The involvement of ZNF281 in other
human diseases is unknown. The present study demonstrated that
ZNF281 was downregulated in OS tissue samples from patients and
overexpression of ZNF281 led to the suppressed invasion and
migration of OS cells. Therefore, ZNF281 may be a tumor suppressive
lncRNA in OS.
ROCK1 serves a critical role in the invasion and
migration of OS cells (14,15). Increased rates of OS invasion and
migration were observed after ROCK1 overexpression (14,15). A
number of tumor suppressive miRNAs in OS target ROCK1 to inhibit
cancer progression. For instance, miR-150 directly targets ROCK1 in
OS to inhibit the invasion, proliferation and migration of cancer
cells (16). Wang et al
(10), have demonstrated that
miR-144 suppresses the proliferation and metastasis of OS cells by
targeting ROCK1. The present study demonstrated the downregulation
of ROCK1 after miR-144 overexpression in OS cells, further
confirming the targeting of ROCK1 by miR-144.
Τhe NF-κB signaling pathway serves oncogenic roles
in the majority of types of cancers (17). NF-κB exerts its roles in cancer
biology by interacting with numerous oncogenic or tumor-suppressive
factors, such as miRNAs (18,19). A
recent study has demonstrated that NF-κB may interact with miR-144
to affect cell development (20).
Τhe findings of the present study demonstrated that ZNF281
upregulated miR-144 to downregulate ROCK1. ROCK1 can inactivate
NF-κB to suppress OS (8). Therefore,
NF-κB may mediate the interaction between ZNF281 and miR-144.
Τhe present study had several limitations. Firstly,
the interaction mechanism of ROCK1, ZNF281 and miR-144 was only
investigated in vitro. In vivo studies are required
to verify and elucidate this mechanism. Secondly, only one cell
line was included in the present study. Future studies with other
cell lines are needed to verify the findings of the present study.
Thirdly, the effects of chemotherapy on ZNF281 were not
investigated.
In conclusion, ZNF281 overexpression in a U2OS cell
line may serve a tumor suppressive role by downregulating ROCK1
through the upregulation of miR-144 to suppress cancer cell
invasion and migration.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YS and TY designed the experiments. YS and ZT
performed experiments. JW analyzed data. TY drafted the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Honghui Hospital Affiliated to Xi'an Jiaotong University (Xi'an,
China; approval no. 32556HHXU20110322). All patients signed an
informed consent form.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
International osteosarcoma incidence patterns in children and
adolescents, middle ages and elderly persons. Int J Cancer.
125:229–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gambarotti M, Dei Tos AP, Vanel D, Picci
P, Gibertoni D, Klein MJ and Righi A: Osteoblastoma-like
osteosarcoma: High-grade or low-grade osteosarcoma? Histopathology.
74:494–503. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Misaghi A, Goldin A, Awad M and Kulidjian
AA: Osteosarcoma: A comprehensive review. SICOT J. 4:122018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sutow WW, Sullivan MP, Fernbach DJ, Cangir
A and George SL: Adjuvant chemotherapy in primary treatment of
osteogenic sarcoma. A southwest oncology group study. Cancer.
36:1598–1602. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang Y, Han L, He Z, Li X, Yang S, Yang J,
Zhang Y, Li D, Yang Y and Yang Z: Advances in limb salvage
treatment of osteosarcoma. J Bone Oncol. 10:36–40. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Longhi A, Pasini A, Cicognani A, Baronio
F, Pellacani A, Baldini N and Bacci G: Height as a risk factor for
osteosarcoma. J Pediatr Hematol Oncol. 27:314–318. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Broadhead ML, Clark J, Myers DE, Dass CR
and Choong P: The molecular pathogenesis of osteosarcoma: A review.
Sarcoma. 2011:9592482011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi J, Wu X, Surma M, Vemula S, Zhang L,
Yang Y, Kapur R and Wei L: Distinct roles for ROCK1 and ROCK2 in
the regulation of cell detachment. Cell Death Dis. 4:e4832013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kale VP, Hengst JA, Desai DH, Amin SG and
Yun JK: The regulatory roles of ROCK and MRCK kinases in the
plasticity of cancer cell migration. Cancer Lett. 361:185–196.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang W, Zhou X and Wei M: MicroRNA-144
suppresses osteosarcoma growth and metastasis by targeting ROCK1
and ROCK2. Oncotarget. 6:10297–10308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li XT, Li JC, Feng M, Zhou YX and Du ZW:
Novel lncRNA-ZNF281 regulates cell growth, stemness and invasion of
glioma stem-like U251s cells. Neoplasma. 66:118–127. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tanaka K and Ozaki T: New TNM
classification (AJCC eighth edition) of bone and soft tissue
sarcomas: JCOG bone and soft tissue tumor study group. Jpn J Clin
Oncol. 49:103–107. 2018. View Article : Google Scholar
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang S, Ren T, Jiao G, Huang Y, Bao X,
Zhang F, Liu K, Zheng B, Sun K and Guo W: BMPR2 promotes invasion
and metastasis via the RhoA-ROCK-LIMK2 pathway in human
osteosarcoma cells. Oncotarget. 8:58625–58641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ren T, Zheng B, Huang Y, Wang S, Bao X,
Liu K and Guo W: Osteosarcoma cell intrinsic PD-L2 signals promote
invasion and metastasis via the RhoA-ROCK-LIMK2 and autophagy
pathways. Cell Death Dis. 10:2612019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li CH, Yu TB, Qiu HW, Zhao X, Zhou CL and
Qi C: miR-150 is down-regulated in osteosarcoma and suppresses cell
proliferation, migration and invasion by targeting ROCK1. Oncol
Lett. 13:2191–2197. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karin M, Cao Y, Greten FR and Li ZW:
NF-kappaB in cancer: From innocent bystander to major culprit. Nat
Rev Cancer. 2:301–310. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma X, Becker BLE, Barker JR and Li Y:
MicroRNAs in NF-kappaB signaling. J Mol Cell Biol. 3:159–166. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Boldin MP and Baltimore D: MicroRNAs, new
effectors and regulators of NF-κB. Immunol Rev. 246:205–220. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yan X, Cui J, Liu X and Xu T: microRNA-144
regulates the NF-κB signaling in miiuy croaker via targeting IL1β.
Dev Comp Immunol. 96:47–50. 2019. View Article : Google Scholar : PubMed/NCBI
|