Introduction
Gastrointestinal cancer is one of the major
contributors to global cancer prevalence and cancer-associated
mortality (1,2). In addition, gastric and colorectal
cancers are the most common gastrointestinal cancers in China
(1,2). With respect to gastric cancer, early
disease could be eradicated by surgical resection, while the
majority of patients with gastric cancer do not recognize any of
the clinical symptoms until the disease has progressed to an
advanced stage (3–5). This situation largely contributes to
the poor prognosis of patients with gastric cancer. It has been
reported that the 5-year survival rate is ~60% in patients with
non-advanced cancer, while it declines to ~30% in patients with
advanced stage cancer (6). In
terms of the colorectal cancer, its incidence is raised throughout
the past three decades, especially in the young population
(7). Furthermore, it is still
important to investigate biomarkers or a model that can boost the
efficiency of prognosis in colorectal cancer management (8). Therefore, identification of
biomarkers that could assist with identifying early disease,
guiding treatment and optimizing disease prognosis is essential in
these two types of cancer.
Kinesin family member 14 (KIF14), a member of the
kinesin-3 family, is a regulator of cell mitosis, which is similar
to other kinesins (9). KIF14 is
involved in numerous essential biological activities in organisms,
such as the regulation of cytokinesis and myelination (10–12).
Notably, an increasing number of studies have examined KIF14 and
its mechanistic role and potential clinical value in several types
of cancer, including cervical, lung and prostate cancer (13–15).
In terms of the role of KIF14 expression in gastrointestinal
cancer, only a few studies have reported this issue. For instance,
Previous studies indicated that KIF14 is positively associated with
aggravating disease features and its high expression is associated
with worse survival in patients with gastric cancer and pancreatic
adenocarcinoma (16,17). Additionally, upregulation of KIF14
has also been found to be associated with the elevated tumor stage
and pathological tumor grade of patients with hepatocellular
carcinoma (HCC) (18). However,
the role of KIF14 in gastrointestinal cancer requires further
in-depth investigation.
Therefore, the present study aimed to investigate
the association between KIF14 expression and disease-free survival
(DFS) and overall survival (OS) times in patients with
gastrointestinal cancer.
Materials and methods
Patients
The present study retrospectively reviewed 101
patients with gastrointestinal cancer [36 patients with gastric
cancer consisting of 16 (44.4%) males and 20 (55.6%) females and 65
patients with colorectal cancer consisting of 40 (61.5%) males and
25 (38.5%) females] who were treated at the General Hospital of
Ningxia Medical University (Ningxia, China) and the First People's
Hospital Affiliated to Shanghai Jiaotong University (Shanghai,
China) between January 2008 and December 2010. The screening
criteria for all patients included: i) Diagnosis of
gastrointestinal cancer by surgery and postoperative pathology; ii)
had complete clinical characteristics and follow-up information;
and iii) had available cancer specimens to perform
immunohistochemistry (IHC) and reverse transcription-quantitative
PCR (RT-qPCR). Patients were excluded if, according to the medical
records, they met the following criteria: i) Had a history of other
cancers or malignancies at diagnosis; and ii) received preoperative
chemotherapy or radiotherapy. The study was approved by the
Internal Review Boards of General Hospital of Ningxia Medical
University (Ningxia, China) and the First People's Hospital
Affiliated to Shanghai Jiaotong University (approval no. 2011-268;
Shanghai, China). All patients or their families provided written
informed consent.
Data collection
By reviewing the medical documents, the clinical
characteristics, including age, sex, current smoking and drinking
status, tumor location, tumor size, pathological grade, T stage,
lymph node metastasis (LNM), distant metastases and TNM stage, were
collected (19,20). Follow-up was conducted every 6
months via either clinic visits, telephone calls or letters, and
survival data were collected to calculate DFS and OS times.
IHC analysis
Tumor tissue was available in all patients, while
adjacent tissue was also available in 49 patients, including 19
patients with gastric cancer and 30 patients with colorectal
cancer. IHC analysis was used to evaluate KIF14 protein expression
in the cancer and adjacent specimens. The methods used were the
same as previously described (21). The slices of cancer and adjacent
specimens were incubated with KIF14 polyclonal antibody (1:200
dilution; cat. no. BL358; Bethyl Laboratories, Inc.) at 4°C
overnight, then the slices were incubated with biotinylated goat
anti-rabbit Envision + System-HRP (1:500 dilution; cat. no. E432;
Dako; Agilent Technologies, Inc.) at 37°C for 15 min.
Diaminobenzidine and hematoxylin were used for staining and
counterstaining, respectively. PBS instead of the primary antibody
was used as the negative control. The results of IHC staining were
evaluated with a light microscope and were independently graded by
two pathologists who were blinded to the clinical and pathological
information of the patients. The staining intensity score and
density score were used to assess the KIF14 protein expression in
specimens using a semi-quantitative scoring method as described
previously (22). Briefly, the
intensity score of IHC staining ranged between 0 and 3, and the
range of the density score was 0–4. The total IHC score was 12,
which was calculated by multiplying the intensity score and the
density score, and the final IHC score was the average value of the
scores of the two pathologists.
RT-qPCR
The mRNA expression levels of KIF14 were determined
using RT-qPCR. Total RNA was extracted from fresh tumor tissue with
TRIzol® LS (cat. no. A33253; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Then, RNA was
reverse transcribed using an ABI P RISM 7900HT thermocycler (cat.
no. 4317596; Thermo Fisher Scientific, Inc.) with PrimeScript RT
Master Mix (Perfect Real Time) (cat. no. RR036A; Takara Bio, Inc.)
according to the manufacturer's instructions. qPCR was performed
with cDNA using iTaq Universal SYBR® Green Supermix
(cat. no. 1725120; Bio-Rad Laboratories, Inc.). The following PCR
primers were used: β-actin (control) forward,
5′-GTGGGGCGCCCCAGGCACCA-3′ and reverse,
5′-CTCCTTAATGTCACGCACGATTTC-3′ (540-bp fragment); and KIF14
forward, 5′-TGGAACACCCTGACACGA-3′ and reverse,
5′-GAGCCAGTCTGACCATAAGC-3′. Quantification of gene expression was
performed using a Real Plex RT-PCR System (Eppendorf) and the
following thermocycling conditions were used: Initial denaturation
at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C
for 20 sec, annealing at 54°C for 15 sec and elongation at 72°C for
20 sec, with a final extension at 72°C for 10 min. Relative KIF14
expression was calculated using the 2−ΔΔCq method
(23).
Definitions
A final IHC score ≤3 was classified as low KIF14
protein expression, while an IHC score >3 was classified as high
KIF14 protein expression. The exact value of 1.000 was included in
the colorectal cancer patients' group. The median relative mRNA
expression levels of KIF14 were used to classify the samples into
high (>1.000) and low (<1.000) mRNA expression groups.
Statistical analysis
SPSS v21.0 software (IBM Corp.) and GraphPad Prism
v7.02 (GraphPad Software, Inc.) were used for statistical analysis
and generation of the graphs. Normally distributed quantitative
data are presented as the mean ± SD. Non-normally distributed
quantitative data are presented as the median (25–75th quantiles).
Qualitative data are presented as the count (percentage).
Associations between clinical characteristics and KIF14 expression
were evaluated using a χ2 test or the Fisher's exact
test. Kaplan-Meier curves and the log-rank test were used to
analyze the survival data and KIF14 expression. Prognostic factors
were determined using a Cox's proportional hazard regression model.
Significant variables (P<0.05) in the univariate analysis were
included in multivariate Cox's regression analysis. P<0.05 was
considered to indicate a statistically significant difference. Each
experiment was repeated in three duplicates.
Results
Characteristics of patients with
gastrointestinal cancer
The mean age of the 101 enrolled patients with
gastrointestinal cancer was 60.3±6.6 years. There were 45 (44.1%)
female patients and 56 (54.9%) male patients. There were 65 (63.7%)
and 36 (35.3%) patients who had colorectal or gastric cancer,
respectively. In addition, the numbers of patients with a tumor
size ≤5 or >5 cm were 57 (55.9%) and 44 (43.1%), respectively.
The numbers of patients with pathological grade 1, 2 and 3 were 24
(23.5%), 45 (44.1%) and 32 (31.4%), respectively. Furthermore,
there were 55 (53.9%) patients with LNM and 46 (45.1%) patients
without LNM. The number of patients who had no distant metastases
was 90 (88.2%) and the number of patients who had distant
metastasis was 11 (10.8%). Other clinical characteristics
including: current smoking and drinking status, T stage and TNM
stage are shown in Table I.
 | Table I.Association of clinical
characteristics with KIF14 expression. |
Table I.
Association of clinical
characteristics with KIF14 expression.
|
|
| KIF14 protein
expression |
|
| KIF14 mRNA
expression |
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Variables | All patients, n
(%) | Low, n (%) | High, n (%) | χ2 | P-value | Low, n (%) | High, n (%) | χ2 | P-value |
|---|
| Age |
|
|
| 0.590 | 0.442 |
|
| 0.524 | 0.469 |
| ≤60
years | 42 (41.2) | 16 (38.1) | 26 (61.9) |
|
| 19 (45.2) | 23 (54.8) |
|
|
| >60
years | 59 (57.8) | 27 (45.8) | 32 (54.2) |
|
| 31 (52.5) | 28 (47.5) |
|
|
| Sex |
|
|
| 0.556 | 0.456 |
|
| 2.222 | 0.136 |
|
Female | 45 (44.1) | 21 (46.7) | 24 (53.3) |
|
| 26 (57.8) | 19 (42.2) |
|
|
|
Male | 56 (54.9) | 22 (39.3) | 34 (60.7) |
|
| 24 (42.9) | 32 (57.1) |
|
|
| Current smoking
status |
|
|
| 0.171 | 0.679 |
|
| 0.685 | 0.408 |
| No | 73 (71.6) | 32 (43.8) | 41 (56.2) |
|
| 38 (52.1) | 35 (47.9) |
|
|
|
Yes | 28 (27.5) | 11 (39.3) | 17 (60.7) |
|
| 12 (42.9) | 16 (57.1) |
|
|
| Current drinking
status |
|
|
| 0.027 | 0.870 |
|
| 0.479 | 0.489 |
| No | 62 (60.8) | 26 (41.9) | 36 (58.1) |
|
| 29 (46.8) | 33 (53.2) |
|
|
|
Yes | 39 (38.2) | 17 (43.6) | 22 (56.4) |
|
| 21 (53.8) | 18 (46.2) |
|
|
| Tumor location |
|
|
| 0.080 | 0.777 |
|
| 0.819 | 0.365 |
|
Colorectal | 65 (63.7) | 27 (41.5) | 38 (58.5) |
|
| 30 (46.2) | 35 (53.8) |
|
|
|
Gastric | 36 (35.3) | 16 (44.4) | 20 (55.6) |
|
| 20 (55.6) | 16 (44.4) |
|
|
| Tumor size |
|
|
| 0.088 | 0.766 |
|
| 0.008 | 0.930 |
| ≤5
cm | 57 (55.9) | 25 (43.9) | 32 (56.1) |
|
| 28 (49.1) | 29 (50.9) |
|
|
| >5
cm | 44 (43.1) | 18 (40.9) | 26 (59.1) |
|
| 22 (50.0) | 22 (50.0) |
|
|
| Pathological
grade |
|
|
| 4.359 | 0.111 |
|
| 4.617 | 0.105 |
| Grade
1 | 24 (23.5) | 13 (54.2) | 11 (45.8) |
|
| 16 (66.7) | 8 (33.3) |
|
|
| Grade
2 | 45 (44.1) | 21 (46.7) | 24 (53.3) |
|
| 22 (48.9) | 23 (51.1) |
|
|
| Grade
3 | 32 (31.4) | 9 (28.1) | 23 (71.9) |
|
| 12 (37.5) | 20 (62.5) |
|
|
| Distant
metastases |
|
|
| 7.302 | 0.007 |
|
| 12.102 | 0.001 |
| No | 90 (88.2) | 43 (47.8) | 47 (52.2) |
|
| 50 (55.6) | 40 (44.4) |
|
|
|
Yes | 11 (10.8) | 0 (0.0) | 11 (100.0) |
|
| 0 (0.0) | 11 (100.0) |
|
|
| T stage |
|
|
| 22.091 | <0.001 |
|
| 25.301 | <0.001 |
| T1 | 15 (14.7) | 14 (93.3) | 1 (6.7) |
|
| 15 (100.0) | 0 (0.0) |
|
|
| T2 | 23 (22.5) | 10 (43.5) | 13 (56.5) |
|
| 13 (56.5) | 10 (43.5) |
|
|
| T3 | 42 (41.2) | 15 (35.7) | 27 (64.3) |
|
| 17 (40.5) | 25 (59.5) |
|
|
| T4 | 21 (20.6) | 4 (19.0) | 17 (81.0) |
|
| 5 (23.8) | 16 (76.2) |
|
|
| LNM |
|
|
| 3.184 | 0.076 |
|
| 8.343 | 0.004 |
| No | 46 (45.1) | 24 (52.2) | 22 (47.8) |
|
| 30 (65.2) | 16 (34.8) |
|
|
|
Yes | 55 (53.9) | 19 (34.5) | 36 (65.5) |
|
| 20 (36.4) | 35 (63.6) |
|
|
| TNM stage |
|
|
| 21.487 | <0.001 |
|
| 44.662 | <0.001 |
| I | 16 (15.7) | 13 (81.2) | 3 (18.8) |
|
| 16 (100.0) | 0 (0.0) |
|
|
| II | 34 (33.3) | 19 (55.9) | 15 (44.1) |
|
| 24 (70.6) | 10 (29.4) |
|
|
|
III | 35 (34.3) | 8 (22.9) | 27 (77.1) |
|
| 8 (22.9) | 27 (77.1) |
|
|
| IV | 16 (15.7) | 3 (18.8) | 13 (81.2) |
|
| 2 (12.5) | 14 (87.5) |
|
|
KIF14 expression
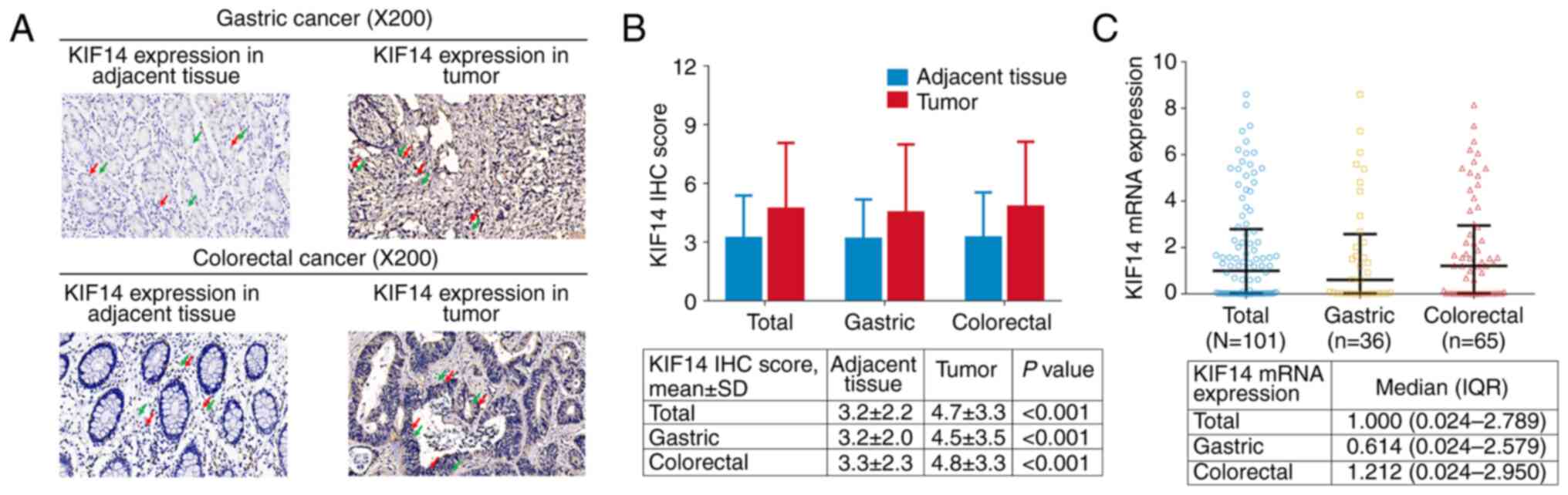
The IHC staining of KIF14 in gastric and colorectal
cancer tissues and the adjacent tissue is shown in Fig. 1A. KIF14 protein expression was
mainly located in the cell nucleus and cytoplasm in both gastric
cancer (upper panel) and colorectal cancer (lower panel) tissues.
In addition, the KIF14 IHC score was elevated in tumor tissue of
all patients (4.7±3.3 vs. 3.2±2.2; P<0.001), patients with
gastric cancer (4.5±3.5 vs. 3.2±2.0; P<0.001) and patients with
colorectal cancer (4.8±3.3 vs. 3.3±2.3; P<0.001) compared with
adjacent tissue (Fig. 1B). The
median KIF14 mRNA expression in all patients, patients with gastric
cancer and patients with colorectal cancer was 1.000 (0.024-2.789),
0.614 (0.024-2.579) and 1.212 (0.024-2.950), respectively (Fig. 1C).
Association of KIF14 expression with
clinicopathological characteristics
In all patients with gastrointestinal cancer, KIF14
protein expression was associated with T stage (P<0.001),
distant metastases (P=0.007) and TNM stage (P<0.001) (Table I). KIF14 protein expression was not
associated with age, sex, current smoking and drinking status,
tumor location, tumor size, pathological grade, or LNM (all
P>0.05). KIF14 mRNA expression was associated with T stage
(P<0.001), LNM (P=0.004), distant metastases (P=0.001) and TNM
stage (P<0.001) (Table I).
KIF14 mRNA expression was not associated with age, sex, current
smoke, current drink, tumor location, tumor size, or pathological
grade (All P>0.05).
Association between KIF14 expression
and survival time
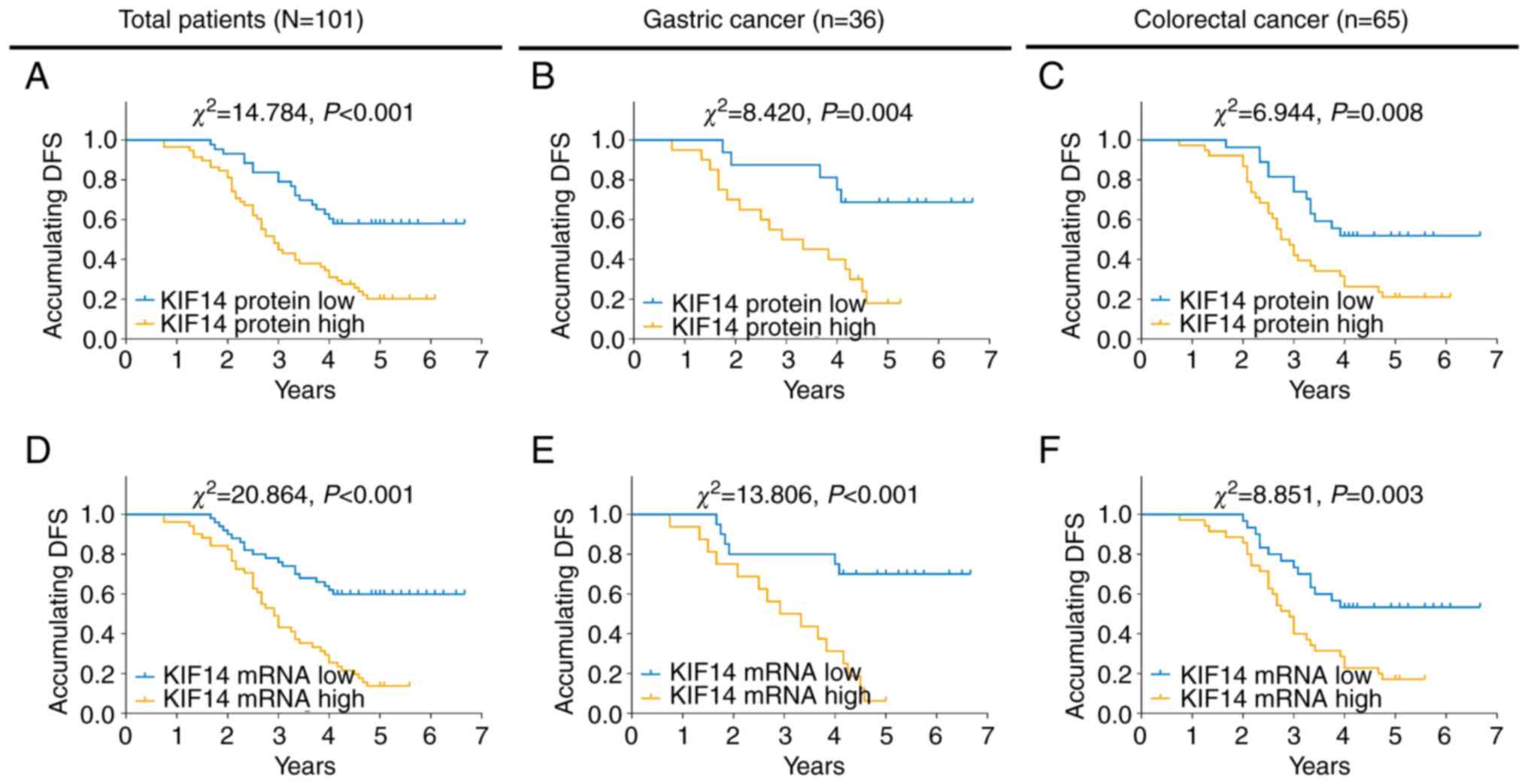
High KIF14 protein expression was associated with
worse DFS time in all patients with gastrointestinal cancer
(P<0.001; Fig. 2A), patients
with gastric cancer (P=0.004; Fig.
2B) and patients with colorectal cancer (P=0.008; Fig. 2C). In addition, high KIF14 mRNA
expression was also associated with DFS time in the aforementioned
patients (all P<0.01; Fig.
2D-F).
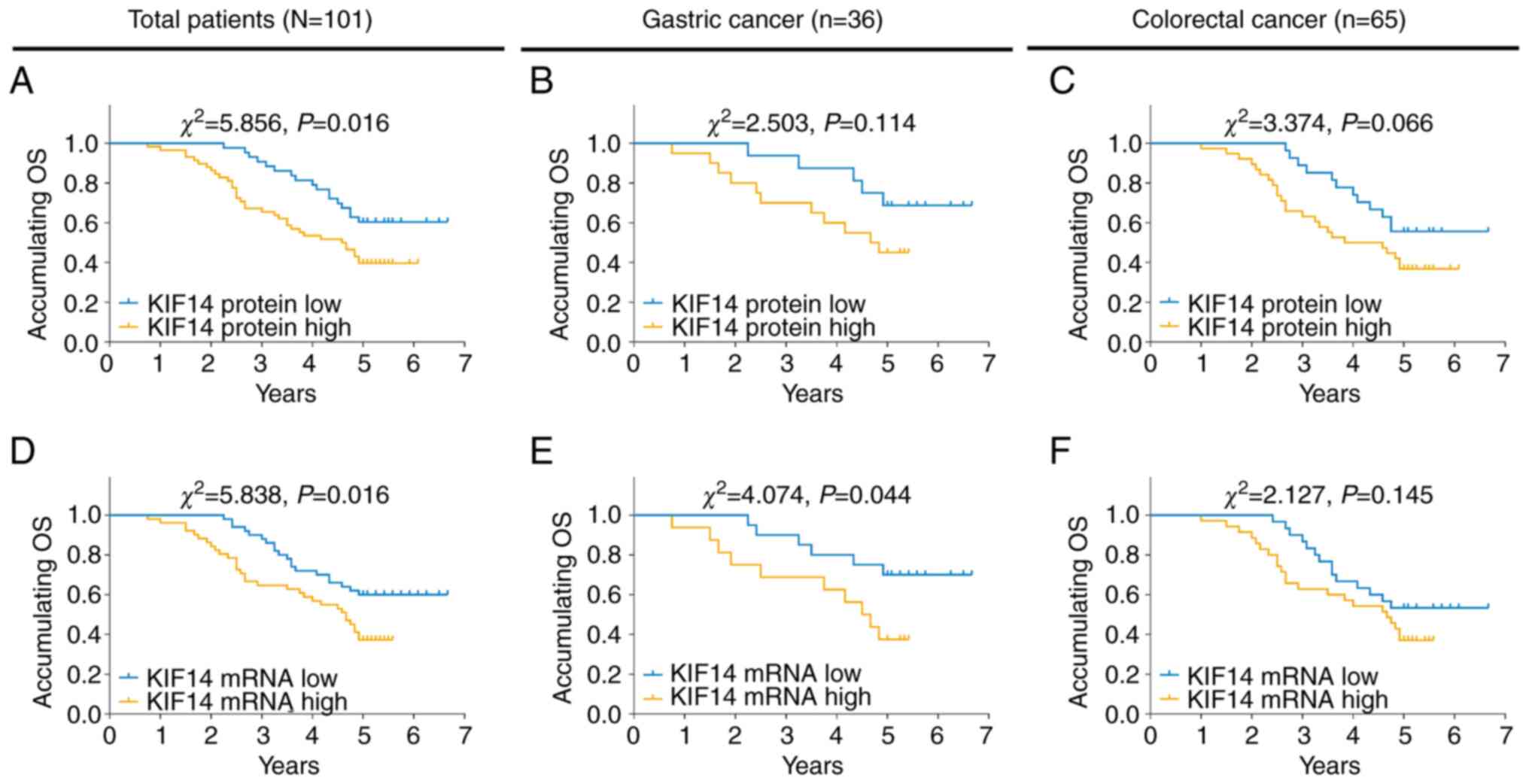
High KIF14 protein expression was associated with
worse OS time in all patients with gastrointestinal cancer
(P=0.016; Fig. 3A) but not in
patients with gastric cancer (P=0.114; Fig. 3B) or patients with colorectal
cancer (P=0.066; Fig. 3C). In
addition, high KIF14 mRNA expression was associated with worse OS
time in all patients with gastrointestinal cancer (P=0.016;
Fig. 3D) and patients with gastric
cancer (P=0.044; Fig. 3E) but not
in patients with colorectal cancer (P=0.145; Fig. 3F).
Independent prognostic factors
Multivariate Cox's proportional hazards regression
analysis revealed that in all patients with gastrointestinal
cancer, high KIF14 protein expression (P=0.007) was an independent
prognostic factor for unfavorable DFS time, along with TNM stage
(III/IV vs. I/II; P=0.010; Table
II). TNM stage (III/IV vs. I/II; P=0.003) could independently
predict shorter OS time in all patients with gastrointestinal
cancer (Table III).
 | Table II.Factors affecting disease-free
survival according to Cox's proportional hazards regression
analysis. |
Table II.
Factors affecting disease-free
survival according to Cox's proportional hazards regression
analysis.
|
|
|
| 95% CI |
|---|
|
|
|
|
|
|---|
| Variables | P-value | HR | Lower | Upper |
|---|
| Univariate Cox's
regression analysis |
|
|
|
|
| KIF14 protein (high
vs. low) |
<0.001a | 2.770 | 1.602 | 4.789 |
| Age (>60 years
vs. ≤60 years) | 0.122 | 0.678 | 0.414 | 1.109 |
| Sex (male vs.
female) | 0.904 | 1.031 | 0.628 | 1.694 |
| Current smoking
status (yes vs. no) | 0.381 | 1.268 | 0.746 | 2.154 |
| Current drinking
status (yes vs. no) | 0.753 | 0.923 | 0.558 | 1.525 |
| Tumor location
(Gastric vs. Colorectal) | 0.415 | 0.804 | 0.476 | 1.358 |
| Tumor size (>5
cm vs. ≤5 cm) | 0.732 | 0.917 | 0.558 | 1.506 |
| Pathological grade
(Grade 3 vs. Grade 1/Grade 2) | 0.277 | 0.742 | 0.434 | 1.270 |
| Distant metastases
(yes vs. no) |
<0.001a | 6.696 | 3.190 | 14.054 |
| T stage (T3/T4 vs.
T1/T2) | 0.035a | 1.801 | 1.042 | 3.111 |
| LNM (yes vs.
no) | 0.679 | 0.902 | 0.552 | 1.473 |
| TNM stage (III/IV
vs. I/II) |
<0.001a | 1.702 | 1.274 | 2.275 |
| Multivariate Cox's
regression analysis |
|
|
|
|
| KIF14
protein (high vs. low) | 0.007a | 2.189 | 1.240 | 3.864 |
| TNM
stage (III/IV vs. I/II) | 0.010a | 1.519 | 1.104 | 2.091 |
 | Table III.Factors affecting overall survival
according to Cox's proportional hazards regression analysis. |
Table III.
Factors affecting overall survival
according to Cox's proportional hazards regression analysis.
|
|
|
| 95% CI |
|---|
|
|
|
|
|
|---|
| Variables | P-value | HR | Lower | Upper |
|---|
| Univariate Cox's
regression analysis |
|
|
|
|
| KIF14 protein (high
vs. low) | 0.018a | 2.010 | 1.125 | 3.591 |
| Age (>60 years
vs. ≤60 years) | 0.623 | 0.871 | 0.504 | 1.507 |
| Sex (male vs.
female) | 0.699 | 0.898 | 0.521 | 1.549 |
| Smoke (yes vs.
no) | 0.305 | 1.355 | 0.758 | 2.420 |
| Drink (yes vs.
no) | 0.403 | 0.784 | 0.443 | 1.388 |
| Tumor location
(Gastric vs. Colorectal) | 0.332 | 0.747 | 0.414 | 1.347 |
| Tumor size (>5
cm vs. ≤5 cm) | 0.638 | 0.876 | 0.505 | 1.519 |
| Pathological grade
(Grade 3 vs. Grade 1/Grade 2) | 0.110 | 0.599 | 0.320 | 1.122 |
| Distant metastases
(yes vs. no) |
<0.001a | 19.843 | 8.014 | 49.136 |
| T stage (T3/T4 vs.
T1/T2) | 0.250 | 1.406 | 0.787 | 2.510 |
| LNM (yes vs.
no) | 0.413 | 0.797 | 0.462 | 1.373 |
| TNM stage (III/IV
vs. I/II) | 0.001a | 1.805 | 1.289 | 2.528 |
| Multivariate Cox's
regression analysis |
|
|
|
|
| TNM stage (III/IV
vs. I/II) | 0.003a | 1.711 | 1.195 | 2.451 |
Discussion
As a crucial member of the kinesin family, KIF14 has
been reported to be a regulator of tumor development and
progression in previous studies and could also be a potential
biomarker in the management of patients with cancer (24,25).
A prior study revealed that KIF14 inhibition using small
interfering (si)RNA could repress proliferation and enhance
apoptosis in medulloblastoma cells, while KIF14 inhibition using
short hairpin RNA could suppress cell viability, colony formation,
invasion, migration and tumor sphere formation in medulloblastoma
cells (18). Furthermore, KIF14
expression is upregulated in patients with medulloblastoma and is
associated with unfavorable progression-free survival and poor OS
time (26). Another study
identified KIF14 as a tumor enhancer in HCC, since knockdown of
KIF14 expression reduced the acquired chemoresistance to sorafenib
by decreasing AKT activation and ETS proto-oncogene 1,
transcription factor expression in the HCC cells, and siRNA
targeting KIF14 reduced tumor growth of sorafenib-resistant HCC
cells in a mice model in vivo (27). Furthermore, in ovarian cancer
cells, upregulated KIF14 expression enhanced proliferation and
colony formation; however, downregulated KIF14 expression promoted
apoptosis and decreased colony formation (28). In addition, KIF14 was upregulated
in tumor tissues and could predict unfavorable outcomes in patients
with ovarian cancer (29). These
aforementioned studies indicated that in numerous types of cancer,
KIF14 might be a promoter of cancer progression, and could be a
potential biomarker for prognosis (21,24–29).
With respect to the role of KIF14 in
gastrointestinal cancer, research is limited. A previous study
reported that KIF14 enhanced gastric cancer cell proliferation,
migration and invasion by positively regulating the AKT signaling
pathway (30). In addition, KIF14
expression was positively associated with tumor stage, TNM stage
and tumor metastasis, and its high expression levels could
independently predict poor survival in patients with gastric cancer
(16). In colorectal cancer, a
previous study found that KIF14 enhanced colorectal cancer cell
proliferation via the activation of AKT signaling, and was also
targeted by miR-200c (30). In
addition, in pancreatic adenocarcinoma, KIF14 was highly expressed
in tumor tissues compared with normal tissues, and this coincided
with unfavorable prognosis (17).
Furthermore, elevated KIF14 expression has also been observed in
tumor tissues of patients with esophageal squamous cell carcinoma
compared with in normal tissues (31). Additionally, upregulation of KIF14
has been found to be associated with the tumor stage and
pathological tumor grade of patients with HCC (18). Notably, to the best of our
knowledge, the role of KIF14 in patients with gastrointestinal
cancer has not been investigated; therefore, the present study
investigated the role of KIF14 in patients with this type of
cancer. It was found that KIF14 expression was associated with
advanced pathological differentiation and tumor stages, and also
predicted worse survival time. It was hypothesized that these
results could be attributed to the following reasons: i) As a
carcinogenesis gene, KIF14 reflected the proliferation speed of
cells, while the proliferation speed of tumor cells was higher than
that of the normal cells, thus, the KIF14 expression was elevated
in tumor tissues compared with adjacent tissues (17,18,31);
ii) KIF14 can promote malignant behavior in cancer cells, such as
enhancement of cancer cell proliferation, by interacting with
related factors (30). This might
explain the association between KIF14 expression and
clinicopathological characteristics in patients with
gastrointestinal cancer and the association with unfavorable
survival times (16,26,27,29,30,32);
and iii) in addition, KIF14 expression was associated with DFS
time, but not OS time, suggesting that OS time could be affected by
additional factors, such as treatment after disease relapse. Thus,
this finding might imply that after determining the KIF14
expression in tumor tissues of patients with gastrointestinal
cancer, the treatment strategy might be adjusted accordingly. For
example, if a patient with gastric cancer exhibits high KIF14
expression, then a more intensive treatment regimen should be
considered for their subsequent treatment.
The present study had several limitations. First,
only 101 patients with gastrointestinal were recruited, which made
that sample size was too small to investigate the clinical value of
KIF14. Second, there may be some confounding factors affecting the
effect of KIF14; therefore, multivariate Cox's proportional hazard
analysis was performed to decrease this effect. Third, the patients
enrolled in the present study were from Northwest China, which
might cause patient selection bias.
In conclusion, KIF14 expression was associated with
advanced pathological differentiation and TNM stage, and also
predicted worse survival time in patients with gastrointestinal
cancer. This indicates its potential as a biomarker for
gastrointestinal cancer prognosis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Ningxia Natural Science
Foundation (grant nos. NZ13166 and 2018AAC03146) and the Key
Research Projects of Ningxia Health and Family Planning Commission
(grant no. 2018-NW-020).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
PC and WF contributed to the study design. PC, WF,
YH, FW and NL reviewed the medical documents and collected the
clinical characteristics of patients. PC, WF, YH, FW and NL
contributed to the data acquisition and analysis. PC, WF, YH, FW
and NL wrote the manuscript, and PC and WF critically revised the
manuscript for important intellectual content. PC agrees to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of the work are appropriately
investigated and resolved. PC and WF confirm the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Internal Review Boards
of General Hospital of Ningxia Medical University (Ningxia, China)
and the First People's Hospital Affiliated to Shanghai Jiaotong
University (Shanghai, China). All patients or their families
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
KIF14
|
kinesin family member 14
|
|
DFS
|
disease-free survival
|
|
OS
|
overall survival
|
|
IHC
|
immunohistochemistry
|
|
RT-qPCR
|
reverse transcription quantitative
PCR
|
|
LNM
|
lymph node metastasis
|
|
HCC
|
hepatocellular carcinoma
|
References
|
1
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Ervik M, Lam F, Colombet M, Mery
L, Piñeros M, Znaor A, Soerjomataram I and Bray F: Global Cancer
Observatory: Cancer Today. International Agency for Research on
Cancer; Lyon: 2020, https://gco.iarc.fr/todayDecember 1–2018
|
|
3
|
Wagner AD, Syn NL, Moehler M, Grothe W,
Yong WP, Tai BC, Ho J and Unverzagt S: Chemotherapy for advanced
gastric cancer. Cochrane Database Syst Rev.
8:CD0040642017.PubMed/NCBI
|
|
4
|
Patel TH and Cecchini M: Targeted
therapies in advanced gastric cancer. Curr Treat Options Oncol.
21:702020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bonelli P, Borrelli A, Tuccillo FM,
Silvestro L, Palaia R and Buonaguro FM: Precision medicine in
gastric cancer. World J Gastrointest Oncol. 11:804–829. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lyons K, Le LC, Pham YT, Borron C, Park
JY, Tran CTD, Tran TV, Tran HT, Vu KT, Do CD, et al: Gastric
cancer: Epidemiology, biology, and prevention: A mini review. Eur J
Cancer Prev. 28:397–412. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zahir Ahmed S, Cirocchi N, Saxton E and
Brown MK: Incidence of age migration of colorectal cancer in
younger population: Retrospective single centred-population based
cohort study. Ann Med Surg (Lond). 74:1032142021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carleton M, Mao M, Biery M, Warrener P,
Kim S, Buser C, Marshall CG, Fernandes C, Annis J and Linsley PS:
RNA interference-mediated silencing of mitotic kinesin KIF14
disrupts cell cycle progression and induces cytokinesis failure.
Mol Cell Biol. 26:3853–3863. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gruneberg U, Neef R, Li X, Chan EH,
Chalamalasetty RB, Nigg EA and Barr FA: KIF14 and citron kinase act
together to promote efficient cytokinesis. J Cell Biol.
172:363–372. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cullati SN, Kabeche L, Kettenbach AN and
Gerber SA: A bifurcated signaling cascade of NIMA-related kinases
controls distinct kinesins in anaphase. J Cell Biol. 216:2339–2354.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ahmed SM, Thériault BL, Uppalapati M, Chiu
CW, Gallie BL, Sidhu SS and Angers S: KIF14 negatively regulates
Rap1a-Radil signaling during breast cancer progression. J Cell
Biol. 199:951–967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang W, Shi Y, Li J, Cui W and Yang B:
Up-regulation of KIF14 is a predictor of poor survival and a novel
prognostic biomarker of chemoresistance to paclitaxel treatment in
cervical cancer. Biosci Rep. 36:e003152016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Corson TW, Zhu CQ, Lau SK, Shepherd FA,
Tsao MS and Gallie BL: KIF14 messenger RNA expression is
independently prognostic for outcome in lung cancer. Clin Cancer
Res. 13:3229–3234. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Yuan Y, Liang P, Zhang Z, Guo X,
Xia L, Zhao Y, Shu XS, Sun S, Ying Y and Cheng Y: Overexpression of
a novel candidate oncogene KIF14 correlates with tumor progression
and poor prognosis in prostate cancer. Oncotarget. 8:45459–45469.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang Z, Li C, Yan C, Li J, Yan M, Liu B,
Zhu Z, Wu Y and Gu Q: KIF14 promotes tumor progression and
metastasis and is an independent predictor of poor prognosis in
human gastric cancer. Biochim Biophys Acta Mol Basis Dis.
1865:181–192. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Klimaszewska-Wiśniewska A, Neska-Długosz
I, Buchholz K, Durślewicz J, Grzanka D, Kasperska A, Antosik P,
Zabrzyński J, Grzanka A and Gagat M: Prognostic significance of
KIF11 and KIF14 expression in pancreatic adenocarcinoma. Cancers
(Basel). 13:30172021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li X, Huang W, Huang W, Wei T, Zhu W, Chen
G and Zhang J: Kinesin family members KIF2C/4A/10/11/14/18B/20A/23
predict poor prognosis and promote cell proliferation in
hepatocellular carcinoma. Am J Transl Res. 12:1614–1639.
2020.PubMed/NCBI
|
|
19
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Niederle MB, Hackl M, Kaserer K and
Niederle B: Gastroenteropancreatic neuroendocrine tumours: the
current incidence and staging based on the WHO and European
neuroendocrine tumour society classification: An analysis based on
prospectively collected parameters. Endocr Relat Cancer.
17:909–918. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang T, Shi W, Tian K and Kong Y:
Chaperonin containing t-complex polypeptide 1 subunit 6A correlates
with lymph node metastasis, abnormal carcinoembryonic antigen and
poor survival profiles in non-small cell lung carcinoma. World J
Surg Oncol. 18:1562020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu H, Jin C, Zhu Q, Liu T, Ke B, Li A and
Zhang T: Dysregulated expressions of PTEN, NF-κB, WWP2, p53 and
c-Myc in different subtypes of B cell lymphoma and reactive
follicular hyperplasia. Am J Transl Res. 11:1092–1101.
2019.PubMed/NCBI
|
|
23
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Neska-Długosz I, Buchholz K, Durślewicz J,
Gagat M, Grzanka D, Tojek K and Klimaszewska-Wiśniewska A:
Prognostic impact and functional annotations of KIF11 and KIF14
expression in patients with colorectal cancer. Int J Mol Sci.
22:97322021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao L and Zhang S, Zheng Q and Zhang S:
Dysregulation of KIF14 regulates the cell cycle and predicts poor
prognosis in cervical cancer: A study based on integrated
approaches. Braz J Med Biol Res. 54:e113632021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li KK, Qi Y, Xia T, Chan AK, Zhang ZY,
Aibaidula A, Zhang R, Zhou L, Yao Y and Ng HK: The kinesin KIF14 is
overexpressed in medulloblastoma and downregulation of KIF14
suppressed tumor proliferation and induced apoptosis. Lab Invest.
97:946–961. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu Q, Ren H, Li X, Qian B, Fan S, Hu F,
Xu L and Zhai B: Silencing KIF14 reverses acquired resistance to
sorafenib in hepatocellular carcinoma. Aging (Albany NY).
12:22975–23003. 2020.PubMed/NCBI
|
|
28
|
Thériault BL, Cybulska P, Shaw PA, Gallie
BL and Bernardini MQ: The role of KIF14 in patient-derived primary
cultures of high-grade serous ovarian cancer cells. J Ovarian Res.
7:1232014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thériault BL, Pajovic S, Bernardini MQ,
Shaw PA and Gallie BL: Kinesin family member 14: An independent
prognostic marker and potential therapeutic target for ovarian
cancer. Int J Cancer. 130:1844–1854. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang ZZ, Yang J, Jiang BH, Di JB, Gao P,
Peng L and Su XQ: KIF14 promotes cell proliferation via activation
of Akt and is directly targeted by miR-200c in colorectal cancer.
Int J Oncol. 53:1939–1952. 2018.PubMed/NCBI
|
|
31
|
Han BA, Yang XP, Hosseini DK, Zhang P,
Zhang Y, Yu JT, Chen S, Zhang F, Zhou T and Sun HY: Identification
of candidate aberrantly methylated and differentially expressed
genes in esophageal squamous cell carcinoma. Sci Rep. 10:97352020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen J, Ma C, Zhang Y, Pei S, Du M, Zhang
Y, Qian L, Wang J, Yin L and He X: MiR-154-5p suppresses cell
invasion and migration through inhibiting KIF14 in nasopharyngeal
carcinoma. Onco Targets Ther. 13:2235–2246. 2020. View Article : Google Scholar : PubMed/NCBI
|