Introduction
Accumulating evidence indicates that activating
mutations of the epidermal growth factor receptor (EGFR) are
present in ~50% of patients with advanced non-small cell lung
cancer (NSCLC) of Eastern Asian ethnicity compared with 12–15% in
the Caucasian population (1,2). These
mutations predict sensitivity to first- and second-generation
tyrosine kinase inhibitors (TKIs), such as gefitinib, erlotinib, or
afatinib, and are pivotal molecular biomarkers in the selection of
targeted therapy (3–5). Thus, a sensitive and reliable
laboratory test is critical in the design of personalized therapy
for this subset of patients.
Recent advances in molecular pathology have
introduced highly sensitive technology platforms for detecting EGFR
mutations by reverse transcription-quantitative (RT-q)PCR using
specific probes or amplified refractory mutation system (ARMS),
such as the EGFR PCR Kit or therascreen EGFR RGQ PCR kit provided
by Qiagen GmbH (6). Both kits
characteristically produce flat or linear lines for negative
results, making it difficult to ignore the occasional late
upward-shifted PCR curves beyond the limits of detection (LOD). The
peptide nucleic acid (PNA)-sequencing was designed to construct the
PCR clamp reactions, in which the PNA clamp suppresses the
amplification of wild-type DNA and thus enriches the amplification
of mutant sequences. PNA-mediated PCR clamping is an interesting
technology in the detection of gene mutations. Both simplicity and
versatility make it especially advantageous for large-scale
screening programs. Thiede et al (7) proposed the simple and sensitive method
of PNA-mediated PCR clamping in the detection of mutations in the
ras proto-oncogenes. Sano et al (8) reported that PNA-clamped probe assay is
more sensitive than direct sequencing in the detection of mutations
in samples containing <1% mutant alleles. Our previous study
confirmed its comparable performance in terms of EGFR mutation
detection compared with RNA sequencing but was more sensitive in
malignant pleural effusion than by direct sequencing (9).
In a pilot study performed on a lung cancer cohort
(n=346), 9 equivocal cases with late upward PCR curves were further
investigated using mutant-enriched PCR with PNA-sequencing, of
which the analytical sensitivities were conservatively estimated at
1% or better for both E746_A750del and L858R mutations using cloned
DNA fragments mixed with a wild-type EGFR background. All 20 cases
revealed unequivocally negative results in exons 19 and 21 by
PNA-sequencing, supporting the notion that a flat baseline or
linearly increasing straight line observed in reverse
transcription-quantitative PCR is genuinely negative. The data
verified no false positive and false negative results for
PNA-sequencing (10).
PNA-sequencing showed that 6 of the 9 equivocal cases harbored EGFR
mutations. The results supported the usage of PNA sequencing as a
supplementary method in a clinical diagnostic setting.
Herein, the validation of the clinical value of the
supplementary PNA-sequencing technology in a larger cohort and an
independent referral cohort from other institutes is reported.
Clinical responses to first- or second-generation EGFR-TKIs were
assessed in 17 PNA-positive cases in the recruited cohort (n=1,783)
and 3 PNA-positive cases from the referral cohort (n=1,944).
Materials and methods
Patients and samples
For the retrospective cohort study, the need for
informed consent from the patients was waived by the Institutional
Review Board of National Cheng Kung University Hospital (Tainan,
Taiwan). Informed consent was obtained for the use of patient
tissues but was waived for the use of patient data. A total of
1,783 cases of formalin-fixed, paraffin-embedded (FFPE) NSCLC,
diagnosed between September 2012 and February 2018 in our institute
were included in the present study. Out of 1,783 patients, 847
(47.5%) were males. The median age was 66 years (range, 26–99
years). All cases were diagnosed as stage III or IV adenocarcinoma
and reviewed independently by two pathologists. The clinical
information of the patient cohort was independently reviewed by two
clinicians. Any disagreement in the reviewing of samples was
resolved by a third author. Basic demographic information, smoking
status, stage at presentation, EGFR mutation type, and treatment
details were obtained from the medical records. The response of the
tumor to EGFR TKI was evaluated by chest radiography and computed
tomography (CT) every 2–4 and 8–12 weeks after treatment,
respectively. Treatment response was assessed according to the
Response Evaluation Criteria in Solid Tumors (RECIST) v1.1
(11). Progression of disease was
evaluated on scan imaging (11). In
addition, clinical deterioration or death from any cause without
evidence of disease progression was also defined as progression in
the calculation of progression-free survival (PFS). Objective
response rate (ORR) was defined as the achievement of either a
complete or partial response.
An independent lung cancer cohort of 1,944 cases was
also collected from other referral hospitals between September 2012
and February 2018. Out of 1,944 patients, 953 (49.0%) were male.
The median age was 67 years (range, 24–98 years). Referral
hospitals included: The Department of Surgery, E-DA hospital;
Department of Oncology, Tainan Hospital, Ministry of Health and
Welfare; Department of Oncology, Tainan Municipal Hospital; and
Department of Oncology, Dalim, Tzu Chi Hospital.
The independent lung cancer cohort from other
hospitals was recruited to reduce potential selection bias. All
cases were diagnosed as stage III or IV adenocarcinoma. Only 3
cases with EGFR mutations from E-DA hospital had follow-up
data.
FFPE DNA extraction
FFPE tissues were used for molecular diagnosis.
Hematoxylin and Eosin-stained histological sections were reviewed
by a pathologist who marked the tumor area (>20%) for
macrodissection. DNA was then extracted using a QIAcube automated
extractor (Qiagen GmbH) using the QIAamp DNA FFPE tissue kit
(Qiagen GmbH) according to the manufacturer's instructions.
EGFR mutation analysis
The EGFR mutations were analyzed using an EGFR PCR
Kit (EGFR RUO Kit; Qiagen GmbH) (n=256) and therascreen EGFR RGQ
PCR Kit (EGFR IVD Kit; Qiagen GmbH) (n=3,471). Both diagnostic kits
combine Scorpion probe/primers and ARMS technologies to detect the
mutations by RT-qPCR. The assay was performed according to the
manufacturer's instructions. This assay system was developed for
the detection of the most commonly reported EGFR mutations,
including 19 deletions in exon 19, 3 insertions in exon 20, and
point mutations of G719X (in exon 18), S768I (in exon 20), L858R
and L861Q (in exon 21).
EGFR IVD Kit assessing a T790M (exon 20) mutation,
an important TKI-resistant mutation (12,13),
was used for clinical analysis. Analysis was performed using the
Rotor-Gene Q series built-in software version 2.0.3 (Build 2) for
the EGFR RUO Kit and EGFR IVD Kit (Qiagen GmbH). The EGFR PCR Kit
(EGFR RUO Kit) was first introduced to our laboratory for EGFR
mutations, and subsequently replaced by the therascreen EGFR RGQ
PCR Kit (EGFR IVD Kit) to improve the detection of T790M (exon 20).
The EGFR mutation rate was 54.4 and 54.9% for the EGFR RUO Kit and
IVD Kit, respectively based on the results. No significant
differences between the methods were observed. Real-time curves
were generated using FAM-labeled probes for exon 2 control and each
mutation in separate tubes. To calculate a ΔCq value for each
mutation reaction, the following equation was used: [Mutation
Cq]-[Control Cq]=ΔCq. Manufacturer-supplied ΔCq thresholds were
used as the limit of detection (LOD) to call a mutation (≤ΔCT
threshold is positive for mutation).
During the study period, a total of 34 and 22 cases
showing upward-shifted amplification curves in the late cycles
beyond the LOD were discovered in our hospital and other
institutes, respectively. They were submitted for further
investigation using direct sequencing and PNA-sequencing.
PNA-sequencing
For PNA-sequencing of EGFR exons 19 and 21,
PNA was used to construct the PCR clamp reactions, in which the PNA
clamp suppresses the amplification of wild-type DNA, and thus
increases the preferential amplification of mutant sequences
(10). PNA oligos were synthesized
by PANAGENE Inc. The PNA clamp primer for exons 19 and 21 was
designed to be homologous to the wild-type allele at codons 746–751
and 855–860, respectively (RefSeq accession no. NM_005228.3).
The analytical sensitivity of PNA-sequencing was
estimated at ~1% for both E746_A750del and L858R mutations using
cloned DNA fragments serially diluted with A549 cells containing
wild-type EGFR. They were mixed with ratios from 1:1 to 1:1,000 in
the prior pilot study (10).
PNA-sequencing was performed in a mixture of PCR
primers (10 mmol/l each), 100 mmol/l PNA oligos, 25 ng genomic DNA
(gDNA), and polymerase mix (Super Therm Gold MasterMix; Bionovas
Biotechnology). The PCR amplicons were purified and then subjected
to bidirectional sequencing.
Clinical response to EGFR-TKIs
All of the patients received a CT scan of the chest
before initiation of the treatment and every 12 weeks thereafter to
evaluate the tumor response. Brain imaging and bone scans were
performed if associated symptoms occurred in patients. The primary
endpoint was progression-free survival (PFS). The secondary
endpoints included disease control rate, overall response rate, and
overall survival (OS). The EGFR mutation status was analyzed by
PNA-sequencing.
PFS was calculated from the date of TKI treatment
until the date of radiological progression according to the RECIST
v1.1 guidelines or death, with censoring at the date of last
follow-up in the event that the patient did not progress. The
overall response rate was calculated as the percentage of patients
who exhibited a partial response or complete response in the first
image study after the introduction of the TKI treatment, while the
disease control rate was defined as the percentage of patients who
exhibited a partial response, complete response, or stable disease.
Furthermore, the duration of OS was defined as the period from the
beginning date of TKI treatment until the date of death. If
follow-up was incomplete or interrupted at the end of the study,
the nurse practitioner was asked to call their families to clarify
the status of the clinical response.
Major changes in the status according to the RECIST
v1.1 guidelines related to imaging include the following: i) Number
of target lesions; ii) assessment of pathologic lymph nodes; iii)
status of disease progression; iv) clarification of unequivocal
progression of non-target lesions; and v) hybrid imaging with F-18
fludeoxyglucose positron emission tomography/magnetic resonance
imaging (FDG PET/MRI) that correlated with the results of a CT scan
in the detection of new lesions. The progression of the disease was
evaluated using scan imaging. Primary outcomes were OS and PFS;
secondary outcomes were overall ORR based on the RECIST v 1.1
guidelines.
Chemotherapy
Of 17 NSCLC patients with low levels of EGFR
mutation discovered in our hospital, three patients did not receive
TKIs due to presence of a brain metastasis (n=2) or as a
conservative strategy following after lobectomy. The remaining
patients with activating EGFR mutations (exon 19 deletions and exon
21 L858R received TKIs. A first-generation EGFR-TKI (gefitinib and
erlotinib) is the standard treatment for patients with locally
advanced or metastatic NSCLC, in particular for those patients
harboring EGFR mutations. Afatinib is an oral irreversible
second-generation EGFR TKI. This drug was developed for patients
showing resistance to the first-generation EGFR TKIs. If resistance
is still observed, Osimertinib is a potent irreversible third
generation EGFR-TKI targeting EGFR mutations with very little
effect on wild-type EGFR (14,15).
If chemotherapy is considered (for example, in case
of EGFR exon 20 insertion), patients should receive pemetrexed
(Alimta®) 500 mg/m2 combined with carboplatin
(area under the concentration-time curve of 5 mg/ml/min,) both
administered on day 1 of a 21-day cycle. The treatment should be
continued for 4 cycles or until unacceptable toxicity or disease
progression appears, in which case bevacizumab may be administered
as a salvage chemotherapy (16,17).
Statistical analysis
Patient characteristics (age, sex, smoking history,
stage and histology) were tabulated in relation to mutation status.
A Fisher's exact test was used to analyze the relationship between
patient characteristics and EGFR mutations. P<0.05 was
considered to indicate a statistically significant difference.
The PFS after TKI therapy was calculated from the
first day of TKI treatment until the earliest sign of disease
progression or death from any cause. OS for the entire cohort was
calculated from the date of diagnosis to the time of death or last
follow-up. The follow-up data was censored on October 30, 2022.
Kaplan-Meier curves were used to estimate the probabilities at each
time point when an event occurred and the probability of survival
at each point in time. A log-rank test was used to identify factors
affecting the OS of the entire cohort, and multivariate analysis
was performed using Cox regression analyses. SPSS version 17.0
(SPSS, Inc.) was used for statistical analysis.
Results
Patient characteristics
A total of 3,727 patients (NCKUH, 1,783; other
institutes, 1,944) were enrolled in the present study. The patient
characteristics (age and sex) and histological profile did not
differ significantly between our institute and the other institutes
(Table SI). For the 3,727
patients, the median age was 66.5 years (range, 20–99 years). EGFR
mutation status was not associated with patient age (P>0.999)
but was more frequent in females than in males (60.1 vs. 39.9%;
P<0.05). The histology of the tumors included adenocarcinoma
(89.3%), adenosquamous carcinoma (1.4%), squamous cell carcinoma
(4.0%), and NSCLC-NOS (5.3%). The rate of EGFR mutation was
significantly higher than that of wild-type EGFR in patients with
adenocarcinoma (60 vs. 40%; P<0.05) but did not differ in terms
with regard to smoking habits and clinical stage. Around one-fifth
(22.5%) of the specimens came from surgical resection and the
others (77.5%) were small biopsy or cell block specimens. Among
these patients, 2,075 cases (55.7%) had EGFR mutations, with exon
19 deletions in 831 cases (40%), L858R mutation in 1,001 cases
(48.2%), and other mutations in 243 cases (11.7%) (Table I). Interestingly, 68 cases (3.3%)
had T790M mutation in combination with other mutations analyzed by
the EGFR IVD Kit.
 | Table I.Summary of the characteristics of the
patients based on the EGFR mutation status. |
Table I.
Summary of the characteristics of the
patients based on the EGFR mutation status.
| Characteristic | EGFR mutant | EGFR wild-type | P-value |
|---|
| Age, years |
|
|
|
| Mean ±
SD | 66.5±11.6 | 66.0±12.7 | >0.05 |
|
Median | 67 | 66 |
|
|
Range | 26-99 | 20-98 |
|
| Sex, n (%) |
|
|
|
| Male | 827 (43.4) | 1,078 (56.6) | <0.05 |
|
Female | 1248 (68.5) | 574 (31.5) |
|
| Histological type, n
(%) |
|
|
|
|
Adenocarcinoma | 1,999 (60.0) | 1,331 (40.0) | <0.05 |
|
Adenosquamous carcinoma | 4 (47.1) | 27 (52.9) |
|
| Squamous
cell carcinomas | 13 (8.8) | 135 (91.2) |
|
|
NSCLC-NOS | 39 (19.7) | 159 (80.3) |
|
| Smoking history, n
(%) |
|
|
|
| Never
smoker | 1,452 (56.1) | 1,135 (43.9) | >0.05 |
|
Smoker | 623 (54.6) | 517 (45.4) |
|
| Stage, n (%) |
|
|
|
| III | 15 (37.5) | 25 (62.5) | >0.05 |
| IV | 2,060 (55.9) | 1,627 (44.1) |
|
| Tumor sample, n
(%) |
|
|
|
| Surgical
specimen | 481 (57.4) | 357 (42.6) | >0.05 |
| Small
biopsy or cell block specimen | 1,594 (55.2) | 1,295 (44.8) |
|
| Mutation Type |
|
|
|
| Exon 19
deletion | 831 (40.0) |
|
|
| Exon 21
L858R | 1,002 (48.3) |
|
|
| Other
(including double mutation) | 172 (8.3) |
|
|
| Exon 20
T790M | 2 (0.1) |
|
|
| Exon 20
T790M combined with other mutations | 68 (3.3) |
|
|
EGFR mutation analysis
Between September 2012 and February 2018, 34 cases
were categorized as equivocal from among the 1,783 lung cancer
cases analyzed in our institute (Table
II). Of these, 6 cases were reported in our prior pilot study
(2). To validate the performance of
PNA sequencing technology, an independent lung cancer cohort of
1,944 cases was collected from other institutes between September
2012 and February 2018.
 | Table II.Summary of the equivocal samples and
EGFR mutation rates. |
Table II.
Summary of the equivocal samples and
EGFR mutation rates.
| Cohort | Cases | Equivocal, n
(%) | Detected by
PNA-sequencing, n (%) | Exon 19 deletion,
n | Exon 21
L858R/Pb, n | Exon 21 L861Q,
n |
|---|
| NCKUH | 1,783 | 34 (1.9) | 17 (50.0) | 4 | 13 | 0 |
| Other
hospitals | 1,944 | 22 (1.1) | 14 (63.6) | 4 | 8 | 2 |
| Total
casesa | 3,727 | 56 (1.5) | 31 (55.4) | 8 | 21 | 2 |
In total, EGFR mutations were detected in 2,044 of
3,727 cases of NSCLC (54.8%) using the EGFR RUO Kit or the EGFR IVD
Kit. The control Cq fell within the recommended range of the kits.
The Cq values were calculated and the threshold was set at 0.075.
Then, ΔCq (mutation Cq-control Cq) was calculated. In the Qiagen
EGFR assay, a negative result (wild-type) usually produced an
undoubtedly flat baseline. Additionally, a linearly increasing
straight line may have been observed in certain negative cases. In
certain cases, however, an upward curve appeared in the later
cycles that did not reach the threshold. The result was designated
as negative according to the manufacturer's instructions. In our
laboratory, these cases were reported as equivocal with concerns of
potential false negativity.
Confirmation of the EGFR mutation
status using PNA-sequencing
Of 34 equivocal cases discovered in our hospital, 4
cases were observed in the exon 19 assays and 13 cases in the exon
21 assays. They were submitted for PNA-sequencing and found to
harbor EGFR mutations in 17 cases (50%). These mutations included
13 cases of L858R mutation and 4 cases of exon 19 mutations (two
L746_A750del, one L747_A750delinsP and one E746_P753delinsVP).
Of 22 equivocal cases collected from referral
institutes, 4 cases were found in the exon 19 assays and 10 cases
in the exon 21 assays. PNA sequencing revealed 14 cases (64%)
containing EGFR mutations, including 7 cases of L858R mutation, 1
case of L858P mutation, 2 cases of L861Q mutation, and 4 cases of
exon-19 mutations (L746_A750del, L747_A751del, E746_P752delinsV and
E746_P751delinsVA).
TKI response rate
Of 17 NSCLC patients with low levels of EGFR
mutations (where the amount of mutant DNA was lower than the LOD),
discovered in our hospital, the median age was 69 years and 15
patients (88%) were female (Table
III). Of these, 3 patients (patients no. 6, 9, and 17) did not
receive TKIs due to a brain metastasis (n=2) or a conservative
strategy chosen after lobectomy (n=1). The remaining 14 patients
received TKIs and were evaluated for clinical response, in which 10
cases had an L858R mutation and two had an exon 19 deletion. Of the
14 patients, 7 patients (50%) showed partial response, of which, 6
patients (42.8%) had stable disease and 1 patient exhibited disease
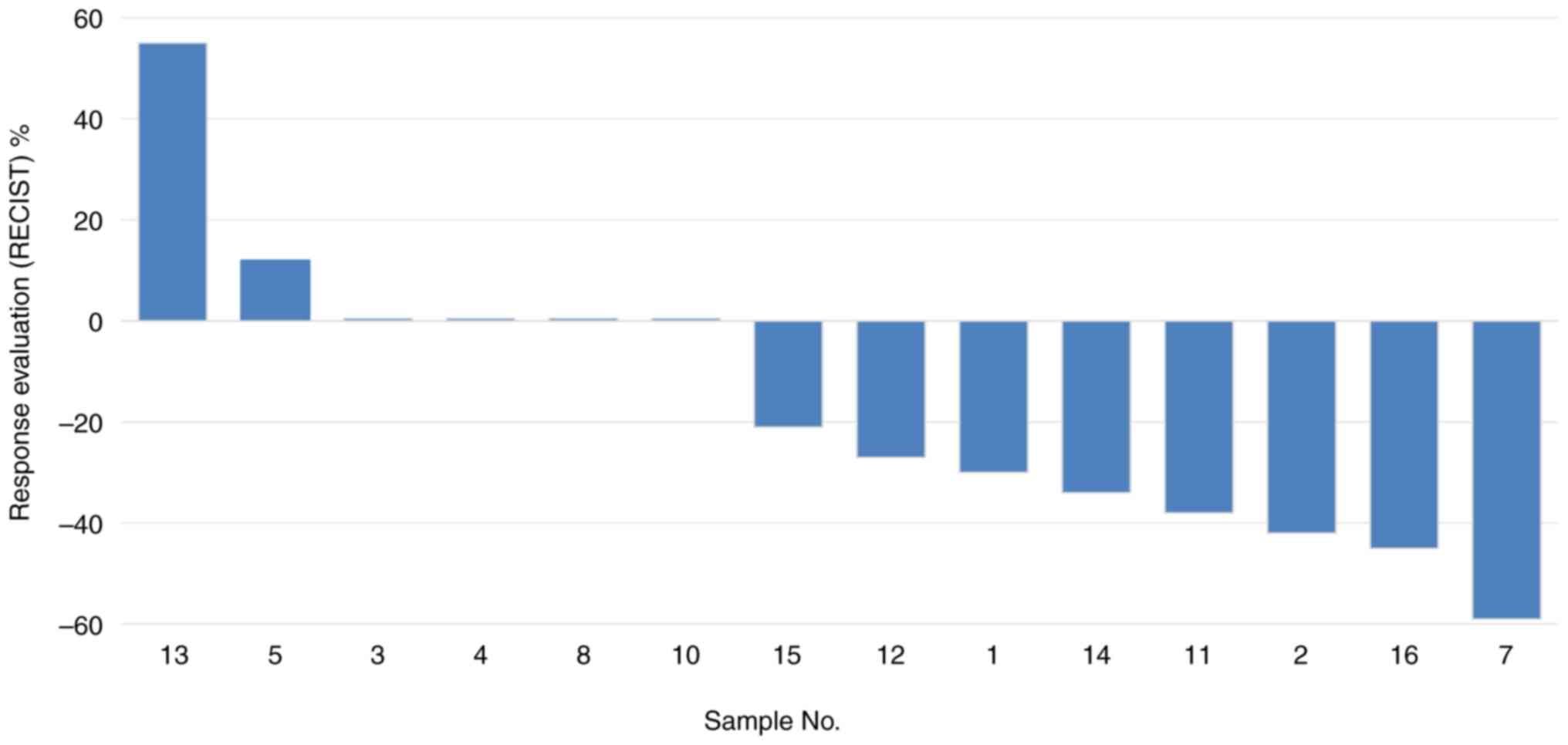
progression. The distribution of reduction in target-lesion size
(waterfall plot) after TKI treatment is shown in Fig. 1.
 | Table III.Summary of equivocal samples and
treatment response. |
Table III.
Summary of equivocal samples and
treatment response.
| Case no. | Sex | Age | Tumor, % | Pathology | Stage | Smoking | TTF-1 | EGFR result | Systemic
therapy | Response
evaluation | RECIST, % | Time since last TKI
use, months | Survival |
|---|
| 1 | F | 90 | 20 | AD | IV | Y | + | L858R | TT | PR | −30 | 50 | No |
| 2 | F | 71 | 20 | AD | IV | N | + | L858R | TT | PR | −42 | 40 | No |
| 3 | F | 70 | 20 | AD | IV | N | + | L858R | TT | SD | 0 | 5 | No |
| 4 | F | 73 | 20 | AD | IV | N | NA | L858R | TT | SD | 0 | 10 | No |
| 5 | F | 84 | 35 | AD | IV | Y | NA | L858R | TT | SD | +12 | 40 | No |
| 6 | F | 52 | 40 | AD | IV | N | + | L858R | Lobectomy | Nonea | None | NA | Yes |
| 7 | F | 40 | 30 | AD | IV | N | + | L858R | TT | PR | −59 | 19 | No |
| 8 | F | 54 | 20 | AD | IV | N | NA | E746_A750del | TT | SD | 0 | 80 | Yes |
| 9 | M | 56 | 70 | NSCLC-NOS | IV | N | + |
E746_S752delinsV | None | Nonea | None | NA | No |
| 10 | F | 52 | 40 | AD | IV | Y | + |
E746_T751delinsVA | TT | SD | 0 | 40 | No |
| 11 | F | 78 | 70 | SCC | IV | N | NA | L858R | TT | PR | −38 | 31 | No |
| 12 | F | 69 | 2 | AD | IV | N | + | L858R | TT | SD | −27 | 51 | No |
| 13 | F | 86 | 10 | AD | IV | N | + | L858R | TT | PD | +55 | 2 | No |
| 14 | F | 81 | 30 | AD | IV | N | NA | L858R | TT | PR | −34 | 30 | No |
| 15 | M | 55 | 20 | AD | IV | N | NA | L858R | TT | SD | −21 | 60 | Yes |
| 16 | F | 60 | 30 | AD | IV | N | + | L858R | TT | PR | −45 | 20 | No |
| 17 | F | 58 | 40 | AD | IV | N | + | L747_T751del, Exon
20 Insertion | None | Nonea | None | NA | No |
Correlation between EGFR mutation
status and clinical response to EGFR TKIs
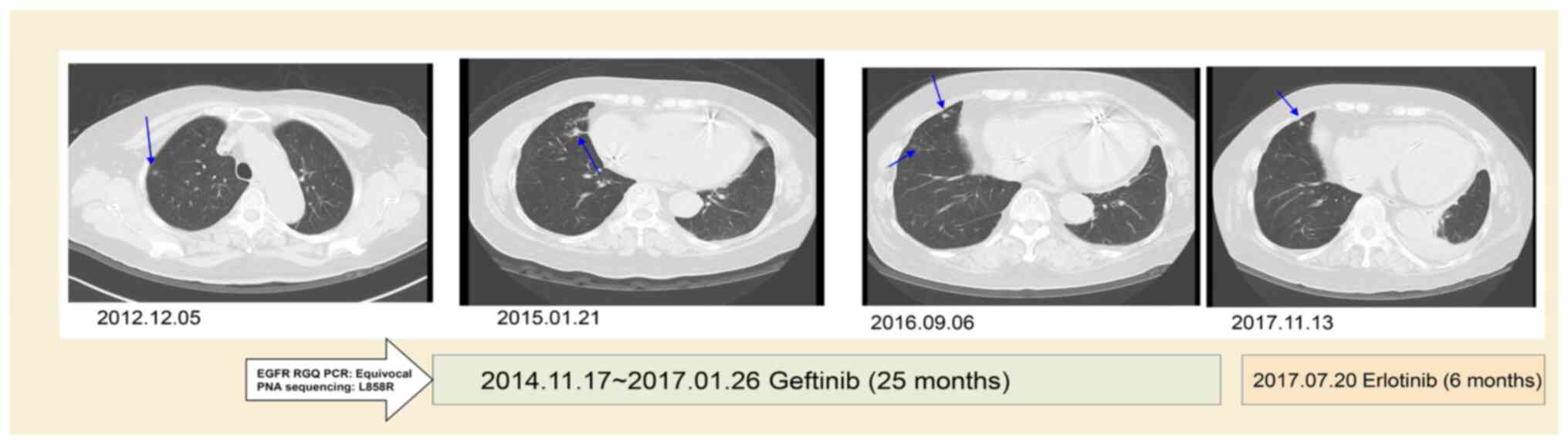
Case 10 was a 78 year female who had stage IIA
squamous cell carcinoma (T2aN1M0) and received video-assisted
thoracic surgery for the right lower lobe of lung and mediastinal
lymph node dissection in 2013. The EGFR RUO kit initially yielded a
negative result, with a late upward shifted curve, but was shown to
have an EGFR L858R mutation by PNA-sequencing in October 2014. The
recurrence of lung cancer with left pleural seeding occurred in
November 2014. The patient first received Gefitinib (Iressa; 250 mg
OD) treatment for 25 months, which showed mild regression of the
left lingual lung. Later, Gefitinib was replaced with Erlotinib
(Tarceva; 150 mg OD) for severe xerosis.
The follow-up CT at 31 months after TKI treatment
showed a reduction of the right lung tumor of ~53% (from 2.8 to 1.3
cm in diameter) with slight regression of left side pleural seeding
(Fig. 2). The patient was lost to
follow-up in March 2018.
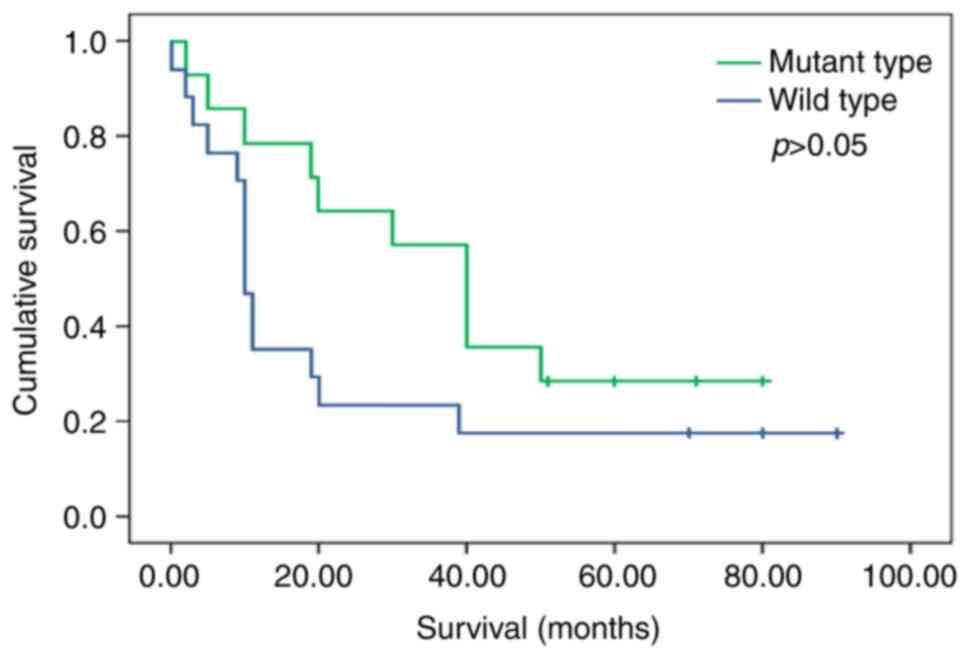
Overall survival of equivocal
cases
A total of 14 patients with EGFR mutations had a
longer 5-year OS (median: 40.0 vs. 10.0 months) after TKI treatment
than those with wild-type EGFR (n=17; P=0.085, log-rank test) using
a univariate analysis model (Fig.
3). The OS of patients with or without EGFR gene mutations was
40.0 months (95% CI, 24.2–33.8) and 10.0 months (95% CI,
14.7–27.3), respectively. The difference in OS was not significant
(P>0.05).
Discussion
The Qiagen kits are trustworthy methods that have
been used daily for diagnosis for years in our practice. The
infrequent occurrence of a late upward shift in the PCR curve
renders them useful only in specific cases. In our experience,
homemade RT-qPCR tests very often produce late upward PCR curves,
but these negative results should be neglected. In contrast, the
Scorpion ARMS system characteristically almost always produces flat
or linear lines for these same negative results. At least three
possible explanations can account for the upward curves in the
later cycles of amplification. First, the amount of mutant DNA may
be just lower than the detection limit of the Qiagen kits. This is
because most of the positive cases are taken from small biopsy
specimens with a low percentage of tumor cells. Second, the
heterogeneous distribution of the low levels of mutant tumor cells
in the specimen. Third, low levels of mutant DNA in these samples
result in sampling bias, resulting in divergent results when
analyzed using different techniques. The primary purpose of this
study was to share our experience on how to pick up the rare
drop-off of a robust system by rechecking cases with late upward
shifts in the PCR curve, which occur infrequently.
In the prior pilot study, it was demonstrated that a
small subset (6/346) of NSCLC lung cancer patients with low levels
of EGFR mutations may have been misclassified by a commercial
RT-qPCR assay system. The apparent reduction of lesion size or
partial response of a tumor to TKI treatment supports the
application of PNA-sequencing as an important supplementary
technology for patients with a low tumor burden (18–21).
The present study validated the performance and
clinical value of the PNA-sequencing platform. Between September
2012 and February 2018, 34 equivocal cases were encountered in
1,783 patients with lung cancer patients in our institute and 17 of
these (50%) were proven to harbor EGFR mutations by PNA-sequencing.
These patients achieved either partial response or stable disease
(92.8% disease control rate) to TKI treatment, which indicated a
subset of patients may benefit from a more sensitive diagnostic
method.
PNA oligos can be used as a sequence-specific PCR
blocker given its higher binding affinity and specificity to the
target DNA without recognition by DNA polymerase. PNA clamping is
inexpensive and convenient to set up in a clinical diagnostic
laboratory (22,23). In the present cohort study, several
uncommon subtypes of EGFR exon-19 deletions were detected,
including L747_A750delinsP, E746_P753delinsVP, L747_A751del,
E746_P752delinsV, and E746_P751delinsVA. The results confirm that
PNA clamping is useful for the detection of rare alleles at
hotspots of sequence variations.
The Qiagen kits were designed to detect 29 somatic
mutations of the EGFR gene, which covers the majority of the
mutational spectra. Approximately 90% of EGFR mutations are either
exon 19 deletions or exon 21 L858R point mutations (24). Both RUO and IVD kits utilize the
Scorpion ARMS technology and typically produce flat or linear lines
in the cases of negative results. The aim of this investigation was
to share our experience with the ‘drop-off’ of an optimized system.
Evidence that cases with a late upward shift in the PCR curve may
benefit from TKI treatment and thus confer a survival advantage to
these patients was presented here. However, PNA-sequencing
technology has its own inherent weaknesses: i) The method was
designed to detect exon 19 deletions and L858R mutations only,
excluding exons 18 and 20; ii) DNA-based PNA-sequencing depends
solely upon the proportion of non-tumor cells. The non-tumor cells
contain considerably lower EGFR expression levels compared with
EGFR overexpression and mutant DNA in the NSCLC cells. As the
wild-type background signal cannot be completely blocked, the
genomic DNA of non-tumor cells inevitably affects the performance
of PNA sequencing (3). The primary
drawback of PNA sequencing is it is extremely sensitive (<1%)
with a higher risk of contamination when handling the post-PCR
material.
In daily practice, ~75% of EGFR mutation analysis is
performed on small biopsies or cell block specimens. Whether
PNA-sequencing technology should be included in the routine
diagnostic algorithm requires further investigation. However,
clinicians should be cognizant of the possibility of low-level EGFR
mutations in the interpretation of companion diagnostic molecular
tests, since testing accuracy is critical in the selection of
patients for optimal therapy and reducing treatment-related
toxicity.
In conclusion, a small subset of NSCLC patients with
a low tumor burden may be misclassified by currently approved tests
and thus delay starting the correct therapy. The evidence
highlights the benefit of a complementary technique to maximize the
potential of precision medicine. The novelty and significance of
this manuscript is to draw attention to the cases with a late
upward shift in the PCR curve during EGFR mutation analysis. The
complementary technique may be of assistance in the identification
of a small subset of NSCLC patients with a low tumor burden
misclassified by currently approved tests and thus delay starting
the correct therapy. The evidence highlights the benefit of this
strategy to maximize the potential of precision medicine.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Zheng Yumin for
providing useful comments and allowing use of material (Department
of Surgery, E-DA Healthcare group Hospital, Kaohsiung, Taiwan) and
Dr Li Yanqian (Department of Oncology, Tainan Hospital, Ministry of
Health and Welfare, Tainan, Taiwan), Dr Li Yangcheng (Department of
Oncology, Tainan Municipal Hospital, Tainan, Taiwan) and Dr Lai
Junming for allowing to use of material (Department of Internal
Medicine, Dalim, Tzu Chi Hospital, Chiayi, Taiwan).
Funding
This work was supported by research grants from the Ministry of
Science and Technology, Taiwan (grant no. MOST 111-2320-B-006-025),
and from the National Cheng Kung University Hospital, Tainan,
Taiwan (grant nos. NCKUH-11208006 and NCKUH-11208013).
Availability of data and materials
The datasets for this manuscript are not publicly
available due to legal restrictions imposed by the government of
Taiwan in relation to the Personal Information Protection Act.
Requests for access to the datasets should be directed to the
corresponding author.
Authors' contributions
CLH and YLC conceived the study. CCL, SCY, and WCS
collected the samples and designed the study. YTY and WLC performed
the experiments. YLC and WLC confirm the authenticity of all the
raw data. NHC and WH contributed to the data interpretation. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of National Cheng Kung University Hospital, Tainan,
Taiwan. In the study design, NSCLC patients diagnosed at the
National Cheng Kung University Hospital between 2012 and 2018 were
retrospectively collected with concurrent analysis of EGFR status
by Qiagen EGFR kits and PNA-sequencing according to an approved
protocol (approval no. A-ER-108-311).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Midha A, Dearden S and McCormack R: EGFR
mutation incidence in non-small-cell lung cancer of adenocarcinoma
histology: A systematic review and global map by ethnicity
(mutMapII). Am J Cancer Res. 5:2892–2911. 2015.PubMed/NCBI
|
|
2
|
Hsu PC, Jablons DM, Yang CT and You L:
Epidermal growth factor receptor (EGFR) pathway, yes-associated
protein (YAP) and the regulation of programmed death-ligand 1
(PD-L1) in non-small cell lung cancer (NSCLC). Int J Mol Sci.
20:38212019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He J, Huang Z, Han L, Gong Y and Xie C:
Mechanisms and management of 3rd-generation EGFR-TKI resistance in
advanced non-small cell lung cancer (Review). Int J Oncol.
59:902021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gelatti ACZ, Drilon A and Santini FC:
Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in
epidermal growth factor receptor (EGFR) mutation-positive non-small
cell lung cancer (NSCLC). Lung Cancer. 137:113–122. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo Y, Song J, Wang Y, Huang L, Sun L,
Zhao J, Zhang S, Jing W, Ma J and Han C: Concurrent genetic
alterations and other biomarkers predict treatment efficacy of
EGFR-TKIs in EGFR-mutant non-small cell lung cancer: A review.
Front Oncol. 10:6109232020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vallée A, Le Loupp AG and Denis MG:
Efficiency of the Therascreen® RGQ PCR kit for the
detection of EGFR mutations in non-small cell lung carcinomas. Clin
Chim Acta. 429:8–11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thiede C, Bayerdörffer E, Blasczyk R,
Wittig B and Neubauer A: Simple and sensitive detection of
mutations in the ras proto-oncogenes using PNA-mediated PCR
clamping. Nucleic Acids Res. 24:983–984. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sano R, Takahashi Y, Nakajima T, Yoshii M,
Kubo R, Takahashi K, Kominato Y, Takeshita H, Yasuda T, Tsuneyama
H, et al: ABO chimerism with a minor allele detected by the peptide
nucleic acid-mediated polymerase chain reaction clamping method.
Blood Transfus. 12:431–434. 2014.PubMed/NCBI
|
|
9
|
Chen YL, Lee CT, Lu CC, Yang SC, Chen WL,
Lee YC, Yang CH, Peng SL, Su WC, Chow NH and Ho CL: Epidermal
growth factor receptor mutation and anaplastic lymphoma kinase gene
fusion: Detection in malignant pleural effusion by RNA or PNA
analysis. PLoS One. 11:e01581252016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen YL, Lu CC, Yang SC, Su WP, Lin YL,
Chen WL, Huang W, Su WC, Chow NH and Ho CL: Verification of
wild-type EGFR status in non-small cell lung carcinomas using a
mutant-enriched PCR on selected cases. J Mol Diagn. 16:486–494.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rotow J and Bivona TG: Understanding and
targeting resistance mechanisms in NSCLC. Nat Rev Cancer.
17:637–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yeo WL, Riely GJ, Yeap BY, Lau MW, Warner
JL, Bodio K, Huberman MS, Kris MG, Tenen DG, Pao W, et al:
Erlotinib at a dose of 25 mg daily for non-small cell lung cancers
with EGFR mutations. J Thorac Oncol. 5:1048–1053. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tan CS, Gilligan D and Pacey S: Treatment
approaches for EGFR-inhibitor-resistant patients with
non-small-cell lung cancer. Lancet Oncol. 16:e447–e459. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ricciardi S, Tomao S and de Marinis F:
Pemetrexed as first-line therapy for non-squamous non-small cell
lung cancer. Ther Clin Risk Manag. 5:781–787. 2009.PubMed/NCBI
|
|
17
|
Okamoto I, Aoe K, Kato T, Hosomi Y,
Yokoyama A, Imamura F, Kiura K, Hirashima T, Nishio M, Nogami N, et
al: Pemetrexed and carboplatin followed by pemetrexed maintenance
therapy in chemo-naive patients with advanced nonsquamous
non-small-cell lung cancer. Invest New Drugs. 31:1275–1282. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jeon SH, Kim HW, Kim BN, Kang N, Yeo CD,
Park CK, Kim YK, Lee YH, Kim TJ, Lee KY, et al: Comparison of PNA
clamping-assisted fluorescence melting curve analysis and PNA
clamping in detecting EGFR mutations in matched tumor tissue, cell
block, pleural effusion and blood of lung cancer patients with
malignant pleural effusion. In Vivo. 33:595–603. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fouz MF and Appella DH: PNA clamping in
nucleic acid amplification protocols to detect single nucleotide
mutations related to cancer. Molecules. 25:7862020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Billaud A, Verriele V, Dauvé J, Chevalier
LM and Morel A: Non-small-cell lung cancer-sensitive detection of
the p.Thr790Met EGFR alteration by preamplification before
PNA-mediated PCR clamping and pyrosequencing. Diagnostics (Basel).
10:5272020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang S, Chen Z, Huang C, Ding C, Li C,
Chen J, Zhao J and Miao L: Ultrasensitive and quantitative
detection of EGFR mutations in plasma samples from patients with
non-small-cell lung cancer using a dual PNA clamping-mediated
LNA-PNA PCR clamp. Analyst. 144:1718–1724. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song JU and Lee J: Peptide nucleic acid
clamping and direct sequencing in the detection of oncogenic
alterations in lung cancer: Systematic review and meta-analysis.
Yonsei Med J. 59:211–218. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Orum H: PCR clamping. Curr Issues Mol
Biol. 2:27–30. 2000.PubMed/NCBI
|
|
24
|
Zhang T, Wan B, Zhao Y, Li C, Liu H, Lv T,
Zhan P and Song Y: Treatment of uncommon EGFR mutations in
non-small cell lung cancer: New evidence and treatment. Transl Lung
Cancer Res. 8:302–316. 2019. View Article : Google Scholar : PubMed/NCBI
|