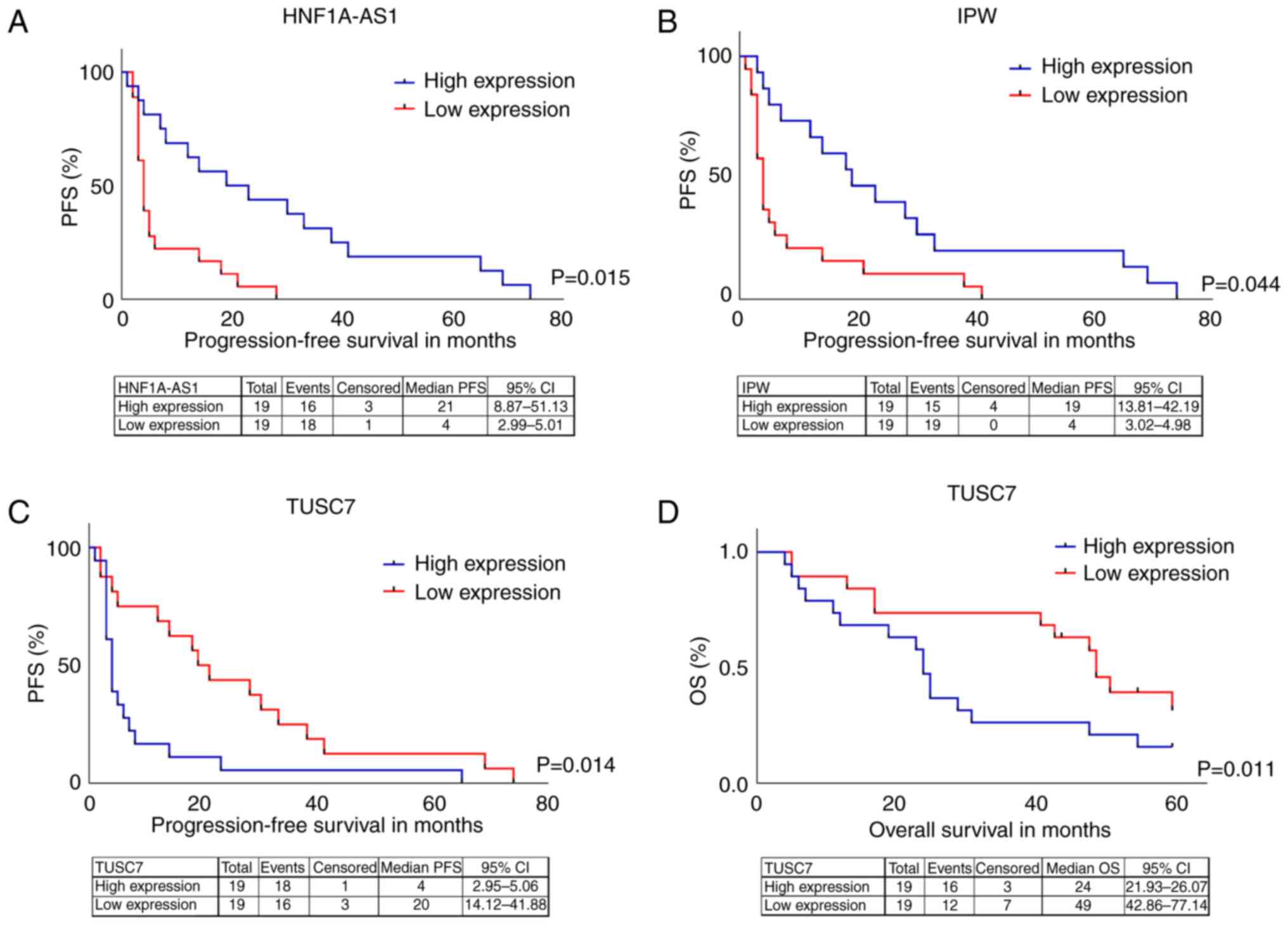
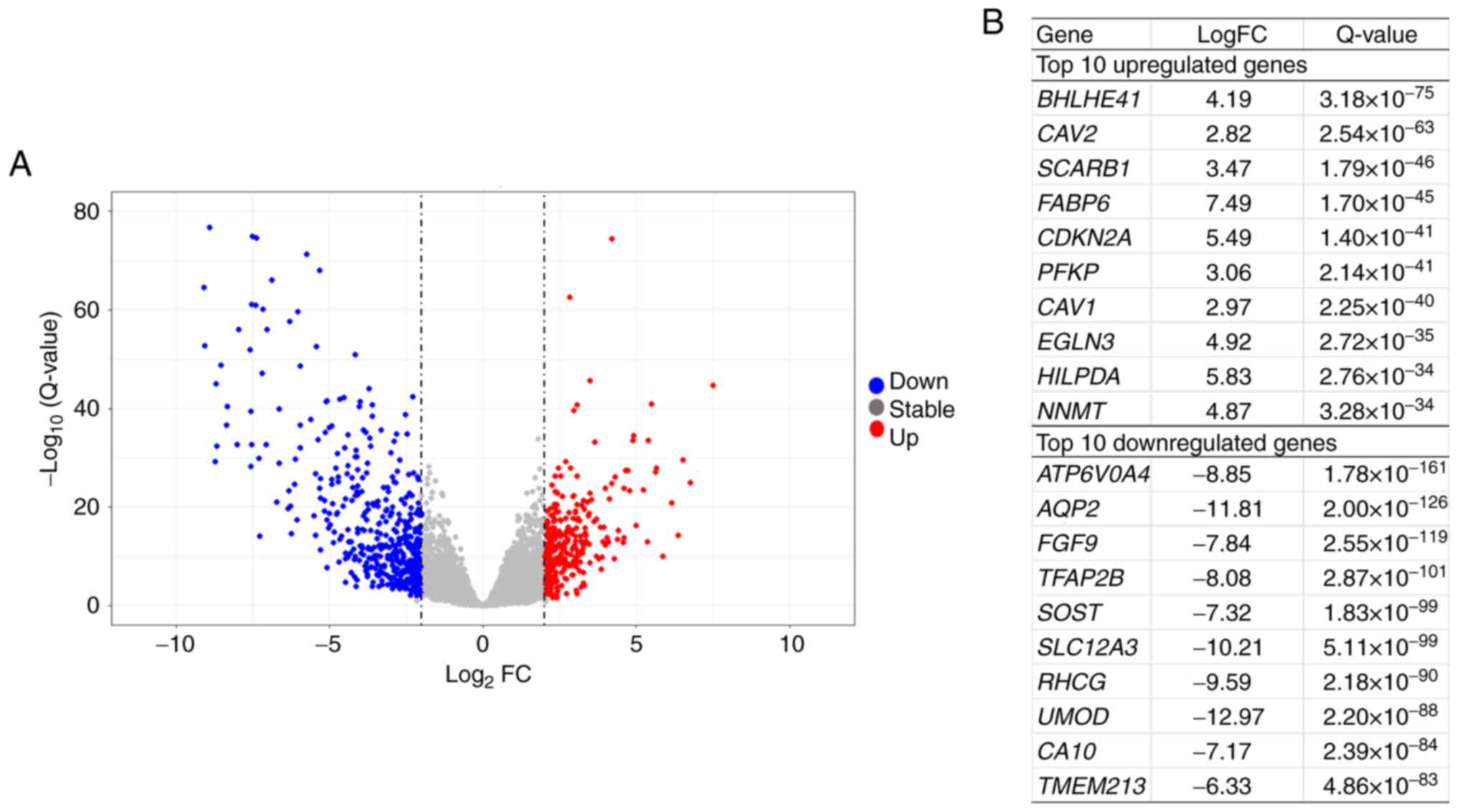
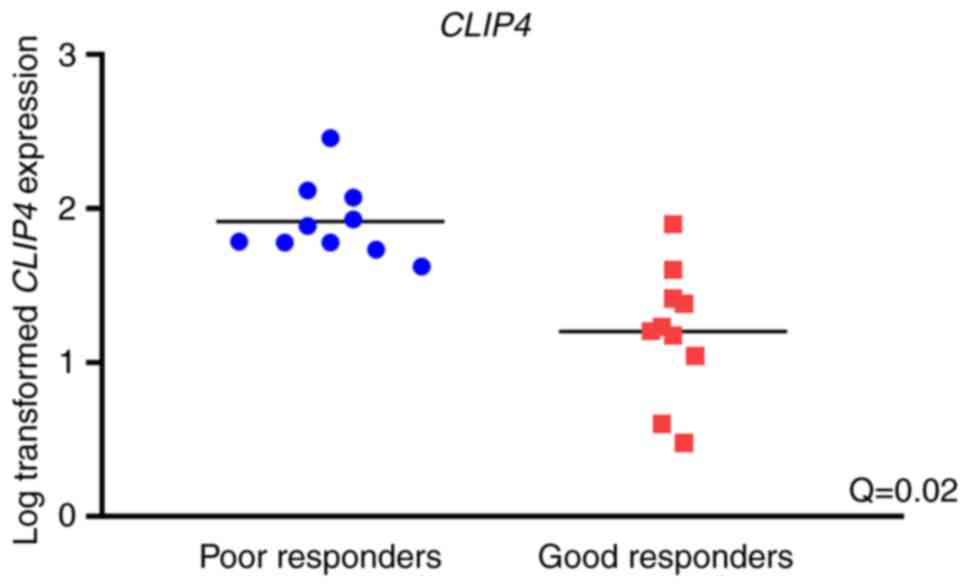
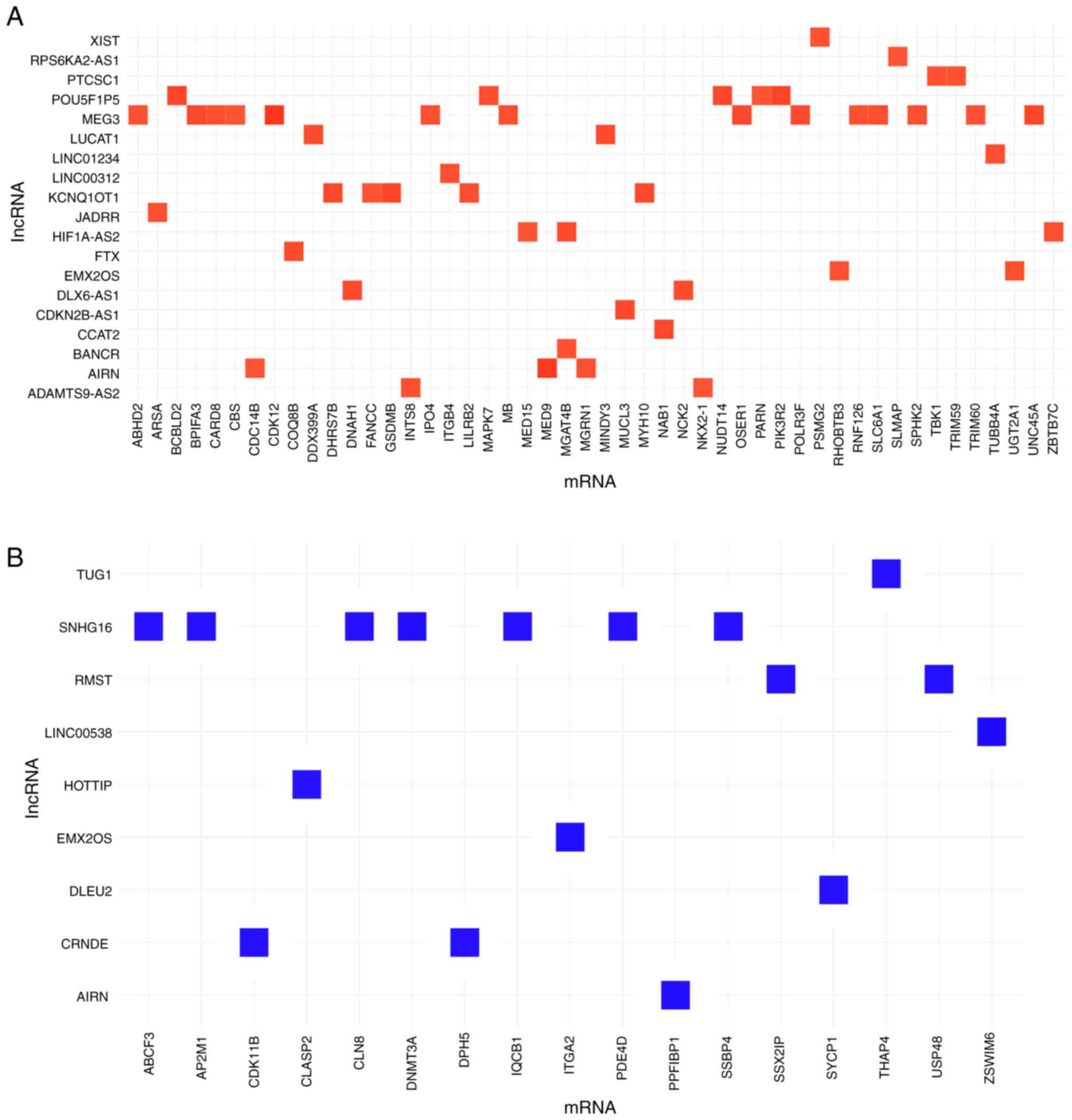
|
1
|
Znaor A, Lortet-Tieulent J, Laversanne M,
Jemal A and Bray F: International variations and trends in renal
cell carcinoma incidence and mortality. Eur Urol. 67:519–530. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Therasse P, Arbuck SG, Eisenhauer EA,
Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van
Oosterom AT, Christian MC and Gwyther SG: New Guidelines to
evaluate the response to treatment in solid tumors. European
Organization for Research and Treatment of Cancer, National Cancer
Institute of the United States, National Cancer Institute of
Canada. J Natl Cancer Inst. 92:205–216. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
National Cancer Institure (NCI), . Clear
Cell Renal Cell Carcinoma-NCI. https://www.cancer.gov/pediatric-adult-rare-tumor/rare-tumors/rare-kidney-tumors/clear-cell-renal-cell-carcinomaFebruary
1–2023
|
|
4
|
Xue J, Chen W, Xu W, Xu Z, Li X, Qi F and
Wang Z: Patterns of distant metastases in patients with clear cell
renal cell carcinoma - - A population-based analysis. Cancer Med.
10:173–187. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dabestani S, Thorstenson A, Lindblad P,
Harmenberg U, Ljungberg B and Lundstam S: Renal cell carcinoma
recurrences and metastases in primary non-metastatic patients: A
population-based study. World J Urol. 34:1081–1086. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eggers H, Schünemann C, Grünwald V,
Rudolph L, Tiemann ML, Reuter C, Anders-Meyn MF, Ganser A and
Ivanyi P: Improving survival in metastatic renal cell carcinoma
(MRCC) patients: Do elderly patients benefit from expanded targeted
therapeutic options? World J Urol. 40:2489–2497. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sheng IY and Ornstein MC: Ipilimumab and
nivolumab as first-line treatment of patients with renal cell
carcinoma: The evidence to date. Cancer Manag Res. 12:4871–4881.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Motzer RJ, Jonasch E, Agarwal N, Alva A,
Baine M, Beckermann K, Carlo MI, Choueiri TK, Costello BA, Derweesh
IH, et al: Kidney cancer, Version 3.2022, NCCN clinical practice
guidelines in oncology. J Natl Compr Canc Netw. 20:71–90. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long Non-Coding RNAs and Its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shao K, Shi T, Yang Y, Wang X, Xu D and
Zhou P: Highly expressed LncRNA CRNDE promotes cell proliferation
through Wnt/β-Catenin signaling in renal cell carcinoma. Tumour
Biol. Oct 6–2016.(Epub ahead of print). View Article : Google Scholar
|
|
11
|
Wang L, Cai Y, Zhao X, Jia X, Zhang J, Liu
J, Zhen H, Wang T, Tang X, Liu Y and Wang J: Down-regulated long
non-coding RNA H19 inhibits carcinogenesis of renal cell carcinoma.
Neoplasma. 62:412–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu Y, Liu J, Zheng Y, You L, Kuang D and
Liu T: Suppressed expression of long Non-coding RNA HOTAIR inhibits
proliferation and tumourigenicity of renal carcinoma cells. Tumour
Biol. 35:11887–11894. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang M, Lu W, Huang Y, Shi J, Wu X, Zhang
X, Jiang R, Cai Z and Wu S: Downregulation of the long noncoding
RNA TUG1 inhibits the proliferation, migration, invasion and
promotes apoptosis of renal cell carcinoma. J Mol Histol.
47:421–428. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang M, Huang T, Luo G, Huang C, Xiao XY,
Wang L, Jiang GS and Zeng FQ: Long Non-Coding RNA MEG3 induces
renal cell carcinoma cells apoptosis by activating the
mitochondrial pathway. J Huazhong Univ Sci Technolog Med Sci.
35:541–545. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qiao HP, Gao WS, Huo JX and Yang ZS: Long
Non-Coding RNA GAS5 functions as a tumor suppressor in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li M, Wang Y, Cheng L, Niu W, Zhao G, Raju
JK, Huo J, Wu B, Yin B, Song Y and Bu R: Long Non-Coding RNAs in
renal cell carcinoma: A systematic review and clinical
implications. Oncotarget. 8:48424–48435. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Yang F, Chen SJ, Che J and Zheng
J: Upregulation of Long Non-Coding RNA MALAT1 correlates with tumor
progression and poor prognosis in clear cell renal cell carcinoma.
Tumour Biol. 36:2947–2955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Posa I, Carvalho S, Tavares J and Grosso
AR: A Pan-cancer analysis of MYC-PVT1 Reveals CNV-Unmediated
deregulation and poor prognosis in renal carcinoma. Oncotarget.
7:47033–47041. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Liu J, Bai H, Dang Y, Lv P and Wu
S: Long Intergenic Non-Coding RNA 00152 promotes renal cell
carcinoma progression by epigenetically suppressing P16 and
negatively regulates MiR-205. Am J Cancer Res. 7:312–322.
2017.PubMed/NCBI
|
|
20
|
Xiao H, Bao L, Xiao W, Ruan H, Song Z, Qu
Y, Chen K, Zhang X and Yang H: Long Non-Coding RNA Lucat1 is a poor
prognostic factor and demonstrates malignant biological behavior in
clear cell renal cell carcinoma. Oncotarget. 8:113622–113634. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song EL, Xing L, Wang L, Song WT, Li DB,
Wang Y, Gu YW, Liu MM, Ni WJ, Zhang P, et al: LncRNA ADAMTS9-AS2
inhibits cell proliferation and decreases chemoresistance in clear
cell renal cell carcinoma via the MiR-27a-3p/FOXO1 axis. Aging
(Albany NY). 11:5705–5725. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y,
Chen W, Liu F, Sun W, Li XF, et al: Exosome-Transmitted LncARSR
promotes sunitinib resistance in renal cancer by acting as a
competing endogenous RNA. Cancer Cell. 29:653–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhai W, Sun Y, Guo C, Hu G, Wang M, Zheng
J, Lin W, Huang Q, Li G, Zheng J and Chang C: LncRNA-SARCC
suppresses renal cell carcinoma (RCC) progression via altering the
androgen receptor(AR)/MiRNA-143-3p signals. Cell Death Differ.
24:1502–1517. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Saad OA, Li WT, Krishnan AR, Nguyen GC,
Lopez JP, McKay RR, Wang-Rodriguez J and Ongkeko WM: The Renal
clear cell carcinoma immune landscape. Neoplasia. 24:145–154. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Roldán FL, Izquierdo L, Ingelmo-Torres M,
Lozano JJ, Carrasco R, Cuñado A, Reig O, Mengual L and Alcaraz A:
Prognostic gene expression-based signature in clear-cell renal cell
carcinoma. Cancers (Basel). 14:37542022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Flaifel A, Xie W, Braun DA, Ficial M,
Bakouny Z, Nassar AH, Jennings RB, Escudier B, George DJ, Motzer
RJ, et al: PD-L1 expression and clinical outcomes to cabozantinib,
everolimus and sunitinib in patients with metastatic renal cell
carcinoma: Analysis of the randomized clinical trials METEOR and
CABOSUN. Clin Cancer Res. 25:6080–6088. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qu Y, Lin Z, Qi Y, Qi Y, Chen Y, Zhou Q,
Zeng H, Liu Z, Wang Z, Wang J, et al: PAK1 expression determines
poor prognosis and immune evasion in metastatic renal cell
carcinoma patients. Urol Oncol. 38:293–304. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Motzer RJ, Bacik J, Murphy BA, Russo P and
Mazumdar M: Interferon-Alfa as a comparative treatment for clinical
trials of new therapies against advanced renal cell carcinoma. J
Clin Oncol. 20:289–296. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fiala O, Finek J, Poprach A, Melichar B,
Kopecký J, Zemanova M, Kopeckova K, Mlcoch T, Dolezal T, Capkova L
and Buchler T: Outcomes according to MSKCC risk score with focus on
the Intermediate-Risk Group in metastatic renal cell carcinoma
patients treated with first-line sunitinib: A Retrospective
analysis of 2390 Patients. Cancers (Basel). 12:8082020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
European Medicines Agency (EMA), . Sutent.
European Medicines Agency; Amsterdam: 2021, https://www.ema.europa.eu/en/medicines/human/EPAR/sutentMay
23–2023
|
|
31
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (Version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schroeder A, Mueller O, Stocker S,
Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M
and Ragg T: The RIN: An RNA integrity number for assigning
integrity values to RNA measurements. BMC Mol Biol. 7:32006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-Time PCR Experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Babraham Bioinformatics, . FastQC: A
Quality Control Tool for High Throughput Sequence Data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/November
2–2021
|
|
36
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Statistical Soc Series B
(Methodological). 57:289–300. 1995. View Article : Google Scholar
|
|
37
|
Ensembl, . Human (GRCh38.p13). http://www.ensembl.org/Homo_sapiens/Info/IndexMay
25–2023
|
|
38
|
Bray NL, Pimentel H, Melsted P and Pachter
L: Near-optimal probabilistic RNA-Seq quantification. Nat
Biotechnol. 34:525–527. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
McCarthy DJ, Chen Y and Smyth GK:
Differential expression analysis of multifactor RNA-Seq experiments
with respect to biological variation. Nucleic Acids Res.
40:4288–4297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bonferroni CE: Il Calcolo Delle
Assicurazioni Su Gruppi Di Teste. Studi in onore del Professore
Salvatore Ortu Carboni. 1935.
|
|
41
|
Gillespie M, Jassal B, Stephan R, Milacic
M, Rothfels K, Senff-Ribeiro A, Griss J, Sevilla C, Matthews L,
Gong C, et al: The reactome pathway knowledgebase 2022. Nucleic
Acids Res. 50:D687–D692. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
GTEx Portal: TUSC7. https://www.gtexportal.org/home/gene/TUSC7October
12–2022
|
|
43
|
Ren W, Chen S, Liu G, Wang X, Ye H and Xi
Y: TUSC7 acts as a tumor suppressor in colorectal cancer. Am J
Transl Res. 9:4026–4035. 2017.PubMed/NCBI
|
|
44
|
Cong M, Li J, Jing R and Li Z: Long
Non-Coding RNA tumor suppressor candidate 7 functions as a tumor
suppressor and inhibits proliferation in osteosarcoma. Tumour Biol.
37:9441–9450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zheng BH, He ZX, Zhang J, Ma JJ, Zhang HW,
Zhu W, Shao ZM and Ni XJ: The biological function of
TUSC7/MiR-1224-3p axis in triple-negative breast cancer. Cancer
Manag Res. 13:5763–5774. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Darb-Esfahani S, Denkert C, Stenzinger A,
Salat C, Sinn B, Schem C, Endris V, Klare P, Schmitt W, Blohmer JU,
et al: Role of TP53 mutations in triple negative and HER2-Positive
breast cancer treated with neoadjuvant Anthracycline/Taxane-Based
chemotherapy. Oncotarget. 7:67686–67698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
GTEx Portal: HNF1A-AS1. https://www.gtexportal.org/home/gene/HNF1A-AS1February
1–2023
|
|
48
|
Liu Y, Zhao F, Tan F, Tang L, Du Z, Mou J,
Zhou G and Yuan C: HNF1A-AS1: A Tumor-associated long non-coding
RNA. Curr Pharm Des. 28:1720–1729. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang G, An X and Zhao H, Zhang Q and Zhao
H: Long non-coding RNA HNF1A-AS1 promotes cell proliferation and
invasion via regulating MiR-17-5p in Non-Small cell lung cancer.
Biomed Pharmacother. 98:594–599. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu L, Chen Y, Li Q and Duan P: LncRNA
HNF1A-AS1 modulates non-small cell lung cancer progression by
targeting MiR-149-5p/Cdk6. J Cell Biochem. 120:18736–18750. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang Y, Shi J, Luo J, Liu C and Zhu L:
Regulatory mechanisms and potential medical applications of
HNF1A-AS1 in cancers. Am J Transl Res. 14:4154–4168.
2022.PubMed/NCBI
|
|
52
|
Zhou X, Fan YH, Wang Y and Liu Y:
Prognostic and clinical significance of long non-coding RNA
HNF1A-AS1 in solid cancers: A systematic review and meta-analysis.
Medicine (Baltimore). 98:e182642019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Shi Y, Zhang Q, Xie M, Feng Y, Ma S, Yi C,
Wang Z, Li Y, Liu X, Liu H, et al: Aberrant Methylation-mediated
decrease of LncRNA HNF1A-AS1 contributes to malignant progression
of laryngeal squamous cell carcinoma via EMT. Oncol Rep.
44:2503–2516. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ding CH, Yin C, Chen SJ, Wen LZ, Ding K,
Lei SJ, Liu JP, Wang J, Chen KX, Jiang HL, et al: The
HNF1α-Regulated LncRNA HNF1A-AS1 reverses the malignancy of
hepatocellular carcinoma by enhancing the phosphatase activity of
SHP-1. Mol Cancer. 17:632018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dang Y, Lan F, Ouyang X, Wang K, Lin Y, Yu
Y, Wang L, Wang Y and Huang Q: Expression and clinical significance
of long Non-Coding RNA HNF1A-AS1 in human gastric cancer. World J
Surg Oncol. 13:3022015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kanduri C: Long Noncoding RNAs: Lessons
from genomic imprinting. Biochim Biophys Acta. 1859:102–111. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
BioGPS, . IPW (Imprinted in Prader-Willi
Syndrome). http://biogps.org/#goto=genereport&id=3653October
12–2022
|
|
58
|
Ma B, Li Y and Ren Y: Identification of a
6-lncRNA prognostic signature based on microarray Re-annotation in
gastric cancer. Cancer Med. 9:335–349. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tang SJ, You GR, Chang JT and Cheng AJ:
Systematic analysis and identification of dysregulated panel
LncRNAs contributing to poor prognosis in Head-neck cancer. Front
Oncol. 11:7317522021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
GeneCards - The Human Gene Database.
CLIP4. https://www.genecards.org/cgi-bin/carddisp.pl?gene=CLIP4&keywords=CLIP4February
2–2023
|
|
61
|
Park JS, Pierorazio PM, Lee JH, Lee HJ,
Lim YS, Jang WS, Kim J, Lee SH, Rha KH, Cho NH and Ham WS: Gene
expression analysis of aggressive clinical T1 stage clear cell
renal cell carcinoma for identifying potential diagnostic and
prognostic biomarkers. Cancers (Basel). 12:E2222020. View Article : Google Scholar
|
|
62
|
Ahn J, Han KS, Heo JH, Bang D, Kang YH,
Jin HA, Hong SJ, Lee JH and Ham WS: FOXC2 and CLIP4: A potential
biomarker for synchronous metastasis of ≤7-Cm clear cell renal cell
carcinomas. Oncotarget. 7:51423–51434. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gong A, Zhao X, Pan Y, Qi Y, Li S, Huang
Y, Guo Y, Qi X, Zheng W and Jia L: The LncRNA MEG3 mediates renal
cell cancer progression by regulating ST3Gal1 transcription and
EGFR sialylation. J Cell Sci. 133:jcs2440202020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
He H, Dai J, Zhuo R, Zhao J, Wang H, Sun
F, Zhu Y and Xu D: Study on the mechanism behind LncRNA MEG3
affecting clear cell renal cell carcinoma by regulating
MiR-7/RASL11B signaling. J Cell Physiol. 233:9503–9515. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Cheng T, Shuang W, Ye D, Zhang W, Yang Z,
Fang W, Xu H, Gu M, Xu W and Guan C: SNHG16 promotes cell
proliferation and inhibits cell apoptosis via regulation of the
MiR-1303-p/STARD9 Axis in clear cell renal cell carcinoma. Cell
Signal. 84:1100132021. View Article : Google Scholar : PubMed/NCBI
|