Introduction
Remarkable advancements have occurred in the last
few decades regarding cancer diagnostic techniques and treatment
approaches, leading to enhanced patient survival rates and overall
well-being. Nevertheless, cancer remains a substantial cause of
morbidity and mortality worldwide, imposing a considerable strain
on healthcare systems and economies (1). The tumor microenvironment (TME) plays
a crucial function in the initiation and advancement of human
malignancies and encompasses diverse cell types, including a
substantial fraction of infiltrating immune cells (2). The interactions between immune
components and stromal elements in TMEs remain unclear. The
identification of tumor-immune cell interactions was made possible
by the emergence of immune therapeutic vaccines and checkpoint
blockade (3). Immunotherapy has
emerged as a successful method of treating different forms of
cancer by reinvigorating the immune system of the body (4). In contrast to conventional cancer
therapies, immunotherapy utilizes checkpoint-blocking drugs such as
anti-CTLA-4, anti-PD-1 and anti-PD-L1 to treat cancer (5). Consequently, it is imperative to
comprehensively understand various immuno-phenotypes and
authenticate novel therapeutic targets in the realm of cancer
treatment.
Metadherin (MTDH), also known as astrocyte elevated
gene-1 (AEG-1) and lysine-rich CEACAM1 co-isolated, is an integral
protein of 64 kDa. MTDH was initially cloned from primary human
fetal astrocytes as a transcript induced by human immunodeficiency
virus 1 (6). MTDH plays a
significant role in the development and progression of various
cancer types including hepatocellular carcinoma (HCC), breast
cancer (BRCA), prostate cancer, gastric cancer, renal cancer,
colorectal cancer, non-small cell lung cancer, esophageal squamous
cell carcinoma and glioma. Its overexpression has been linked to
promotion of cancer invasion, angiogenesis, autophagy and formation
of metastases. In HCC, MTDH was shown to enhance cell invasion and
migration, leading to increased metastatic potential. In BRCA, MTDH
expression is associated with poor prognosis and decreased overall
survival (OS) rates. In prostate cancer, MTDH promotes
angiogenesis, facilitating the growth and spread of the tumor. In
gastric cancer, MTDH contributes to tumor progression and
resistance to chemotherapy. Overall, the expression of MTDH in
various cancer types is a common factor in promoting cancer
aggressiveness and the formation of metastases. Understanding the
mechanisms by which MTDH influences these processes may provide new
insights for targeted therapies and improving patient outcomes in
these malignancies (7–11). MTDH downregulation leads to a
decrease in cell proliferation and an increase in apoptosis
(12). Conversely, overexpression
of MTDH in invasive BRCA is indicative of a negative prognosis
(13,14). Additionally, MTDH is known to
enhance resistance to both chemotherapy and tamoxifen (15–18).
However, the assessment of MTDH in prior investigations is
currently restricted to only a handful of malignancies; as a
result, the overall clinical implications and biological functions
of MTDH in cancer remain ambiguous and demand additional
elucidation.
The present study aimed to thoroughly examine the
expression pattern of MTDH in various types of cancer by utilizing
publicly available transcriptional and clinical data. Additionally,
the authors performed comprehensive analyses on discrepancies in
mutations, protein levels, prognostic significance and biological
functions associated with MTDH. Furthermore, the relationship
between MTDH and infiltration of immune cells, microsatellite
instability (MSI), tumor mutational burden (TMB), immune-related
genes and immune checkpoint genes in TMEs were assessed. Moreover,
in the present study, the potential of MTDH as an immunotherapy
target for diverse forms of cancer was appraised through the
utilization of immunotherapy cohorts.
Materials and methods
Data source and availability
The possible impact of the MTDH gene on cancer was
investigated by utilization of various databases. The UCSC Xena
database (https://xena.ucsc.edu/) provided RNA
expression and clinical data from The Cancer Genome Atlas (TCGA)
and Genotype-Tissue Expression (GTEx) (19). Information regarding DNA copy number
and methylation was obtained from the cBioPortal database
(https://www.cbioportal.org/). Reactome
database (reactome.org/) was used for enrichment analysis.
CancerSEA (biocc.hrbmu.edu.cn/CancerSEA/home.jsp) was used to
comprehensively analyze MTDH function in pan-cancer at single-cell
resolution. The expression data were standardized by converting
them to log2 (x+0.001). Comparisons of MTDH expression profiles
among different tumor types and adjacent normal tissues were
conducted using TIMER2 (20).
Cell lines and cell culture
To verify the expression of MTDH in BRCA and kidney
cancer, the normal breast cell line MCF10A, the BRCA cell lines
MCF7, BT549 and SK-BR3, the normal kidney cell line 293T, and the
kidney cancer cell ACHN and 786-O were obtained from Procell Life
Science & Technology Co., Ltd. (https://www.procell.com.cn/). The cell lines were
maintained in Dulbecco's Modified Eagle's Medium supplemented with
10% fetal bovine serum (Procell Life Science & Technology Co.,
Ltd.) and 1% penicillin-streptomycin at 37°C in a 5% CO2
atmosphere.
Collection of pathological
samples
Between August 2021 and April 2023, a collective of
20 BC tissues, 18 kidney cancer tissues, along with their
respective normal tissue samples, were obtained from Xingtai
People's Hospital (Xingtai, China). The breast cancer samples were
all female, aged 28–65 years, while the kidney cancer patients
included 11 men and 7 women, aged 36–68 years. Tissue samples were
frozen in liquid nitrogen and stored in a refrigerator at −80°C.
Tissue sections were 7 µm thick. Approval for the current
investigation was obtained from the Medical Ethics Committee of
Xingtai People's Hospital [approval no. 2021(031)] and the research
was conducted following the guidelines of the Declaration of
Helsinki (as revised in 2013). Written informed consent was
obtained from all patients.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Cells and tissue were used to isolate total RNA with
the TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) as per the manufacturer's instructions. The
first-strand cDNA was synthesized using the Takara PrimeScript RT
reagent kit (Takara Bio, Inc.) according to the manufacturer's
instructions. The RT-qPCR assay was carried out using the SYBR
Premix Ex Taq (Takara Bio, Inc.) following the manufacturer's
protocol. The following primer pairs were used for qPCR: MTDH
forward, 5′-AAATGGGCGGACTGTTGAAGT-3′ and reverse,
5′-CTGTTTTGCACTGCTTTAGCAT-3′; and GAPDH forward
5′-GTCTCCTCTGACTTCAACAGCG-3′ and reverse
5′-ACCACCCTGTTGCTGTAGCCAA-3′. GAPDH was utilized as a reference for
relative quantification in the experiment. The following
thermocycling conditions were used for qPCR: Initial denaturation
at 95°C for 5 min; followed by 40 cycles of denaturation at 95°C
for 5 sec, 60°C for 30 sec and 72°C for 30 sec, with a final
extension step at 72°C for 2 min. The relative mRNA expression was
calculated using the comparative cycle threshold
(2-ΔΔCq) method (21).
Protein level analysis of MTDH in
multiple cancers
The Human Protein Atlas (HPA) database (https://www.proteinatlas.org/) was employed to examine
the protein levels of MTDH in both human tumor and normal tissues.
Additionally, the string database (https://string-db.org/) was utilized to construct the
protein-protein interaction (PPI) network associated with MTDH
(22). Furthermore, the Metascape
(https://metascape.org/) database was utilized for
conducting Gene Ontology (GO) enrichment analysis (23).
Evaluation of genetic alterations in
MTDH
TMB was calculated using Perl scripts based on the
total number of somatic mutations per million bases. MSI scores
were calculated based on DNA-seq data from TCGA (https://www.cancer.gov/ccg/research/genome-sequencing/tcga).
The correlation between MTDH expression and TMB or MSI was assessed
using Spearman's test by utilizing the ‘cor.test’ tool package of R
software (https://www.r-project.org; v.3.6.3).
Radar plots showing the correlations were created using the radar
chart function of the ‘fmsb’
(cran.r-project.org/web/packages/fmsb/index.html) package in R.
Relationship between MTDH expression
and survival prognosis
To analyze the link between survival outcomes and
MTDH mRNA expression levels, the present analysis employed both the
Kaplan-Meier analysis and the Cox proportional hazards model. The
‘maxstat’ (https://cran.r-project.org/web/packages/maxstat/)
and ‘survival’ (https://cran.r-project.org/web/packages/survival/) R
packages (24) were utilized for
this investigation. To establish the most suitable thresholds, the
‘maxstat’ R package was applied for the computation. The optimal
thresholds were determined using the ‘maxstat’ R package.
Tumor immune microenvironment and MTDH
expression
Data from genes linked to various immune-related
pathways such as chemokines, receptors, MHC, immunosuppressants,
immuno-stimulants, as well as immune checkpoint pathways were
extracted from each cancer sample. The gene expression data was
then used to calculate the tumor stroma score of each patient using
the ‘ESTIMATE’ R package (17).
Additionally, infiltration scores of immune-related cells in
patients were evaluated using the EPIC, Timer and quanTIseq methods
from the ‘IOBR’ R package (25).
Statistical analysis
Data were analyzed using SPSS 23.0 (IBM Corp.) and
GraphPad Prism 8 (GraphPad Software, Inc.). Pearson correlation
coefficients were utilized to conduct the correlation analysis
between MTDH and all genes based on TCGA data. Subsequently,
MTDH-correlated genes were selected for gene set enrichment
analysis (12). To make group
comparisons, unpaired Student's t-test, paired Student's t-test,
Mann-Whitney U-test, or one-way ANOVA were employed. One-way ANOVA
was followed by Bonferroni's post hoc test. Each experiment was
replicated thrice and the data are presented as the mean ± standard
deviation. To determine the relationship between MTDH expression
levels and patient survival, univariate Cox analysis and
Kaplan-Meier (KM) plotter was used, followed by the log-rank test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Gene expression of MTDH
TIMER 2.0 analysis revealed that the expression
levels of MTDH were significantly increased in BRCA,
cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal
carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC),
kidney renal clear cell carcinoma (KIRC), liver hepatocellular
carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell
carcinoma (LUSC), rectum adenocarcinoma (READ) and stomach
adenocarcinoma (STAD). By contrast, the expression of MTDH was
significantly lower in thyroid cancer (THCA) and uterine corpus
endometrial carcinoma (UCEC) compared with normal controls
(Fig. 1A).
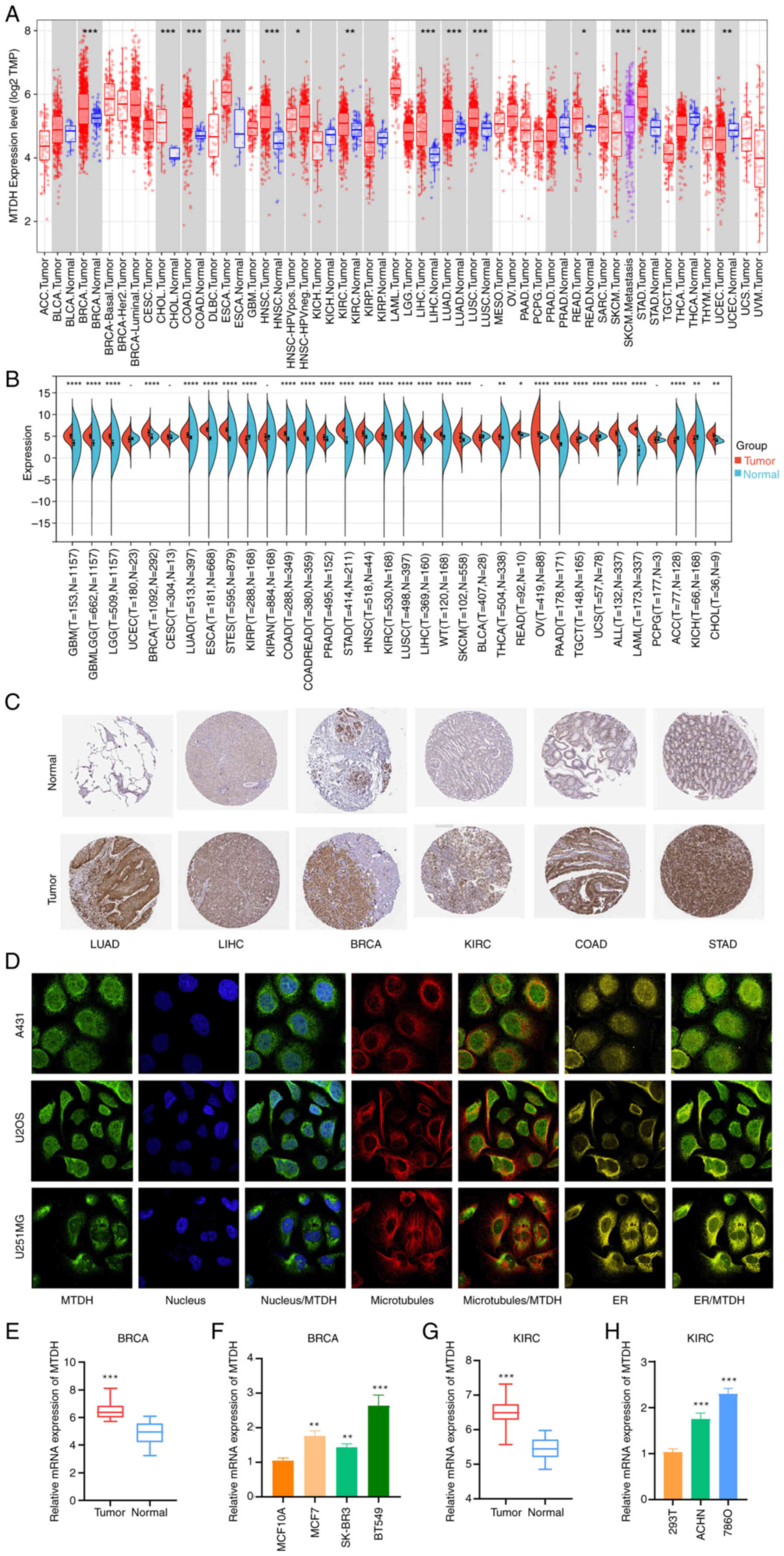 | Figure 1.Differential expression of MTDH in
pan-cancer. (A) TIMER shows the level of MTDH expression in The
Cancer Genome Atlas tumors and nearby tissues, if available. (B)
Expression of MTDH in normal and cancerous tissues. (C)
Representative immunohistochemistry images from the HPA database
showing MTDH protein expression in LUAD, LIHC, BRCA, KIRC, COAD and
STAD tumor and normal tissues. (D) Immunofluorescence staining of
the subcellular location of MTDH in the HPA database. (E)
Comparison of MTDH mRNA expression levels between normal breast
tissue and tumor tissues. (F) Relative expression levels of MTDH
mRNA in BRCA cells and a normal breast cell line. (G) Comparison of
MTDH mRNA expression levels between normal kidney tissue and tumor
tissues. (H) Relative expression levels of MTDH mRNA in KIRC cells
and a normal kidney cell line. *P<0.05, **P<0.01 and
***P<0.001, ****P<0.0005 vs. normal. MTDH, metadherin; HPA,
Human Protein Atlas; LUAD, lung adenocarcinoma; LIHC, liver
hepatocellular carcinoma; BRCA, breast cancer; KIRC, kidney renal
clear cell carcinoma; COAD, colon adenocarcinoma; STAD, stomach
adenocarcinoma; ER, endoplasmic reticulum. |
After including the normal tissues of the GTEx
dataset as controls, the difference in MTDH expression between
normal and tumor tissues was further evaluated. It was found that
glioblastoma multiforme (GBM), lower grade glioma (LGG), GBMLGG,
BRCA, LUAD, ESCA, stomach and esophageal carcinoma (STES), COAD,
COADREAD, prostate adenocarcinoma (PRAD), STAD, HNSC, KIRC, LUSC,
LIHC, Wilms' tumor, skin cutaneous melanoma (SKCM), THCA, READ,
ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma
(PAAD), acute lymphoblastic leukemia, acute myeloid leukemia (LAML)
and CHOL showed higher expression in the tumor tissues. In contrast
with that in the control tissues, MTDH expression was decreased in
tenosynovial giant cell tumor (TGCT), uterine carcinosarcoma (UCS),
adrenocortical cancer (ACC) and kidney chromophobe (KICH) tissues
(Fig. 1B; P<0.05).
Immunohistochemistry images for MTDH protein
expression in tumor and normal tissues were extracted from the HPA
database and analyzed. MTDH protein was overexpressed in LUAD,
LIHC, BRCA, KIRC, COAD and STAD, suggesting that MTDH might play an
oncogenic role in the development of these types of cancers
(Fig. 1C).
MTDH subcellular localization was obtained by
immunofluorescence localization of the nuclei, microtubules and
endoplasmic reticulum in A-431, U-2OS and U-251 MG cells. MTDH was
located not only in microtubules and cytoplasm but also in the
nuclei (Fig. 1D). RT-qPCR analysis
revealed that in both BRCA and KIRC, the expression of MTDH was
higher in cancerous tissues or cells than in their corresponding
normal tissues or cells (Fig.
1E-H).
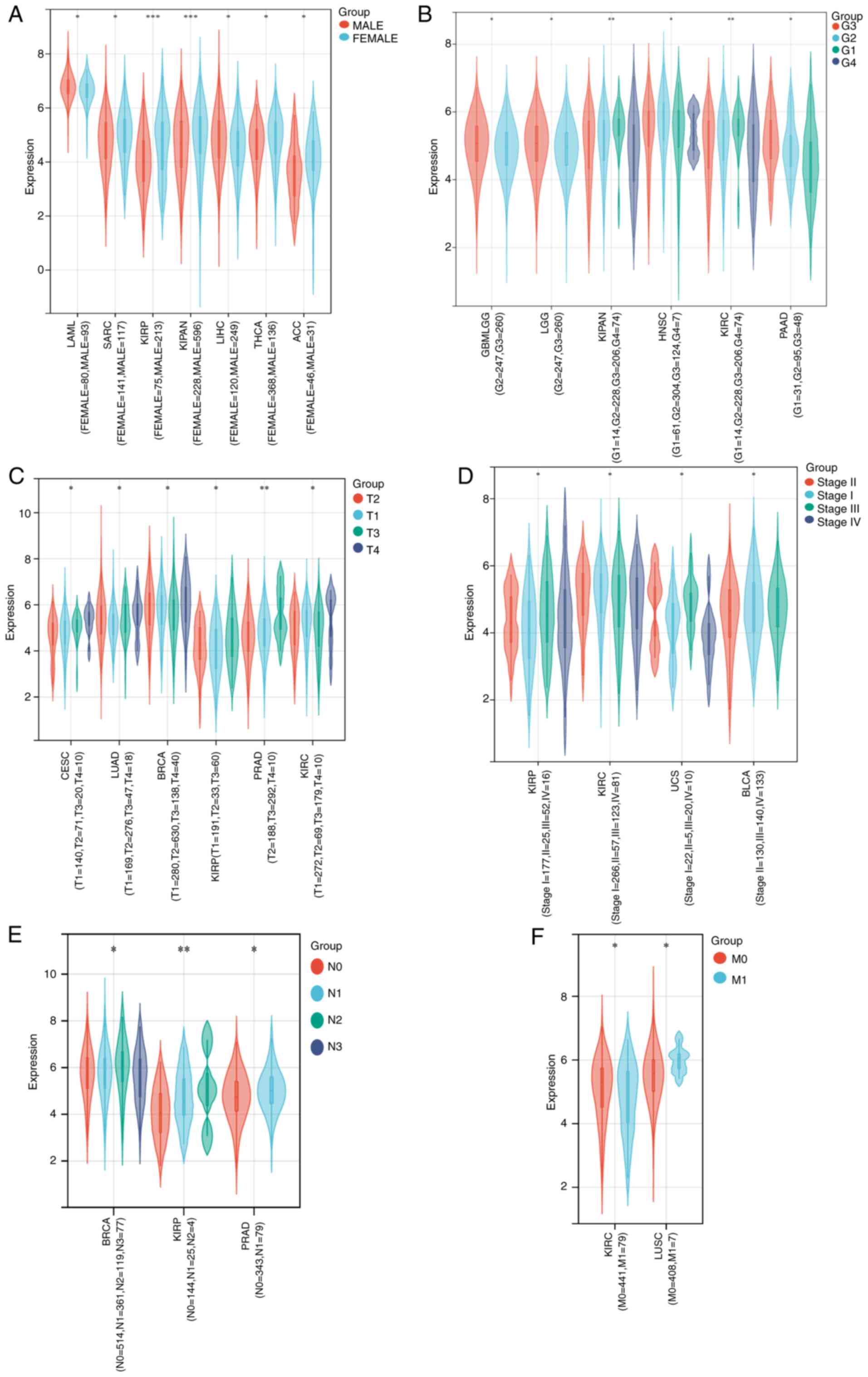
Associations between MTDH expression
and clinicopathologic variables
As illustrated in Fig.
2, high expression of MTDH was significantly associated with
sex in LAML (P=0.02), SARC (P=0.03), kidney renal papillary cell
carcinoma (KIRP; P=0.00012), KICH + KIRC + KIRP (KIPAN)
(P=0.00036), LIHC (P=0.02), THCA (P=0.02) and ACC (P=0.04)
(Fig. 2A); histological grade in
GBMLGG (P=0.04), LGG (P=0.04), KIPAN (P=0.0094), HNSC (P=0.03),
KIRC (P=0.0094) and PAAD (P=0.01) (Fig.
2B); tumor stage in cervical squamous cell carcinoma and
endocervical adenocarcinoma (CESC; P=0.03), LUAD (P=0.02), BRCA
(P=0.03), KIRP (P=0.01), PRAD (P=0.0011) and KIRC (P=0.01)
(Fig. 2C); pathologic stage in KIRP
(P=0.05), KIRC (P=0.03), UCS (P=0.01) and bladder urothelial
carcinoma (BLCA; P=0.02) (Fig. 2D);
N stage in BRCA (P=0.01), KIRP (P=0.0071) and PRAD (P=0.01)
(Fig. 2E); and M stage in KIRC
(P=0.04) and LUSC (P=0.02) (Fig.
2F).
DNA methylation analysis of MTDH
The present study conducted a comparison between the
methylation levels of the MTDH promoter in normal tissues and
primary tumor tissues. By utilizing TCGA dataset, 12 tumor types
[BLCA, BRCA, LUAD, THCA, HNSC, KIRC, sarcoma (SARC), LIHC, LUSC,
PAAD, PRAD and UCEC] were analyzed as depicted in Fig. 3A. The present analysis discovered
notable differences in methylation levels within the MTDH promoter
across various types of tumors and their corresponding
non-cancerous tissues. Significantly elevated methylation levels
were observed in SARC, PAAD, LUSC and KIRC tumor samples compared
with their respective normal tissue counterparts. On the other
hand, the methylation level of the MTDH promoter was higher in
normal tissues compared with tumor tissues in LIHC, LUAD, HNSC,
BLCA, BRCA, UCEC, THCA and PRAD. The P-values for all the
aforementioned comparisons were P<0.05. Subsequently, a detailed
analysis of various typical RNA methylation patterns of the MTDH
gene was conducted using R software, as demonstrated in Fig. 3B. ACC, OV, BRCA and GBM revealed a
positive correlation between MTDH expression and prevalent RNA
methylation types including M6A, M5C and M1A.
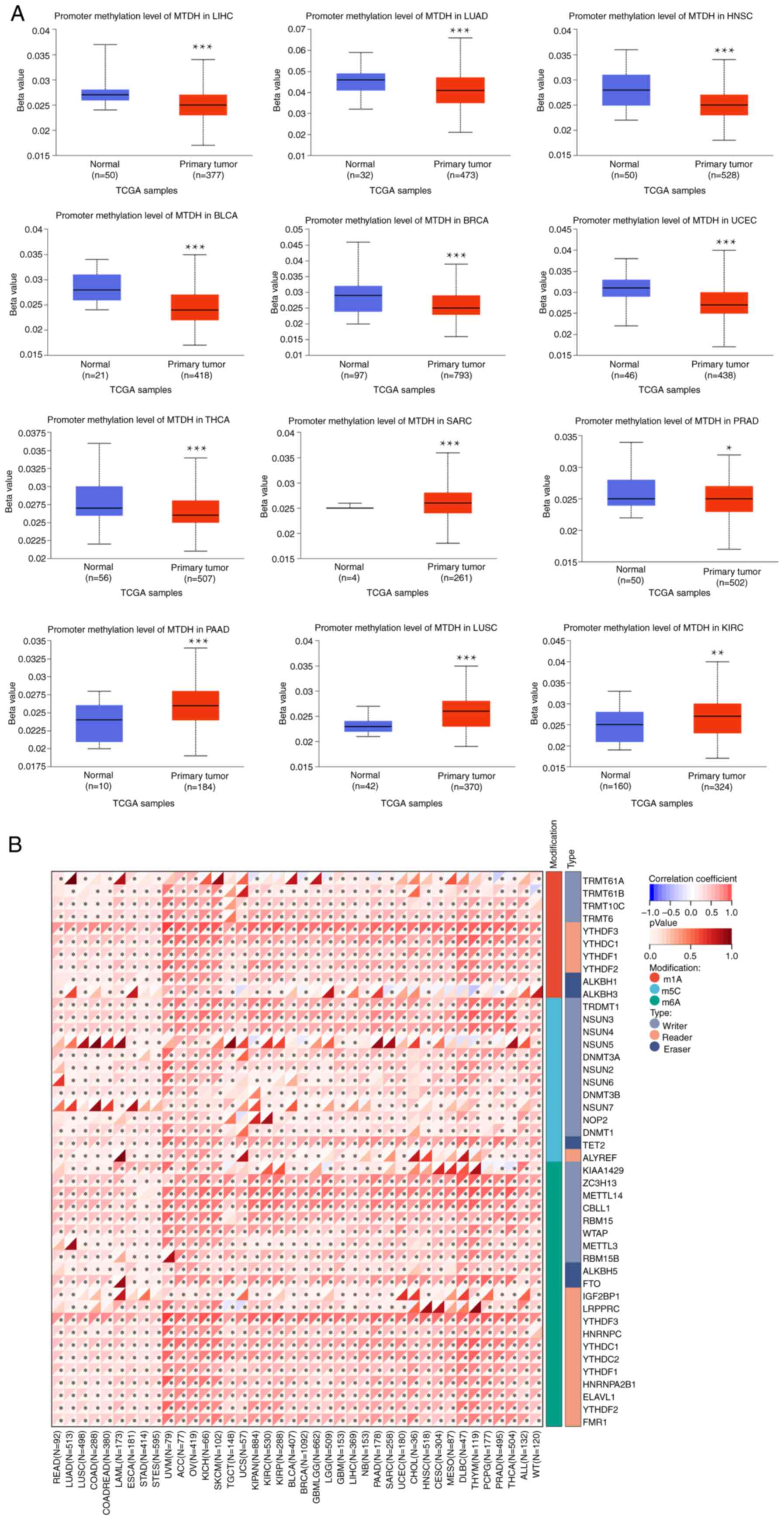 | Figure 3.Association between MTDH with
methylation and methyltransferase. (A) The promoter methylation
level of MTDH in LIHC, LUAD, HNSC, BLCA, BRCA, UCEC, THCA, SARC,
PRAD, PAAD, LUSC and KIRC. (B) The correlation between MTDH
expression and m1A, m5C and m6A regulatory genes. *P<0.05 and
***P<0.001. MTDH, metadherin; LIHC, liver hepatocellular
carcinoma; LUAD, lung adenocarcinoma; HNSC, head and neck squamous
cell carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast
cancer; UCEC, uterine corpus endometrial carcinoma; THCA, thyroid
cancer; SARC, sarcoma; PRAD, prostate adenocarcinoma; PAAD,
pancreatic adenocarcinoma; LUSC, lung squamous cell carcinoma;
KIRC, kidney renal clear cell carcinoma. |
Relationship between MTDH expression
and prognosis in multiple cancers
To thoroughly evaluate the association between MTDH
expression and prognosis in patients with cancer, the correlation
of MTDH with survival-related factors such as OS, progression-free
survival and disease-specific survival (DSS) was examined across 33
different types of cancer. This analysis was conducted through the
use of univariate Cox analysis and KM techniques.
OS
The present study demonstrated significant
associations between MTDH expression and OS for 6 types of cancer,
including GBMLGG [P<0.001, hazard ratio (HR)=1.735], PAAD
(P=0.003, HR=1.857), BRCA (P=0.002, HR=1.687), LGG (P<0.001,
HR=1.84), KICH (P=0.005, HR=10.296) and KIRC (P=0.01, HR=0.676)
(Fig. 4A). Kaplan-Meier OS curves
demonstrated a significant positive association between OS and low
expression of MTDH in GBMLGG (P<0.001), PAAD (P=0.004), BRCA
(P=0.002), LGG (P<0.001) and KICH (P=0.028); however, a
significant negative association was observed between OS and low
expression of MTDH in KIRC (P=0.011) (Fig. 4B).
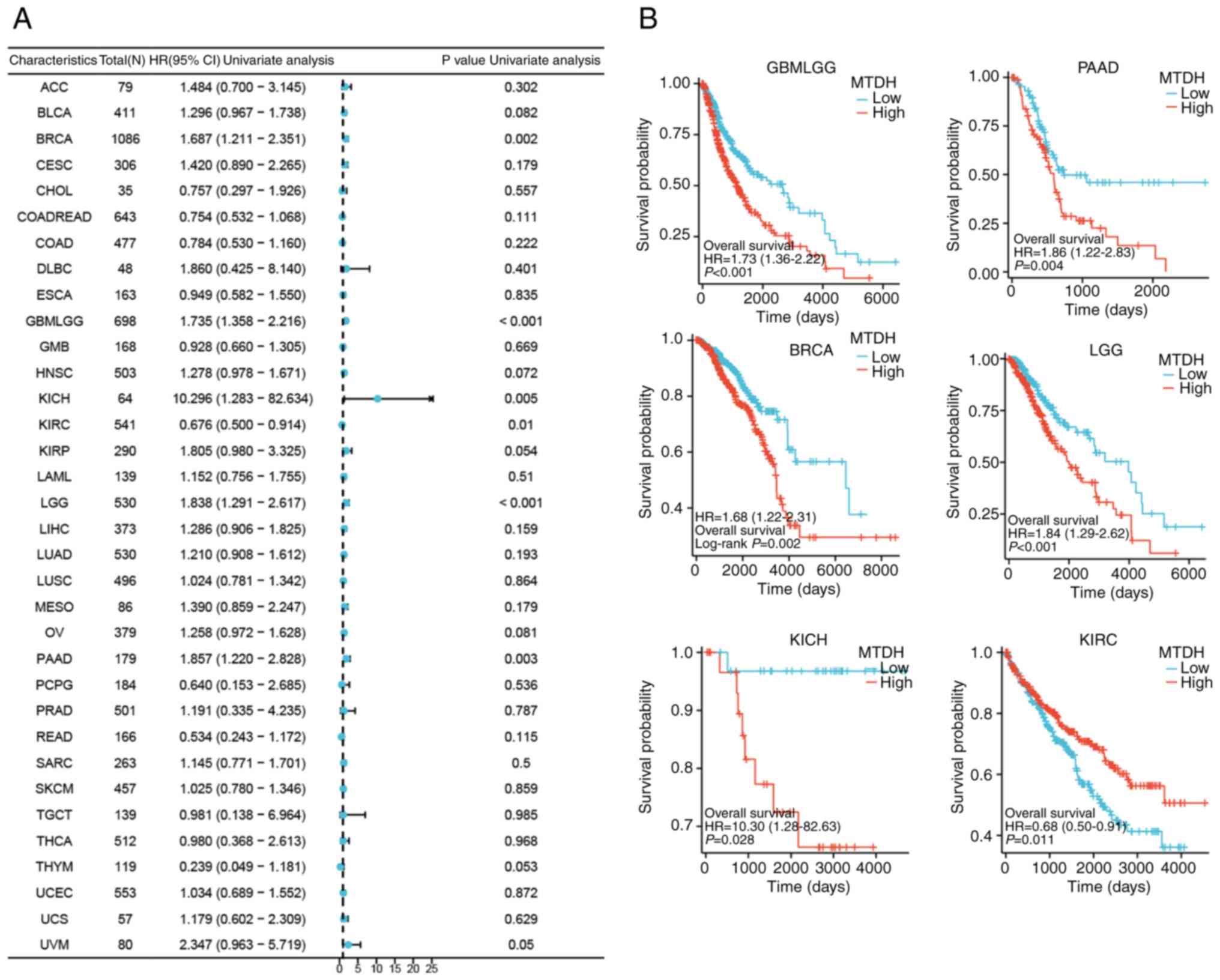 | Figure 4.Association between the expression
level of MTDH and OS. (A) Forest plots showing the relationship
between the expression of MTDH and OS in 33 tumor types from The
Cancer Genome Atlas. (B) Kaplan-Meier curves showing the
association between the expression level of MTDH and OS in KIRC,
PAAD, BRCA, LGG, GBMLGG and KICH. MTDH, metadherin; OS, overall
survival; KIRC, kidney renal clear cell carcinoma; PAAD, pancreatic
adenocarcinoma; BRCA, breast cancer; LGG, lower grade glioma; GBM,
glioblastoma multiforme; KICH, kidney chromophobe; HR, hazard
ratio; CI, confidence interval. |
DSS
MTDH expression was significantly associated with
DSS in BLCA (P=0.017, HR=1.541), BRCA (P=0.042, HR=1.565), COADREAD
(P=0.018, HR=0.581), COAD (P=0.037, HR=0.588), GBMLGG (P<0.001,
HR=1.813), KIRC (P=0.013, HR=0.615), LGG (P<0.001, HR=1.869),
mesothelioma (MESO; P=0.02, HR=2.202) PAAD (P=0.016, HR=1.784) and
uveal melanoma (UVM; P=0.049, HR=2.597) (Fig. 5A). Kaplan-Meier curves of DSS
demonstrated that high expression of MTDH was significantly
associated with a favorable prognosis in COAD (P=0.040), COADREAD
(P=0.020) and KIRC (P=0.013), and was significantly associated with
unfavorable prognosis in PAAD (P=0.016), MESO (P=0.020), GBMLGG
(P<0.001), LGG (P<0.001), BRCA (P=0.045), BLCA (P=0.018) and
UVM (P=0.049) (Fig. 5B).
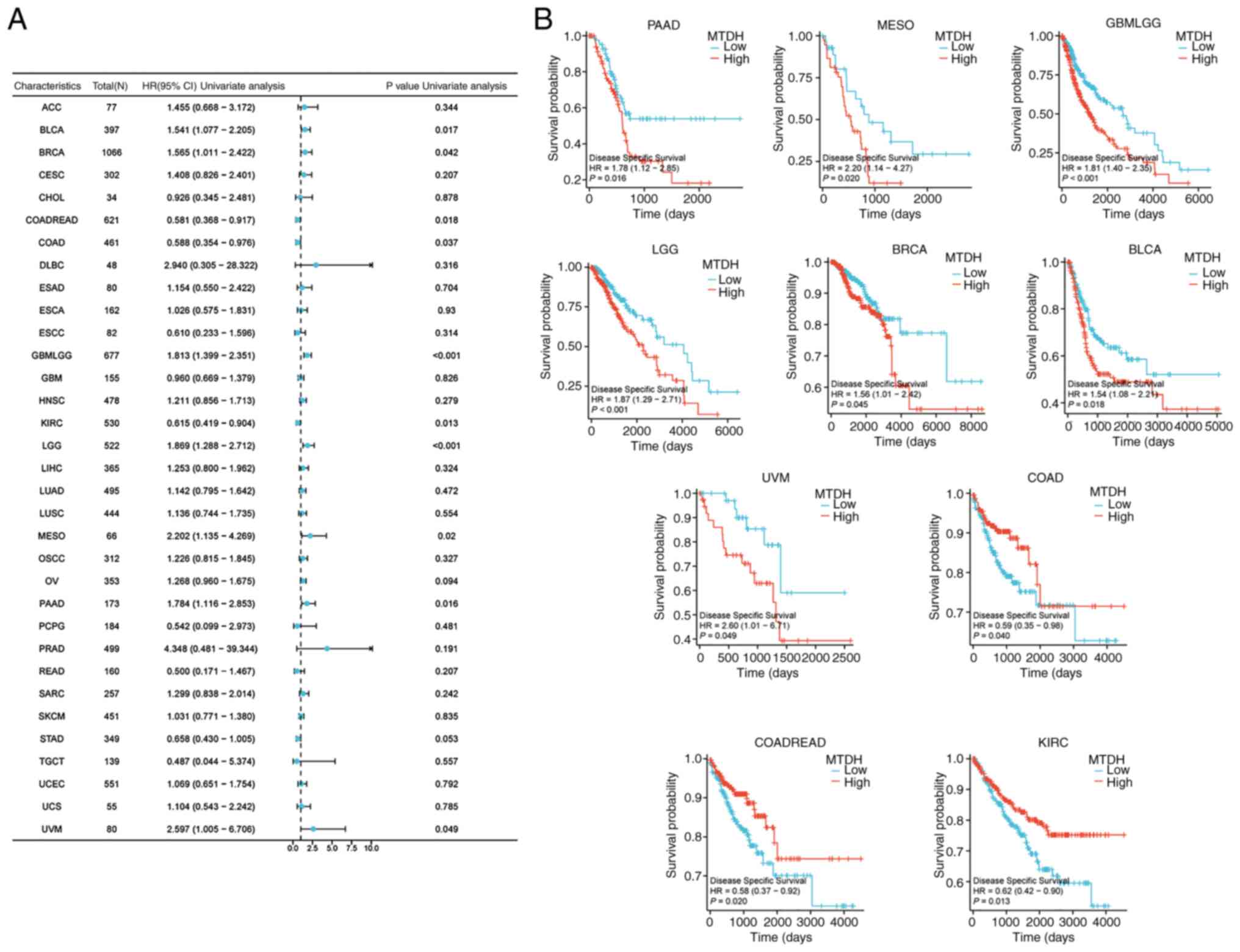 | Figure 5.Association between the expression of
MTDH and DSS. (A) Forest plots showing the relationship between the
expression of MTDH and DSS in 33 tumor types from The Cancer Genome
Atlas. (B) Kaplan-Meier curves demonstrating the association
between the expression of MTDH and DSS in PAAD, MESO, GBMLGG, LGG,
BRCA, BLCA, UVM, COAD, COADREAD and KIRC. MTDH, metadherin; DSS,
disease-specific survival; PAAD, pancreatic adenocarcinoma; MESO,
mesothelioma; GBM, glioblastoma multiforme; LGG, lower grade
glioma; BRCA, breast cancer; BLCA, bladder urothelial carcinoma;
UVM, uveal melanoma; COAD, colon adenocarcinoma; READ, rectum
adenocarcinoma; KIRC, kidney renal clear cell carcinoma; HR, hazard
ratio; CI, confidence interval. |
Progress free interval (PFI)
Furthermore, the present study demonstrated
significant associations between the expression level of MTDH and
PFI in 10 types of cancer: ACC (P=0.024, HR=2.075), BLCA (P=0.006,
HR=1.516), CESC (P=0.042, HR=1.636), GBMLGG (P<0.001, HR=1.616),
KICH (P=0.021, HR=6.168), KIRP (P=0.012, HR=1.998), LGG (P=0.013,
HR=1.418), PAAD (P=0.014, HR=1.628), STAD (P=0.006, HR=0.600) and
UVM (P=0.042, HR=2.229) (Fig. 6A).
Kaplan-Meier curves demonstrated that higher expression level of
MTDH was significantly associated with poor PFI in ACC (P=0.024),
UVM (P=0.042), BLCA (P=0.006), PAAD (P=0.014), LGG (0.013), KIRP
(P=0.012), KICH (P=0.021), GBMLGG (P<0.001) and CESC (P=0.042)
(Fig. 6B).
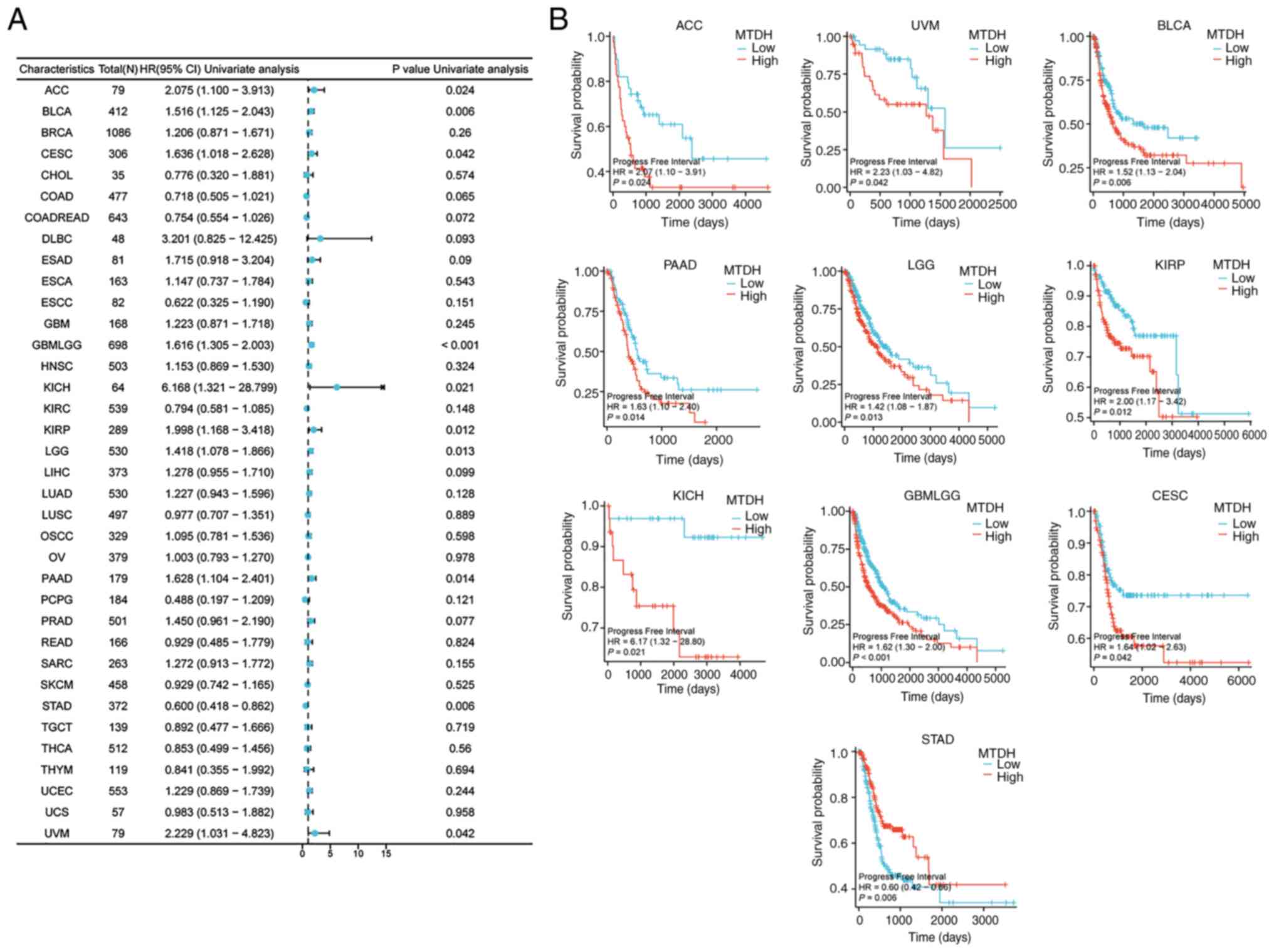 | Figure 6.Association between the expression of
MTDH and PFI. (A) Forest plots showing the relationship between
MTDH expression and PFI in 33 tumor types from The Cancer Genome
Atlas. (B) Kaplan-Meier curves showing the correlation between MTDH
expression and PFI in ACC, UVM, BLCA, PAAD, LGG, KIRP, KICH,
GBMLGG, CESC and STAD. MTDH, metadherin; PFI, progression-free
interval; ACC, adrenocortical cancer; UVM, uveal melanoma; BLCA,
bladder urothelial carcinoma; PAAD, pancreatic adenocarcinoma; LGG,
lower grade glioma; KIRP, kidney renal papillary cell carcinoma;
KICH, kidney chromophobe; GBM, glioblastoma multiforme; CESC,
cervical squamous cell carcinoma and endocervical adenocarcinoma;
STAD, stomach adenocarcinoma. |
Summary of patient prognosis
indicators
The results collectively demonstrated a notable
association between expression of MTDH and prognosis in different
cancer categories, such as BLCA, BRCA, LGG and UVM. These findings
support a possible utility of MTDH as a biomarker for forecasting
patient outcomes.
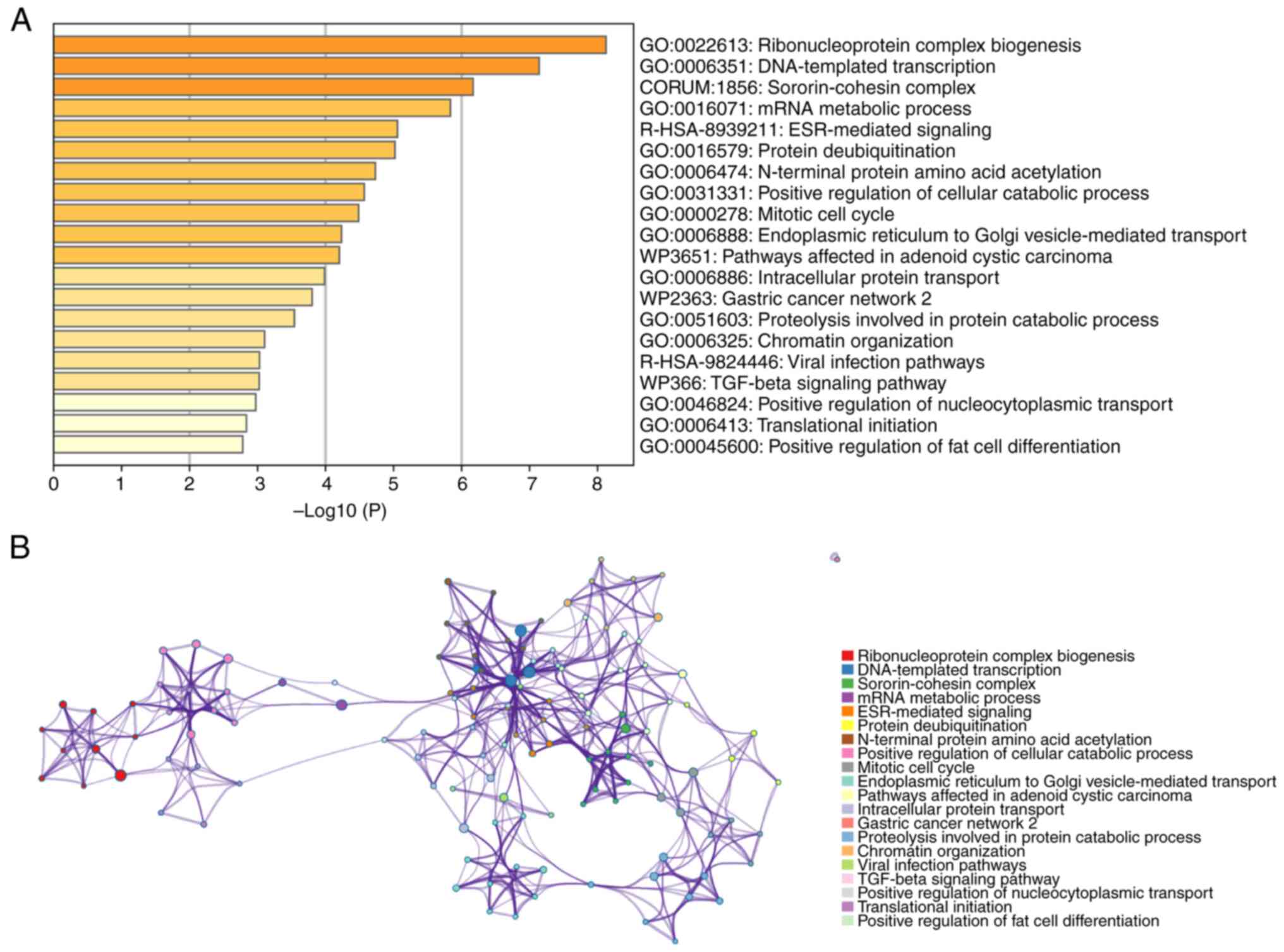
Functional enrichment analysis of MTDH
in pan-cancer
To gain an improved understanding of the functions
and mechanisms of MTDH and the 100 central genes, Metascape was
utilized to analyze GO biological processes (BP) and Reactome gene
sets. The findings indicated that MTDH and its adjacent genes are
primarily associated with biological processes such as assembly of
ribonucleoprotein complexes, DNA-directed transcription and
processing of mRNA. The Reactome pathways that these genes partake
in include ESR-mediated signaling, pathways impacted by adenoid
cystic carcinoma and viral infection pathways (Fig. 7A). Additionally, to explore the
connection between MTDH and various types of cancer, a PPI network
was established using data sourced from the Metascape online
platform (Fig. 7B).
Expression pattern of MTDH at
single-cell levels
The analysis of candidate molecules' functions at
single-cell levels is crucial and can be achieved using single-cell
transcriptome sequencing (25,26).
Subsequently, the association between the expression of MTDH and 14
functional states of cancer was examined using single-cell
sequencing data from CancerSEA (biocc.hrbmu.edu.cn/CancerSEA/).
Positive associations with metastasis were observed in most types
of tumors with MTDH expression (Fig.
8A). In addition, Fig. 8B
displayed the significant correlation between the expression level
of MTDH and differentiation in metastasis, differentiation,
proliferation, inflammation, epithelial-to-mesenchymal transition
(EMT) and angiogenesis in AML, invasion in OV, proliferation in RCC
and stemness and EMT in prostate cancer. While in BC, the
expression of MTDH was positively related to apoptosis, DNA repair
and DNA damage (Fig. 8B).
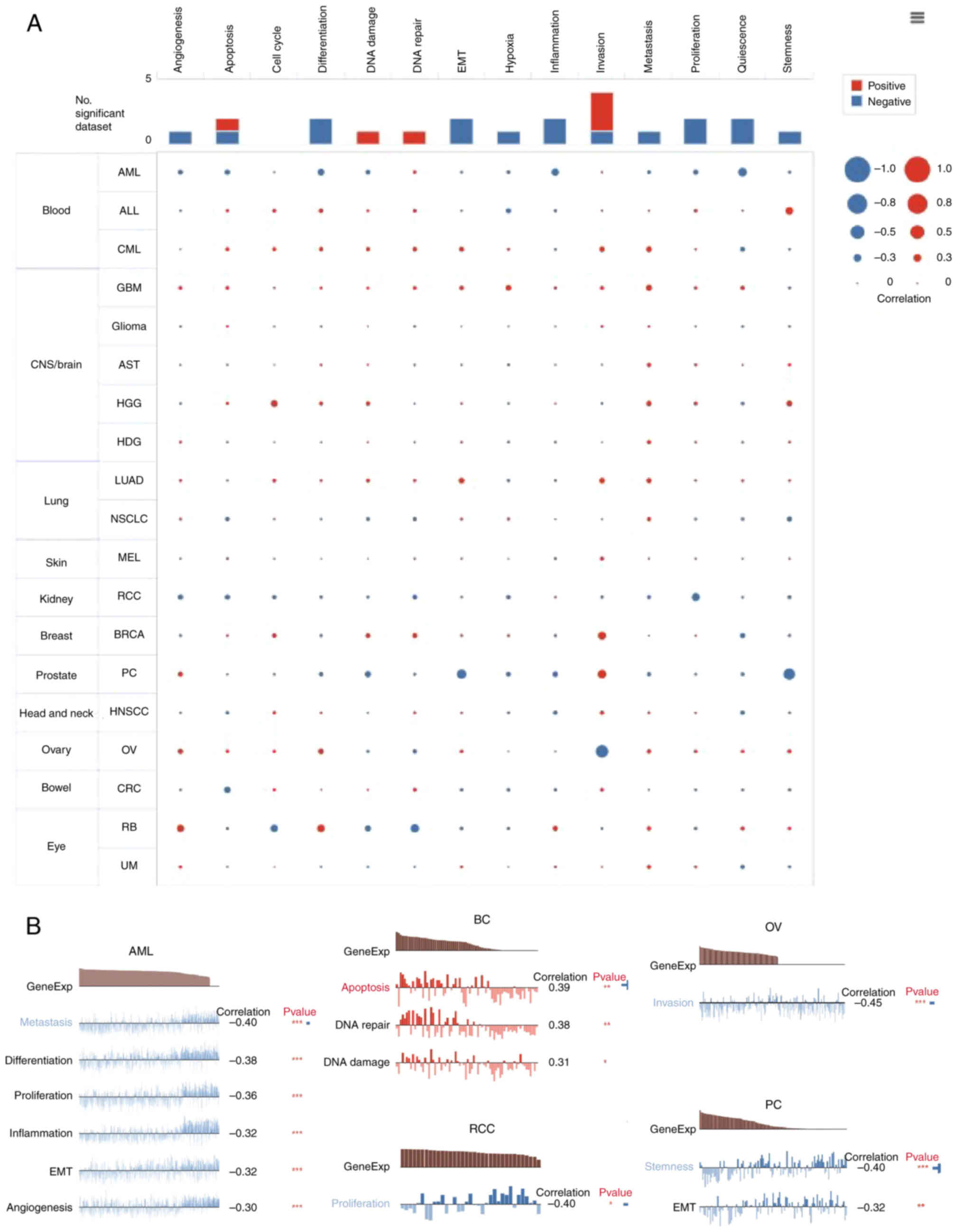 | Figure 8.Function of MTDH in single-cell
functional analysis from the CancerSEA database. (A) Functional
status of MTDH in different human cancers. (B) Correlation analysis
between functional status and MTDH in AML, BC, RCC, OV and PC.
*P<0.05, **P<0.01 and ***P<0.001. MTDH, metadherin; AML,
acute myeloid leukemia; BC, breast cancer; RCC, renal cell
carcinoma; OV, ovarian serous cystadenocarcinoma; PC, prostate
cancer. |
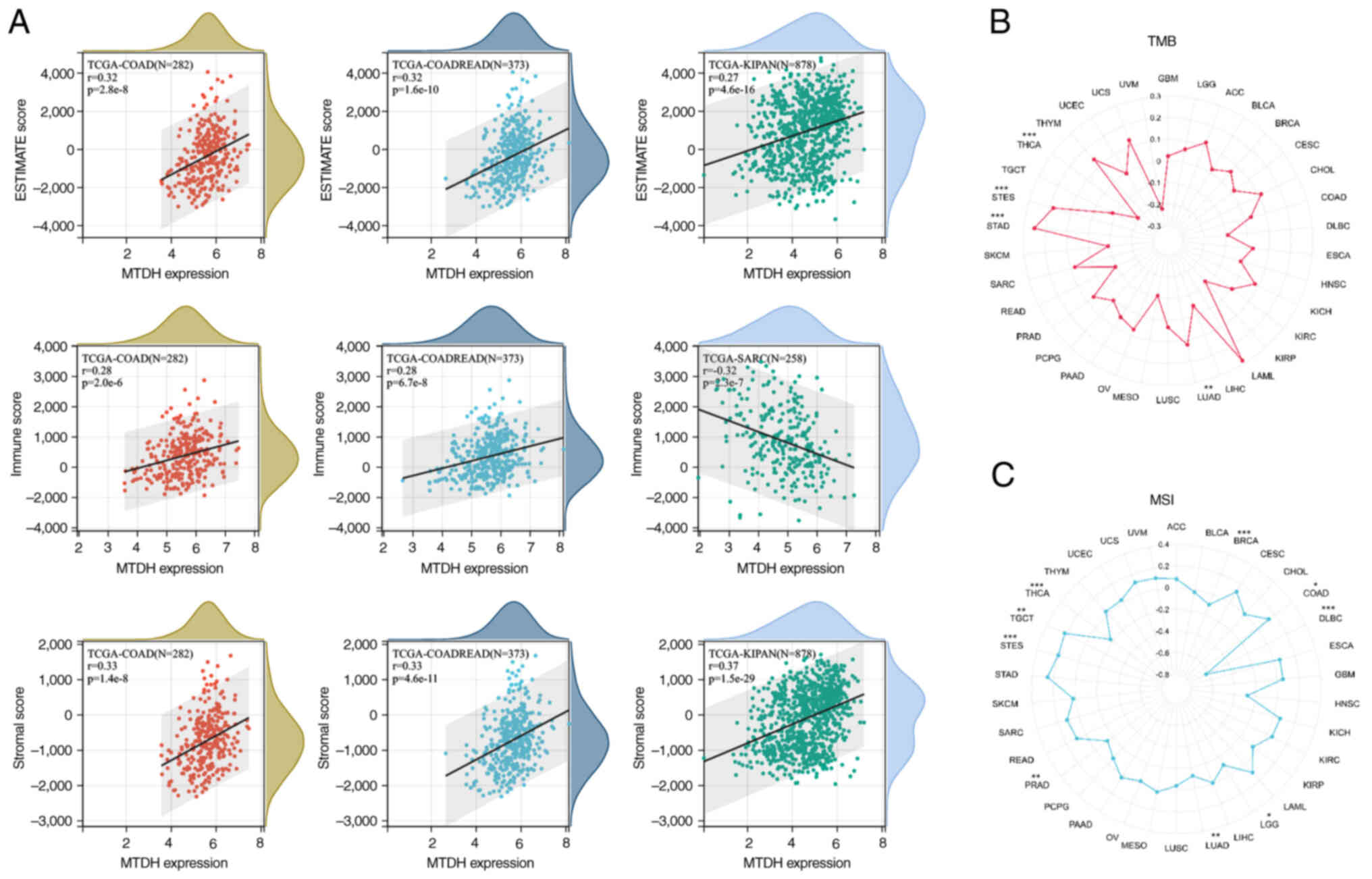
Correlation analysis on TMB and
MSI
The present study investigated the correlation of
TMB/MSI with MTDH expression. The findings demonstrated that there
was a significant positive correlation of MTDH expression with TMB
in LUAD (P=0.006), STAD (P<0.001) and STES (P<0.001), while a
significant negative correlation in THCA (P<0.001) was observed
(Fig. 9B). Moreover, it was found
that MTDH expression was positively correlated with MSI in COAD
(P=0.022), STES (P<0.001) and TGCT (P=0.008) and negatively
correlated with MSI in BRCA (P=0.001), diffuse large B cell
lymphoma (P<0.001), LUAD (P=0.007), PRAD (P=0.002) and THCA
(P<0.001) (Fig. 9C).
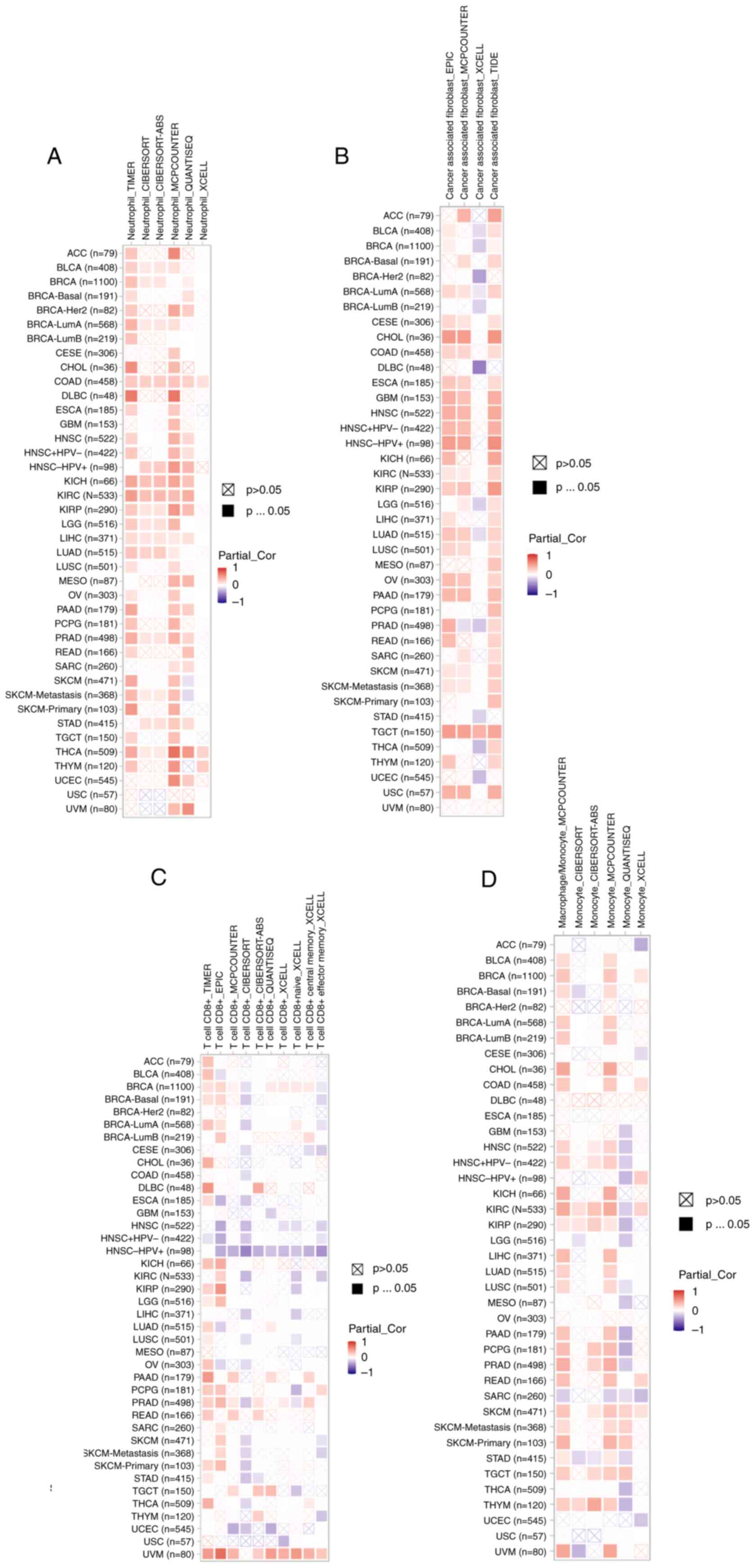
Roles of MTDH on the regulation of
immune cell infiltration
According to recent studies, it has been proven that
immune infiltration plays a crucial role in the onset, advancement
and spread of human malignancies (27–30).
Numerous computational models, including TIMER, EPIC, QUANTISEQ,
XCELL, MCPCOUNTER, CIBERSORT, CIBERSORT-ABS and TIDE, were employed
to investigate the association between MTDH expression and the
infiltration of diverse immune cell populations across various
cancer types. Remarkably, the present study uncovered a significant
positive association between neutrophil infiltration and MTDH
expression specifically in cases of COAD and THCA (Fig. 10A). In TGCT, a strong relationship
was observed between the presence of cancer-associated fibroblasts
and MTDH expression (Fig. 10B).
Furthermore, CD8(+) T cell infiltration in UVM showed a positive
correlation with MTDH expression (Fig.
10C). Additionally, in SKCM, macrophage presence was linked to
higher levels of MTDH expression (Fig.
10D). These results suggested that MTDH could serve as a
promising immune-related indicator for tumor progression.
Pearson's analysis of MTDH expression
and functioning of genes in immune regulation and immune
checkpoints
To demonstrate the potential connections between
MTDH expression and immune status within tumors, an investigation
was carried out analyzing immune-related genes and immune
infiltration patterns in the TME. The aim was to examine the impact
of MTDH on various cancers from an immunological standpoint. The
data from Fig. 11A and B revealed
a correlation between MTDH expression and a wide range of
immunoregulatory and checkpoint genes in UVM, OV, READ, KIPAN and
KIRC. Interestingly, even in PRAD, which is typically considered a
‘cold’ tumor with low immune activity and limited response to
immunotherapy, MTDH expression showed significant associations with
immune-related genes.
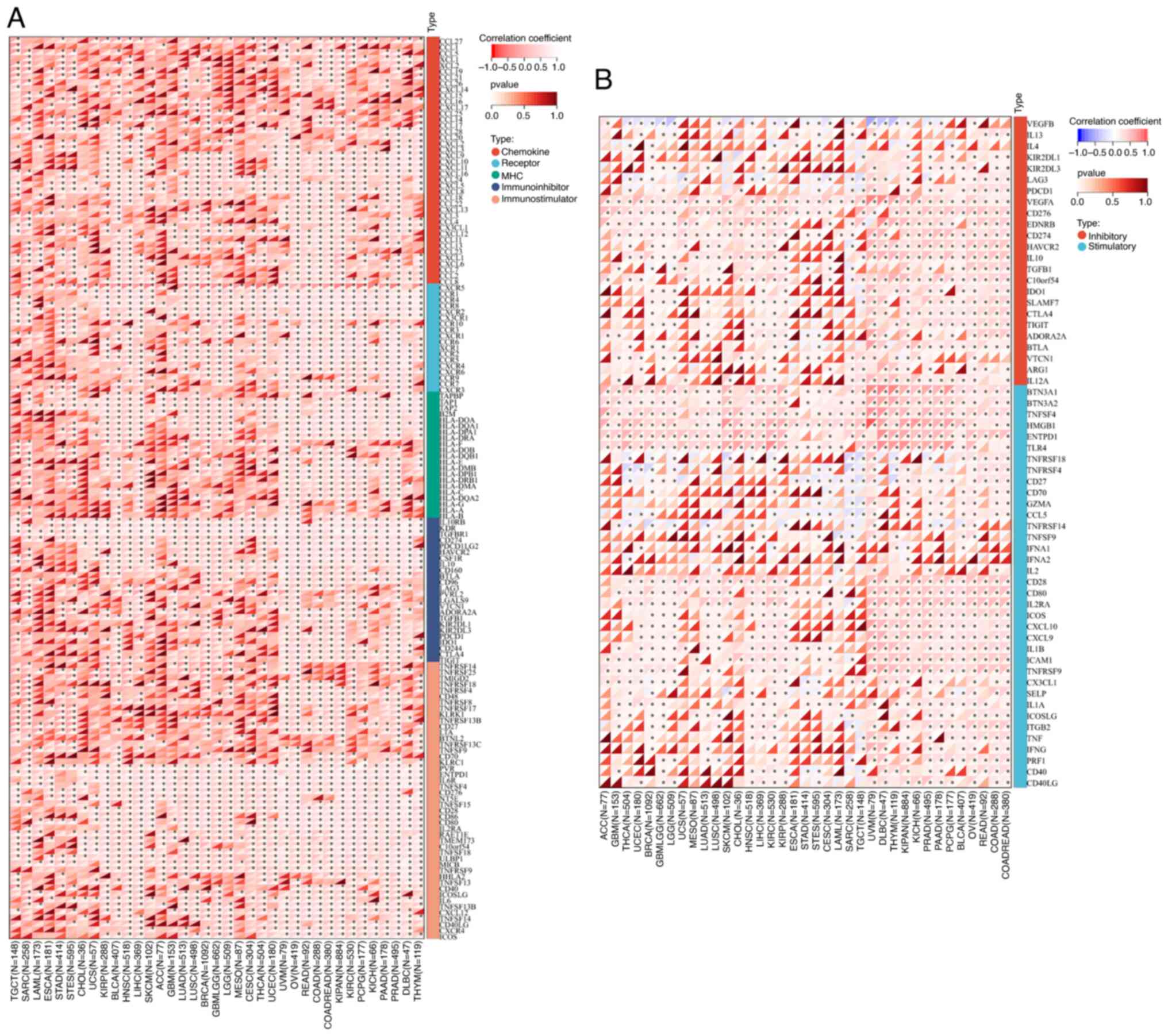 | Figure 11.Correlation between the expression of
MTDH and chemokine, chemokine receptor, MHC, immunostimulatory,
immuno-inhibitory and immune checkpoints. (A) Correlation between
the expression of MTDH and chemokine, chemokine receptor, MHC,
immuno-stimulator, and immuno-inhibitor. (B) Correlation between
the expression of MTDH and immune checkpoints. *P<0.05. MTDH,
metadherin; MHC, major histocompatibility complex. |
Discussion
MTDH, also known as AEG-1, is a key oncoprotein
involved in the advancement of different types of cancers. More
specifically, MTDH plays a critical role in the tumor necrosis
factor alpha-induced protein 2 (TNFAIP2)-induced EMT in urothelial
carcinoma cells (31). Moreover,
MTDH is implicated in the regulation of the NF-kB pathway,
impacting the metastatic and proliferative capabilities of gastric
cancer (32). In the case of BRCA,
the progression of the disease is closely linked to elevated MTDH
expression (33). Notably, various
studies emphasized the significant association between MTDH and EMT
in diverse cancer types, such as head and neck squamous cell
carcinoma, non-small cell lung cancer and nasopharyngeal carcinoma
(34–36).
However, limited data are available regarding the
prognostic significance of MTDH in varying types of solid cancers.
The present study also uncovered that MTDH serves as a detrimental
factor in BRCA, CHOL, COAD, ESCA, HNSC, KIRC, LIHC, LUAD, LUSC,
READ and STAD, whereas acting as a protective element in THCA and
UCEC, indicating that MTDH possessed contrasting roles in different
cancer types. Varied levels of MTDH expression may signify
distinctive underlying mechanisms and functions in disparate tumor
categories. The authors were interested in the mechanism of action
of MTDH in BRCA and kidney cancer because of the differential
expression of MTDH in both BRCA and kidney cancer from previous
studies in the literature and bioinformatics analyses, and the
significant effect on their prognosis (13,37).
Through RT-qPCR analysis, the present study evaluated MTDH
expression in BRCA and renal clear cell carcinoma tissues and cell
lines, confirming that MTDH levels were elevated in these tissues
and cell lines compared with their normal counterparts.
Subsequently, the association between MTDH
expression and sex in pan-cancer was investigated. It was noticed
that LAML and LIHC were higher in men, while SARC, KIRP, KIPAN,
THCA and ACC were higher in women. However, the underlying
mechanism remains to be further explored. In addition, MTDH
expression was positively connected with histological grade in
GBMLGG, LGG, KIPAN, HNSC, KIRC and PAAD. Furthermore, the
expression of MTDH associated with the T stage in CESC, LUAD, BRCA,
KIRP, PRAD and KIRC, N stage in BRCA, KIRP and PRAD, M stage in
KIRC and LUSC, and pathological stage in KIRP, KIRP, UCS and BLCA,
further suggesting that it plays a pivotal role in tumor
development.
To investigate the underlying reasons behind the
high expression of MTDH across multiple types of cancer, the
present study involved the analysis of the methylation of the DNA
promoter as well as RNA modifications including m1A, m2C and m6A
methylation. The present findings revealed patterns of
hypomethylation within the promoter region of MTDH in diverse
cancer tissues. This observation provides some insight into the
potential explanation for the overexpression of MTDH mRNA in these
cancer types. Additionally, it is worth noting that RNA m6A
methylation serves as a crucial mechanism that influences the
regulation of RNA expression. In the current study, a total of 21
genes related to RNA m6A methylation were collected and a
comprehensive correlation analysis was conducted to explore the
relationship between MTDH and these m6A methylation-associated
genes across various types of cancers. The results from this
analysis revealed a significant and consistent association between
MTDH and m6A methylation-related genes in the context of
pan-cancer. This discovery strongly suggested that the mechanism
underlying m6A methylation could have a crucial role in governing
the expression of MTDH in cancerous tissues. Zhang et al
(38) further demonstrated that the
pathway involving MTDH, m6A RNA methylation and EMT might
contribute to the development of immunotherapy resistance in BRCA,
while the present study focused on the validation of MTDH in BRCA
and kidney cancer, as well as the functional clustering of
MTDH-related genes in GO and Kyoto Encyclopedia of Genes and
Genomes, which has its own unique innovation points.
Cox proportional hazards model analysis (including
OS, DSS and PFI) and Kaplan-Meier analysis were conducted to
investigate the prognostic significance of MTDH expression in
pan-cancer. The analysis revealed an association between the high
expression of MTDH in GBMLGG, PAAD, BRCA, LGG, KICH and a positive
impact on OS. Interestingly, the expression of MTDH was negatively
correlated with OS of KIRC, and the mechanism remains to be further
studied. An association was observed between elevated MTDH levels
and unfavorable DSS in PAAD, MESO, GBMLGG, LGG, BRCA, BLCA and UVM.
Additionally, the PFI findings demonstrated that MTDH posed a
significant risk factor for patients with ACC, UVM, BLCA, PAAD,
LGG, KIRP, KICH, GBMLGG and CESC. MTDH overexpression influences
the bleak prognosis of BRCA, and thereby, targeting MTDH was
proposed as a potential therapy for this disease according to a
previous investigation (13). Given
these findings, it was hypothesized that MTDH inhibition could
serve as a promising approach for therapeutic targets in various
tumor types.
There have been substantial advancements and
numerous significant breakthroughs in the field of cancer
immunotherapy over the past few decades. These advancements have
led to improved clinical outcomes for patients with different types
of cancer (39). The application of
immunotherapy in the treatment of tumors has not only enhanced the
overall quality of life but also improved the survival rates
(40). However, it is crucial to
note that the immunosuppressive nature of the TME can promote tumor
progression, invasion and resistance to therapy (41). Tumor cells can evade immune
detection by activating immune checkpoints, leading to a potential
decrease in T cell effectiveness against these cells. The field of
cancer immunotherapy has seen significant advancements with the
development of immune checkpoint inhibitors, offering a promising
strategy. The use of high-throughput sequencing technologies has
led to the identification of numerous new immune checkpoints
(42). The present research set out
to explore the potential of MTDH as an innovative target for
immunotherapy in the TME. The current study findings indicated a
strong link between elevated MTDH levels and estimation, stromal
and immune scores. Moreover, a direct connection between MTDH
expression and both MSI and TMB was noticed. Furthermore, a
thorough evaluation of MTDH along with other immune checkpoints was
carried out. The outcomes displayed a favorable relationship
between MTDH and various immune checkpoints, showing that MTDH
expression correlated with the majority of immunoregulatory genes
and checkpoint genes in UVM, OV, READ, KIPAN, KIRC and PRAD. This
in-depth analysis provided evidence for the potential of MTDH in
signaling the immune microenvironment. Additionally, MTDH
demonstrated potential as a prognostic marker for the immunotherapy
response.
In TMEs, the presence of inflammatory cells can
either facilitate or hinder tumor growth as well as the efficacy of
anti-tumor immunotherapy. It is of utmost importance to comprehend
the role played by these cells in the development of effective
cancer treatments (43). T cells,
an essential component of the defense of the adaptive immune system
against cancer, play a critical role. Regulatory T cells (Tregs),
essential in creating immunosuppressive surroundings, inhibit the
differentiation and activation of CD4(+) helper T cells and CD8(+)
cytotoxic T cells (44). Throughout
the immune response to tumors, Tregs secrete cytokines such as
IL-35, TGF-β and IL-10. These cytokines inhibit the ability of the
body to fight against tumors and support the growth and development
of cancer (45). Tumor-associated
macrophages (TAMs), a crucial component of TMEs, can regulate
inflammation. Additionally, TAMs possess the capability to either
aid, impede, or initiate tumor development through the secretion of
cytokines and modulation of other immune cells (46,47).
Growing research indicates that macrophages associated with tumors,
specifically the M2 subtype, are crucial in fostering an
immune-suppressive TME by aiding in the enlistment of Tregs and
hindering the development and activity of T cells (48,49).
Tumor-associated neutrophils exhibit diverse effects on tumor
immunity based on their subtypes, which can either be
anti-tumorigenic or pro-tumorigenic, as indicated by several
studies (50). Non-neoplastic cells
necessitate the presence of tumor mesenchyme's essential elements
known as cancer-related fibroblasts (CAFs). Their significance lies
in their ability to advance tumor progression and metastasis
through their support to cancer cell growth, invasion and survival.
CAFs employ diverse intricate mechanisms to interact with tumor
cells, such as the secretion of extracellular matrix, growth
factors and cytokines (50). The
present study delved into the correlation between MTDH and the
presence of inflammatory cells in the TME. It was found that MTDH
demonstrates a significant association with various types of
inflammatory cells across different cancer types, including
macrophages, M2 macrophages, T cells, Tregs, CAFs, monocytes,
neutrophils and natural killer cells. These results indicated that
MTDH is closely linked to both tumor cells and the surrounding
stromal cells within the TME. Furthermore, MTDH has been identified
to play a crucial role in several immune-related pathways,
influencing the proliferation, activation and migration of mast
cells, T cells, fibroblasts and macrophages. Overall, the current
findings strongly suggested that MTDH contributes to the
development of an immunosuppressive microenvironment in cancer.
Using CancerSEA, pan-cancer single-cell
investigations on MTDH were conducted. The current analysis
revealed a positive correlation between MTDH and apoptosis, as well
as DNA repair, in certain tumor types. Across different cancer
categories, MTDH was found to stimulate MAPK, PI3K/AKT and
WNT/b-catenin pathways, ultimately encouraging various indicators
of aggressive cancer traits. These characteristics include tumor
expansion, spread, angiogenesis and resilience against chemotherapy
(10,51).
In general, it is important to consider the
limitations of the present study. Initially, it is essential to
note that these findings primarily stem from bioinformatics
analysis. To ascertain the potential function of MTDH, it is
necessary to conduct in vivo and in vitro
experiments. Additionally, it is imperative to acknowledge that
systematic bias may have been introduced by the utilization of
microarray and sequencing data from various databases. Another
limitation is the retrospective nature of the data employed in the
present study. Therefore, for further validation, prospective
studies should be conducted. The relevant experimental studies in
the present study are only for two cancer types, BRCA and kidney
cancer, and the authors will continue to explore the function of
MTDH in other cancer types in the future.
In conclusion, upregulation of MTDH was
significantly associated with prognosis, immune cell infiltration,
mutations of tumor-associated genes and its promoter methylation in
multiple cancers, especially BRCA and renal cancer. MTDH may act as
a novel biomarker for survival and immunotherapy across cancers in
the immediate future.
Acknowledgments
Not applicable.
Funding
The present study was supported by Xingtai Science and
Technology Plan Project (grant no. 2023ZZ104), Xingtai City key
research and development plan (grant no. 2021ZC148) and Scientific
research fund of Health Commission of Hebei (grant no.
20220224).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
LXY, MQH and FTK conceived the research and
supervised the experimental design. SYZ, XLZ and LNJ conducted data
analysis and material input. XWL, LZ and MW wrote and contributed
to the validation of the manuscript and acquisition of data. XWL,
LZ, MW, LXY and FTK interpreted the data. LXY and MQH confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
All procedures performed in the present study
involving human participants were in accordance with The
Declaration of Helsinki (as revised in 2013). The study was
approved by the Institutional Ethics committee of Xingtai People's
Hospital [approval no. 2021(031); Xingtai, China]. Written informed
consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GBM
|
glioblastoma multiforme
|
|
LGG
|
lower grade glioma
|
|
BRCA
|
breast cancer
|
|
LUAD
|
lung adenocarcinoma
|
|
ESCA
|
esophageal carcinoma
|
|
STES
|
stomach and esophageal carcinoma
|
|
COAD
|
colon adenocarcinoma
|
|
PRAD
|
prostate adenocarcinoma
|
|
STAD
|
stomach adenocarcinoma
|
|
HNSC
|
head and neck squamous cell
carcinoma
|
|
KIRC
|
kidney renal clear cell carcinoma
|
|
LUSC
|
lung squamous cell carcinoma
|
|
LIHC
|
liver hepatocellular carcinoma
|
|
SKCM
|
skin cutaneous melanoma
|
|
THCA
|
thyroid cancer
|
|
READ
|
rectum adenocarcinoma
|
|
OV
|
ovarian serous cystadenocarcinoma
|
|
PAAD
|
pancreatic adenocarcinoma
|
|
LAML
|
acute myeloid leukemia
|
|
CHOL
|
cholangiocarcinoma
|
|
ACC
|
adrenocortical cancer
|
References
|
1
|
Arnold M, Rutherford MJ, Bardot A, Ferlay
J, Andersson TM, Myklebust TÅ, Tervonen H, Thursfield V, Ransom D,
Shack L, et al: Progress in cancer survival, mortality, and
incidence in seven high-income countries 1995–2014 (ICBP
SURVMARK-2): A population-based study. Lancet Oncol. 20:1493–1505.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bindea G, Mlecnik B, Tosolini M,
Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T,
Lafontaine L, Berger A, et al: Spatiotemporal dynamics of
intratumoral immune cells reveal the immune landscape in human
cancer. Immunity. 39:782–795. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schumacher TN and Schreiber RD:
Neoantigens in cancer immunotherapy. Science. 348:69–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Topalian SL, Drake CG and Pardoll DM:
Immune checkpoint blockade: A common denominator approach to cancer
therapy. Cancer Cell. 27:450–461. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Becht E, Giraldo NA, Dieu-Nosjean MC,
Sautès-Fridman C and Fridman WH: Cancer immune contexture and
immunotherapy. Curr Opin Immunol. 39:7–13. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Su ZZ, Kang DC, Chen Y, Pekarskaya O, Chao
W, Volsky DJ and Fisher PB: Identification and cloning of human
astrocyte genes displaying elevated expression after infection with
HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid
subtraction hybridization, RaSH. Oncogene. 21:3592–3602. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Blanco MA, Alečković M, Hua Y, Li T, Wei
Y, Xu Z, Cristea IM and Kang Y: Identification of staphylococcal
nuclease domain-containing 1 (SND1) as a Metadherin-interacting
protein with metastasis-promoting functions. J Biol Chem.
286:19982–19992. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kang DC, Su ZZ, Sarkar D, Emdad L, Volsky
DJ and Fisher PB: Cloning and characterization of HIV-1-inducible
astrocyte elevated gene-1, AEG-1. Gene. 353:8–15. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee SG, Kang DC, DeSalle R, Sarkar D and
Fisher PB: AEG-1/MTDH/LYRIC, the beginning: Initial cloning,
structure, expression profile, and regulation of expression. Adv
Cancer Res. 120:1–38. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yoo BK, Emdad L, Lee SG, Su ZZ,
Santhekadur P, Chen D, Gredler R, Fisher PB and Sarkar D: Astrocyte
elevated gene-1 (AEG-1): A multifunctional regulator of normal and
abnormal physiology. Pharmacol Ther. 130:1–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu HY, Liu CX, Han B, Zhang XY and Sun
RP: AEG-1 is associated with clinical outcome in neuroblastoma
patients. Cancer Biomark. 11:115–121. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Subramanian A, Kuehn H, Gould J, Tamayo P
and Mesirov JP: GSEA-P: A desktop application for gene set
enrichment analysis. Bioinformatics. 23:3251–3253. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tokunaga E, Nakashima Y, Yamashita N,
Hisamatsu Y, Okada S, Akiyoshi S, Aishima S, Kitao H, Morita M and
Maehara Y: Overexpression of metadherin/MTDH is associated with an
aggressive phenotype and a poor prognosis in invasive breast
cancer. Breast Cancer. 21:341–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meng X, Brachova P, Yang S, Xiong Z, Zhang
Y, Thiel KW and Leslie KK: Knockdown of MTDH sensitizes endometrial
cancer cells to cell death induction by death receptor ligand TRAIL
and HDAC inhibitor LBH589 co-treatment. PLoS One. 6:e209202011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meng X, Thiel KW and Leslie KK: Drug
resistance mediated by AEG-1/MTDH/LYRIC. Adv Cancer Res.
120:135–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Heo J, Lim CK, Whang DR, Shin J, Jeong SY,
Park SY, Kwon IC and Kim S: Self-deprotonation and colorization of
1,3-bis(dicyanomethylidene)indan in polar media: A facile route to
a minimal polymethine dye for NIR fluorescence imaging. Chemistry.
18:8699–8704. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Emdad L, Das SK, Dasgupta S, Hu B, Sarkar
D and Fisher PB: AEG-1/MTDH/LYRIC: signaling pathways, downstream
genes, interacting proteins, and regulation of tumor angiogenesis.
Adv Cancer Res. 120:75–111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goldman M, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
The UCSC Xena platform for public and private cancer genomics data
visualization and interpretation. bioRxiv. 3264702019.
|
|
20
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48((W1)): W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Szklarczyk D, Gable AL, Nastou KC, Lyon D,
Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al:
The STRING database in 2021: Customizable protein-protein networks,
and functional characterization of user-uploaded gene/measurement
sets. Nucleic Acids Res. 49((D1)): D605–D612. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liao Y, Wang J, Jaehnig EJ, Shi Z and
Zhang B: WebGestalt 2019: Gene set analysis toolkit with revamped
UIs and APIs. Nucleic Acids Res. 47((W1)): W199–W205. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ogłuszka M, Orzechowska M, Jędroszka D,
Witas P and Bednarek AK: Evaluate Cutpoints: Adaptable continuous
data distribution system for determining survival in Kaplan-Meier
estimator. Comput Methods Programs Biomed. 177:133–139. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zeng D, Ye Z, Shen R, Yu G, Wu J, Xiong Y,
Zhou R, Qiu W, Huang N, Sun L, et al: IOBR: multi-omics
immuno-oncology biological research to decode tumor
microenvironment and signatures. Front Immunol. 12:6879752021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He J, Ding H, Li H, Pan Z and Chen Q:
Intra-tumoral expression of SLC7A11 is associated with immune
microenvironment, drug resistance, and prognosis in cancers: A
pan-cancer analysis. Front Genet. 12:7708572021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li L, Yao W, Yan S, Dong X, Lv Z, Jing Q,
Wang Q, Ma B, Hao C, Xue D and Wang D: Pan-cancer analysis of
prognostic and immune infiltrates for CXCs. Cancers (Basel.
13:41532021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stenström J, Hedenfalk I and Hagerling C:
Regulatory T lymphocyte infiltration in metastatic breast cancer-an
independent prognostic factor that changes with tumor progression.
Breast Cancer Res. 23:272021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ren L, Yi J, Yang Y, Li W, Zheng X, Liu J,
Li S, Yang H, Zhang Y, Ge B, et al: Systematic pan-cancer analysis
identifies APOC1 as an immunological biomarker which regulates
macrophage polarization and promotes tumor metastasis. Pharmacol
Res. 183:1063762022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shan L, Lu Y, Song Y, Zhu X, Xiang CC, Zuo
ED and Cheng X: Identification of nine M6A-related long noncoding
RNAs as prognostic signatures associated with oxidative stress in
oral cancer based on data from the cancer genome atlas. Oxid Med
Cell Longev. 2022:95298142022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Niwa N, Tanaka N, Hongo H, Miyazaki Y,
Takamatsu K, Mizuno R, Kikuchi E, Mikami S, Kosaka T and Oya M:
TNFAIP2 expression induces epithelial-to-mesenchymal transition and
confers platinum resistance in urothelial cancer cells. Lab Invest.
99:1702–1713. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jiao Y, Yang H, Qian J, Gong Y, Liu H, Wu
S, Cao L and Tang L: miR-3664-5P suppresses the proliferation and
metastasis of gastric cancer by attenuating the NF-κB signaling
pathway through targeting MTDH. Int J Oncol. 54:845–858.
2019.PubMed/NCBI
|
|
33
|
Luo L, Tang H, Ling L, Li N, Jia X, Zhang
Z, Wang X, Shi L, Yin J, Qiu N, et al: LINC01638 lncRNA activates
MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation
in triple-negative breast cancer. Oncogene. 37:6166–6179. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qin Y, Wang J, Zhu G, Li G, Tan H, Chen C,
Pi L, She L, Chen X, Wei M, et al: CCL18 promotes the metastasis of
squamous cell carcinoma of the head and neck through MTDH-NF-κB
signalling pathway. J Cell Mol Med. 23:2689–2701. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yin Q, Han Y, Zhu D, Li Z, Shan S, Jin W,
Lu Q and Ren T: miR-145 and miR-497 suppress TGF-β-induced
epithelial-mesenchymal transition of non-small cell lung cancer by
targeting MTDH. Cancer Cell Int. 18:1052018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu C, Liu Y and Qin Z: Metadherin
contributes to epithelial-mesenchymal transition and paclitaxel
resistance induced by acidic extracellular pH in nasopharyngeal
carcinoma. Oncol Lett. 15:3858–3863. 2018.PubMed/NCBI
|
|
37
|
Li J, Li C, Li H, Zhang T, Hao X, Chang J
and Xu Y: MicroRNA-30a-5p suppresses tumor cell proliferation of
human renal cancer via the MTDH/PTEN/AKT pathway. Int J Mol Med.
41:1021–1029. 2018.PubMed/NCBI
|
|
38
|
Zhang F, Huang H, Qin Y, Chen C, She L,
Wang J, Huang D, Tang Q, Liu Y, Zhu G and Zhang X: MTDH associates
with m6A RNA methylation and predicts cancer response for immune
checkpoint treatment. iScience. 24:1031022021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang H, Dai Z, Wu W, Wang Z, Zhang N,
Zhang L, Zeng WJ, Liu Z and Cheng Q: Regulatory mechanisms of
immune checkpoints PD-L1 and CTLA-4 in cancer. J Exp Clin Cancer
Res. 40:1842021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Esfahani K, Roudaia L, Buhlaiga N, Del
Rincon SV, Papneja N and Miller WH Jr: A review of cancer
immunotherapy: From the past, to the present, to the future. Curr
Oncol. 27 (Suppl 2):S87–S97. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang H, Luo YB, Wu W, Zhang L, Wang Z,
Dai Z, Feng S, Cao H, Cheng Q and Liu Z: The molecular feature of
macrophages in tumor immune microenvironment of glioma patients.
Comput Struct Biotechnol J. 19:4603–4618. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang H, Wang Z, Dai Z, Wu W, Cao H, Li S,
Zhang N and Cheng Q: Novel Immune Infiltrating cell signature based
on cell pair algorithm is a prognostic marker in cancer. Front
Immunol. 12:6944902021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hiam-Galvez KJ, Allen BM and Spitzer MH:
Systemic immunity in cancer. Nat Rev Cancer. 21:345–359. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Josefowicz SZ, Lu LF and Rudensky AY:
Regulatory T cells: Mechanisms of differentiation and function.
Annu Rev Immunol. 30:531–564. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li C, Jiang P, Wei S, Xu X and Wang J:
Regulatory T cells in tumor microenvironment: New mechanisms,
potential therapeutic strategies and future prospects. Mol Cancer.
19:1162020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen Y, Song Y, Du W, Gong L, Chang H and
Zou Z: Tumor-associated macrophages: An accomplice in solid tumor
progression. J Biomed Sci. 26:782019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Duan Z and Luo Y: Targeting macrophages in
cancer immunotherapy. Signal Transduct Target Ther. 6:1272021.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yin M, Li X, Tan S, Zhou HJ, Ji W, Bellone
S, Xu X, Zhang H, Santin AD, Lou G and Min W: Tumor-associated
macrophages drive spheroid formation during early transcoelomic
metastasis of ovarian cancer. J Clin Invest. 126:4157–4173. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mantovani A, Marchesi F, Malesci A, Laghi
L and Allavena P: Tumour-associated macrophages as treatment
targets in oncology. Nat Rev Clin Oncol. 14:399–416. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Galli F, Aguilera JV, Palermo B, Markovic
SN, Nisticò P and Signore A: Relevance of immune cell and tumor
microenvironment imaging in the new era of immunotherapy. J Exp
Clin Cancer Res. 39:892020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Manna D and Sarkar D: Multifunctional role
of astrocyte elevated gene-1 (AEG-1) in cancer: Focus on drug
resistance. Cancers (Basel). 13:17922021. View Article : Google Scholar : PubMed/NCBI
|