Introduction
Wilms' tumor is the second most common
intra-abdominal cancer in childhood and the fifth most frequent
pediatric malignancy, constituting ~6% of all childhood cancers and
>95% of all renal tumors in the pediatric age group in Europe
(1,2). Furthermore, Wilms' tumor, also known
as nephroblastoma, is among the most common primary malignant
tumors of the kidneys in children, typically originating from
embryonic kidney tissues. Despite significant advancements in
treatment, managing advanced, anaplastic, or recurrent cases
remains challenging due to the lack of curative therapies and
significant long-term effects (3,4),
particularly in chemotherapy and tumor cell growth regulation.
Currently, the treatment of Wilms' tumor globally falls into two
categories: The Children's Oncology Group, primarily in North
America, recommends direct surgical intervention followed by
further treatment based on postoperative pathology and staging
(5); whilst the International
Society of Paediatric Oncology Renal Tumour Study Group,
predominantly in Europe, advocates for preoperative chemotherapy,
followed by surgical excision after tumor reduction, tailored
according to varying risk levels (6).
Chemotherapeutic drugs used in the treatment of
Wilms' tumor primarily act by disrupting cell DNA synthesis and the
cell cycle. Commonly used drugs include vincristine, actinomycin-d,
cyclophosphamide and doxorubicin (7–9);
however, during the administration of these drugs, factors such as
patient age and dosage can lead to late effects such as bowel
obstruction and heart problems (10). Therefore, enhancing tumor cell
sensitivity to chemotherapeutic drugs and reducing dosages are
crucial in Wilms' tumor chemotherapy.
In our previous study, the regulation of unc-13
homolog B (UNC13B) in drug sensitivity was assessed in chronic
lymphoid leukemia. UNC13B was demonstrated to regulate tumor cell
resistance to arsenic trioxide (11). These findings suggest a potential
involvement of UNC13B in chemotherapeutic drug resistance in tumor
cells. The development of chemotherapeutic drug resistance involves
both external and internal factors, including tumor heterogeneity,
the tumor microenvironment and the inactivation of anticancer drugs
(12). Current research about
UNC13B mainly focuses on brain and neural studies, where it serves
a crucial role in synaptic vesicle initiation and fusion (13,14),
potentially influencing neuronal excitability (15). As a regulator of the cell vesicular
system, whether UNC13B mediates mechanisms of chemotherapeutic drug
resistance in tumor cells through vesicle regulation remains
unclear. Thus, the present study aims to assess the role of UNC13B
in regulating drug sensitivity and resistance to doxorubicin in
Wilms' tumor cell lines.
Materials and methods
Cell lines and drug treatment
The human Wilms' tumor 17.94 cell line was purchased
from the Leibniz Institute, Deutsche Sammlung von Mikroorganismen
und Zellkulturen-German Collection of Microorganisms and Cell
Cultures GmbH (cat. no. ACC 741) and cultured in high-glucose DMEM
supplemented with 20% FBS (HyClone; cat. no. SH30088.03) and 1%
penicillin/streptomycin at 37°C in 5% CO2. The human
Ewing sarcoma SK-NEP-1 (cat. no. HTB-48) and human rhabdoid tumor
G401 (cat. no. CRL-1441) cell lines were purchased from the
American Type Culture Collection (ATCC) and maintained in McCoy's
5A modified medium containing 15% FBS (HyClone, SH30088.03). The
human HK-2 normal kidney-derived proximal tubular cell line was
purchased from Procell Life Science & Technology Co., Ltd.
(cat. no. CL-0109) and cultured in MEM supplemented with 10% FBS
(HyClone Laboratories, SH30088.03) and 1% penicillin/streptomycin
at 37°C in 5% CO2. The human kidney rhabdoid tumor
WT-CLS1 cell line was purchased from CLS Cell Lines Services GmBH
(cat. no. 300379) and cultured in IMDM supplemented with 10% FBS
(HyClone Laboratories, SH30088.03) and 1% penicillin/streptomycin
at 37°C in 5% CO2. Authentication of the cell lines used
in the present study was performed using Short Tandem Repeat (STR)
profiling, provided by Pricella Biotechnology Co., Ltd (Elabscience
Bionovation Inc.). This method involved analyzing specific STR loci
to confirm the identity of the cell lines. Comparison of the STR
profile of the samples in the present study against known reference
profiles (source: ATCC) demonstrated a 100% match, ensuring the
authenticity of the cell lines used in the experiments.
The determination of the half-maximal inhibitory
concentration (IC50) was performed as follows: Passaged
cells were counted and seeded at a density of 3,000 cells per well
in a 96-well plate. Following overnight incubation at 37°C, 0.1–5
µM doxorubicin (cat. no. E2516; Selleck Chemicals), 0–500 nM
vincristine sulfate (cat. no. S1241; Selleck Chemicals) or 0.1–5 nM
actinomycin-D (cat. no. S8964; Selleck Chemicals) in DMSO were
added, whilst the negative control group received an equal volume
of DMSO. Each group was set up with 6 replicates. After incubation
for 48 h at 37°C in 5% CO2, 10 µl Cell Counting Kit-8
(CCK-8) reagent (cat. no. C0038; Beyotime Institute of
Biotechnology) was added per well, followed by an additional 4-h
incubation in the culture chamber. The optical density at 450 nm
was measured using a microplate reader. IC50 was
calculated using GraphPad Prism 9.0 software (Dotmatics). After
entering data into GraphPad Prism, a nonlinear regression (curve
fit) analysis was used. The ‘(Inhibitor) vs. normalized
response-Variable slope’ model was chosen. A curve was generated to
fit the data, and the IC50 value was displayed. This
value represented the concentration of the compound at which the
response was half of the maximum effect observed.
Short hairpin (sh)RNA construction and
transfection
The construction of shRNA followed a previously
described method (11). Based on
the nucleotide sequence of UNC13B in the GenBank database
(ncbi.nlm.nih.gov/nuccore/NM_001371189.2, ID no. NM_001371189.2),
17.94 cells were transfected with UNC13B shRNA (shUNC13B) using a
lentivirus vector. The second-generation lentiviral vector system
was used to construct shRNA lentivirus. The plasmid backbone and
negative controls are as follows: lentiCRISPR v2 (cat. no. 52961;
Addgene, Inc.), pCMV-pVSV-G (cat. no. 8454; Addgene, Inc.), and
psPAX2 (cat. no. 12260; Addgene, Inc.). The negative controls are
scramble sequences synthesized by igebio biotech Co., Ltd. UNC13B
shRNA lentiviral vector using lentiCRISPR v2. (cat. no. 52961;
Addgene, Inc.) was constructed. The sequence of the sense strand of
shUNC13B was 5′-CGAGTCCTATGAGTTGCAGAT-3′ and the antisense strand
was 5′-ATCTGCAACTCATAGGACTCG-3′. Cells were also transfected with
non-target shRNA as a negative control (shCtrl; scramble). The
sequence of the sense strand was 5′-CCTAAGGTTAAGTCGCCCTCGC-3′ and
the antisense strand was 5′-GCGAGGGCGACTTAACCTTAGG-3′, connected by
a ‘TCGA’ loop, with an additional 5 thymine (T) at the end.
Scrambled shRNA is designed so that it does not target any specific
mRNA for degradation (16). All
shRNAs were synthesized by igebio biotech Co., Ltd. shRNA was
inserted into the lentiCRISPR v2 plasmid using AgeI and EcoRI
restriction enzyme sites. The plasmid was then transformed into
DH5α cells (Beyotime, D1031S) for amplification. After
amplification, the plasmid was purified using the Endo-Free Plasmid
Midi Kit (Omega Bio-Tek, D6915). The purified plasmid was verified
by sequencing to ensure the correct insert.
Lipofectamine 3,000 (cat. no. L3000015; Invitrogen™;
Thermo Fisher Scientific, Inc.) was used to transfect these shRNAs
into 293T cells (cat no. CL-0005; Procell Life Science &
Technology Co., Ltd.), cultured in DMEM supplemented with 10% FBS
(HyClone Laboratories, SH30088.03) and 1% penicillin/streptomycin
at 37°C in 5% CO2 for 48 h, along with the packaging
plasmids pCMV-pVSV-G (cat. no. 8454; Addgene, Inc.) and psPAX2
(cat. no. 12260; Addgene, Inc.). Transfect with a ratio of 10 µg of
psPAX2 plasmid, 10 µg of lentiCRISPR v2 plasmid containing the
inserted shRNA, and 5 µg of pCMV–VSV-G plasmid into a 10 cm dish of
293T cells at approximately 80% confluence. The transfection
duration was 12 h at 37°C, after which the medium was replaced.
Lentiviral particles were collected and used to infect 17.94 cells
at a multiplicity of infection (MOI) of 10. The duration of
transduction into 17.94 cells is 12 h. Gene expression and
transcription levels were identified 24 h after transduction using
qPCR and western blot methods. Subsequent experiments were
conducted 24–48 h after transduction. To normalize the transfection
efficiency, green fluorescent protein (GFP) cDNA was inserted into
an empty lentiCRISPR v2 to construct a GFP-lentiviral vector. A
transfection experiment was performed on 17.94 cells using
GFP-lentivirus. A total of 24 h after transfection, the cells were
analyzed using a fluorescence microscope.
Overexpression vector and knockout
vector construction and transfection
The full-length UNC13B sequence was synthesized by
General Biosystems, Inc. according to the GenBank database
(ncbi.nlm.nih.gov/nuccore/NM_001371189.2/, ID no. NM_001371189.2)
and inserted into the pcDNA3.1 vector using BamHI and
HindIII restriction enzyme sites. The plasmid is transformed into
DH5α (Beyotime, D1031S) for amplification. Plasmid is then purified
using Endo-Free Plasmid Midi Kit (Omega Bio-Tek, D6915). The
purified plasmid is verified by restriction enzyme digestion and
sequencing to ensure the correct insert.
For overexpression experiments, Transfection was
carried out using Lipofectamine™ 3,000 Transfection Reagent (cat.
no. L3000015; Invitrogen™; Thermo Fisher Scientific, Inc.). 1 µg of
plasmid was used per well in a 12-well plate with a cell density of
over 80%. The transfection duration was 12 h at 37°C, after which
the medium was replaced. Post-transfection, overexpression levels
were assessed after 48 h using western blot for subsequent
experiments. An empty vector pcDNA3.1 was used as the negative
control.
The second-generation lentiviral vector system was
used to construct sgRNA lentivirus. The lentiCRISPR V2 plasmid
(cat. no. 52961; Addgene, Inc.) was used. Two single guide (sg)RNAs
targeting UNC13B, hUNC13B-KO-1: 5′-TGATCAGCCTTCCTGGGAACAGG-3′ and
hUNC13B-KO-2: 5′-TCTTCACATTCTGTACTTTCAGG-3′, were inserted,
respectively, and two plasmids were developed, each containing a
different sgRNA. All sgRNAs were synthesized by igebio biotech Co.,
Ltd. Lipofectamine 3000 was used to co-transfect these sgRNAs into
293T cells (cat no. CL-0005; Procell Life Science & Technology
Co., Ltd.), cultured in DMEM supplemented with 10% FBS and 1%
penicillin/streptomycin at 37 °C in 5% CO2 for 48 h,
along with the packaging plasmids pCMV-pVSV-G (cat. no. 8454;
Addgene, Inc.) and psPAX2 (cat. no. 12260; Addgene, Inc.). Cells
were transfected with a ratio of 10 µg of psPAX2 plasmid, 10 µg of
lentiCRISPR v2 plasmid containing the inserted shRNA, and 5 µg of
pCMV–VSV-G plasmid into a 10 cm dish of 293T cells at approximately
80% confluence. A total of two cas9 plasmids targeting UNC13B were
generated. Both prepared lentiviruses were co-transduced into 17.94
cells at a MOI of 10. The duration of transduction was 12 h, after
which the lentivirus is removed. At 24 h post-transduction, cells
were diluted using a limited dilution method into a 96-well plate
and cultured for 14 days at 37 °C in 5% CO2 to establish
single-cell clones of UNC13B-KO using puromycin (Beyotime, ST551)
at a selection concentration of 4 µg/ml and a maintenance
concentration of 0.8 µg/ml. We used PCR and sequencing methods,
performed by GENEWIZ Biotechnology Co., Ltd., to identify the
amplified single-cell clones, confirming that the target locus had
been successfully edited. Reverse transcription (RT)-quantitative
(q)PCR and western blotting were used to assess the transcription
and expression levels of UNC13B.
RT-qPCR
RT-qPCR for UNC13B was performed as previously
described (11). Briefly, RNA
extraction from samples was performed using TRIzol™ (cat. no.
15596018CN; Invitrogen; Thermo Fisher Scientific, Inc.), and the
concentration and purity of the extracted RNA were measured using a
NanoDrop™ 2000 (Thermo Fisher Scientific, Inc.) to ensure its
suitability for subsequent experimental procedures. The RNA was
reverse transcribed into cDNA using a cDNA synthesis kit
(EasyScript® First-Strand cDNA Synthesis SuperMix,
TransGen Biotech, AE301-03) according to the manufacturer's
instructions. The sequences for UNC13B were as follows: UNC13B
forward, 5′-CCAGCTACACAACTCACTGAGG-3′ and UNC13B reverse,
5′-CTGGTCAGCAAATCCACTGTGG-3′. The sequences for the reference gene
18SN5 were as follows: 18SN5 forward, 5′-ACCCGTTGAACCCCATTCGTGA-3′
and 18SN5 reverse, 5′-GCCTCACTAAACCATCCAATCGG-3′. Hieff qPCR SYBR
Green Master Mix (No Rox) from YEASEN (catalog number 11201ES08)
was used with the Bio-Rad CFX96™ system. The thermocycling
conditions for the qPCR assay were as follows: initial denaturation
at 95°C for 5 min; 40 cycles of 95°C for 10 sec and 60°C for 30
sec; the melt curve stage followed the instrument's default
settings. The following steps were used to validate the results of
the RT-qPCR assays: The efficiency of the qPCR reaction was
determined by running a standard curve with serial dilutions of a
known template; the efficiency of the UNC13B primers was ~95%.
After amplification, a melting curve analysis revealed a single,
sharp peak, indicating the specificity of the PCR amplification. In
each experiment, three technical replicates were performed to
assess the consistency of the results. The 18SN5 gene was used as
the reference gene, and the 2−ΔΔCq method (17) was used for quantification.
Western blot
17.94, SK-NEP-1, G401, WT-CLS1, and HK-2 cells were
lysed using RIPA buffer (Beyotime, P0013B), and protein
concentration was measured using a BCA protein determination kit
(Abcam). For SDS-PAGE, 20 µg of protein was loaded/lane. The
separating gel used was 12%. After electrophoresis, proteins were
electrotransferred to a polyvinylidene fluoride membrane, which was
blocked with 5% BSA (cat. no. V900933; Vetec™; Sigma-Aldrich; Merck
KGaA)/TBST (0.1% Tween 20) at room temperature for 2 h. Rabbit
polyclonal UNC13B (1:1,000; cat. no. NBP2-93337; Novus Biologicals,
Ltd.) and mouse monoclonal lysosomal-associated membrane protein 1
(LAMP1; 1:1,000; cat. no. sc-20011; Santa Cruz Biotechnology, Inc.)
primary antibodies were added at the appropriate dilution and
incubated at 4°C overnight. Corresponding secondary antibodies,
HRP-conjugated goat anti-mouse (1:10,000; cat. no. 7076; Cell
Signaling Technology, Inc.) or anti-rabbit (1:10,000; cat. no.
7074; Cell Signaling Technology, Inc.), were added and incubated at
room temperature for 2 h. HRP-labeled GAPDH (1:5,000; cat. no.
3683; Cell Signaling Technology, Inc.) was used as an internal
reference. Membranes were incubated for 3 min in Supersignal West
Pico Plus Chemiluminescent Substrate (Thermo Scientific, 34580) and
exposed using the iBright FL1000 Imaging System (Thermo Fisher
Scientific Inc.), and iBright Analysis Software (desktop version
5.1.0; Thermo Fisher Scientific Inc.) was used for
semi-quantification.
Flow cytometry analysis
For cell cycle analysis, 17.94 cells were digested
with trypsin, and the trypsin digestion was terminated using
complete culture medium. The cells were washed once with pre-cooled
serum-free medium and fixed with pre-cooled 75% ethanol overnight.
Following a PBS wash and cell precipitation, the cells were stained
with PI staining solution (cat. no. 40710ES03; Shanghai Yeasen
Biotechnology Co., Ltd.) at 37°C for 30 min. Data collection was
performed using a CytoFLEX LX Flow Cytometer (Beckman Coulter,
Inc.), with 40,000 events collected for each sample. FlowJo 10.8.1
(Becton, Dickinson and Company) was used for data analysis.
For apoptosis analysis, an Annexin V-FITC/PI kit
(cat. no. 40302ES60; Shanghai Yeasen Biotechnology Co., Ltd.) was
used according to the manufacturer's instructions. Cells were
digested with trypsin (without EDTA) and centrifuged at 300 × g and
4°C for 5 min. Cells were washed twice with pre-chilled PBS, each
time centrifuging at 300 × g and 4°C for 5 min. Cells were
collected, and 5×105 cells were resuspended in 100 µl of
1X Binding Buffer. A total of 5 µl Annexin V-FITC and 10 µl PI
Staining Solution were then added, and cells were incubated in the
dark at room temperature for 10–15 min. Subsequently, 400 µl of 1X
Binding Buffer was added and kept on ice. Samples were analyzed
within 1 h using flow cytometry. Data collection was performed
using a CytoFLEX LX Flow Cytometer, with 40,000 events collected
for each sample. FlowJo 10.8.1 was used for data analysis.
Indirect immunofluorescence and
lysosome staining
Indirect immunofluorescence was used to detect the
localization of UNC13B in 17.94 cells. Initially, cell slides were
prepared and fixed with 100% methanol at −20°C for 10 min. After
three PBS washes, a blocking solution of 3% BSA (Sigma-Aldrich;
Merck KGaA; cat. no. V900933) + 0.3% Triton™ X-100 in PBS was added
at room temperature for 1 h. The blocking solution was removed, and
the slides were incubated with rabbit UNC13B primary antibodies
(1:200; cat. no. NBP2-93337; Novus Biologicals, Ltd.) at 4°C
overnight. Following PBS washes (3 times for 5 min each), the
slides were incubated with Goat Anti-Rabbit IgG H&L (Alexa
Fluor® 488) secondary antibodies (1:2,000; cat. no.
ab150077; Abcam) at room temperature for 1 h. The slides were then
sealed with a mounting medium containing DAPI (cat. no. P0131;
Beyotime Institute of Biotechnology) and images were captured using
a confocal microscope.
Staining of cells with Lyso-Tracker Red (cat. no.
L8010; Beijing Solarbio Science & Technology Co., Ltd.) was
performed by preparing a final concentration of 50 nM Lyso-Tracker
Red working solution. The cell culture medium was removed, and
cells were incubated with pre-warmed Lyso-Tracker Red staining
working solution at 37°C for 2 h. Following the incubation, the
Lyso-Tracker Red staining solution was removed, and 2 µM Hoechst
33342 was added. The cells were then incubated at 37°C for an
additional 30 min before replacing the medium with fresh cell
culture medium. Observations were performed using a confocal
microscope, with an excitation wavelength of 577 nm and an emission
wavelength of 590 nm during detection. The mean fluorescence
intensity was calculated using ImageJ software (version 1.53q;
National Institutes of Health).
Data analysis
For in vitro experiments, each experiment was
independently repeated ≥3 times. Error bars indicate standard
deviation. The unpaired Student's t-test was used for comparisons
between two groups, whilst one-way ANOVA was used for multiple
comparisons. Dunnett's test was used for post hoc analysis when all
pairwise comparisons involved one specific group being compared
with all other groups in the dataset. Tukey's test was used for
post hoc analysis when comparing all possible pairs of mean.
P<0.05 was considered to indicate a statistically significant
difference. GraphPad 9.0 (Dotmatics) was used for statistical
analysis and image generation.
Results
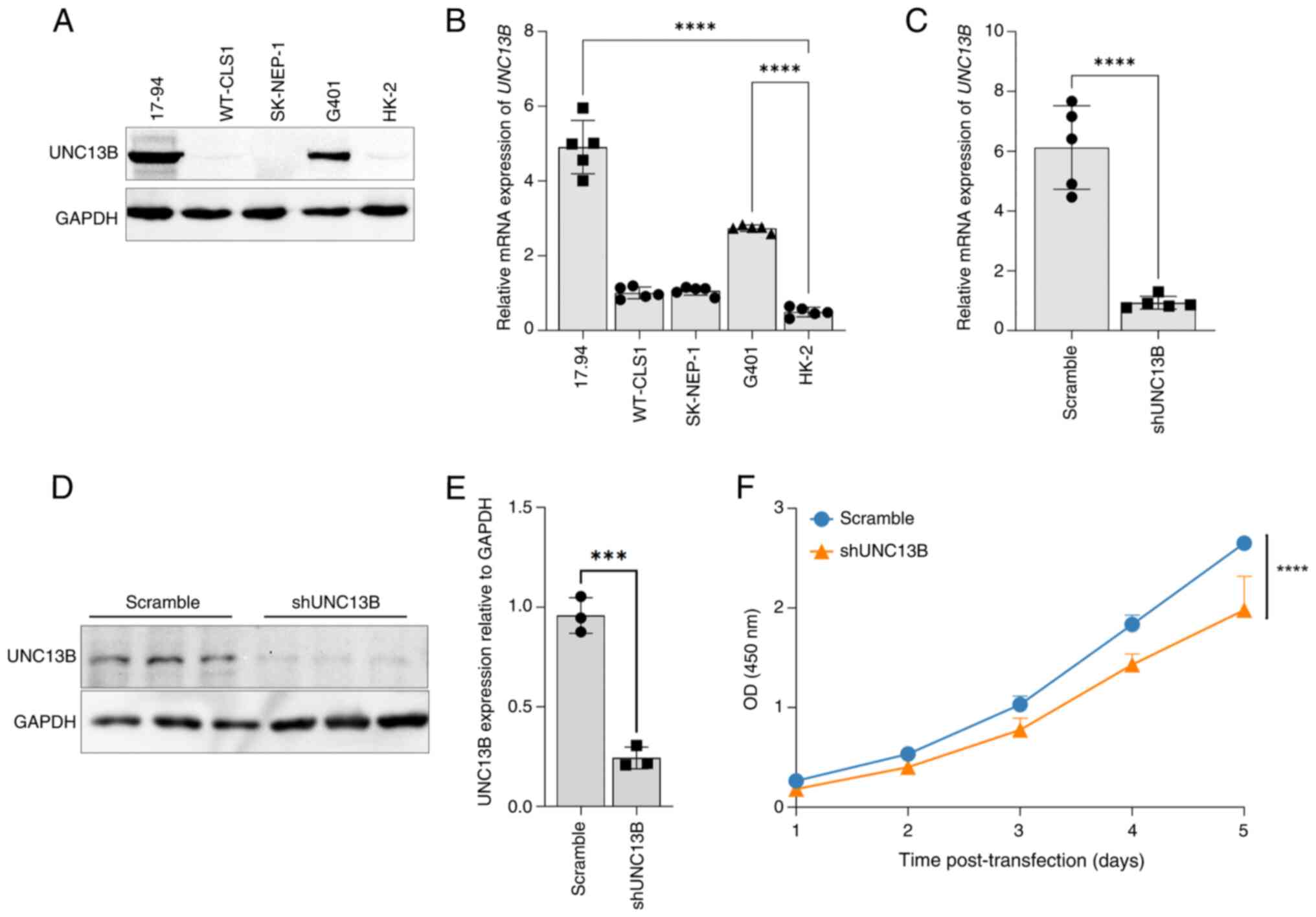
Elevated expression of UNC13B in
Wilms' tumor cell line
Using commercially available 17.94 Wilms' tumor cell
line (18), alongside other renal
tumor cell lines, an Ewing sarcoma (SK-NEP-1) (19), a rhabdoid tumor (G-401) (20), a kidney rhabdoid tumor (WT-CLS1) and
human normal renal cell lines (HK-2), an analysis of UNC13B
transcription and expression levels was performed. The results
revealed a significant increase in both the mRNA and protein
expression levels of UNC13B in the 17.94 cell line compared with
that in the HK-2 cell line (Fig. 1A and
B). The UNC13B expression level in 17.94 cells was also notably
higher compared with that in the other cell lines. Additionally,
there was a clear positive association between mRNA and protein
expression levels (Fig. 1A and B).
Before shRNA knockdown experiment, we inserted GFP cDNA into an
empty lentiCRISPR v2 vector to construct a GFP-lentiviral vector
for verifying transfection efficiency. The GFP-lentivirus was
transfected into 17.94 cells. After 24 h, fluorescence microscopy
analysis showed that over 95% of the cells expressed GFP,
indicating a transfection efficiency of over 95%. In shRNA
knockdown experiments performed in 17.94 cells, a significant
reduction in UNC13B mRNA levels was observed 48 h post-infection
under knockdown condition compared with controls, with a
multiplicity of infection (MOI) of 5 (Fig. 1C). Consequently, an MOI of 5 was
selected for protein level validation post-knockdown, revealing a
significant decrease in UNC13B expression levels compared with
controls (Fig. 1D and E). Moreover,
assessing the proliferation levels of knockdown cells revealed a
significant reduction in cell proliferation following the reduction
of UNC13B levels (Fig. 1F) compared
with controls, which is consistent with findings from our previous
study on UNC13B knockdown in other tumor cells (11).
UNC13B modulates Wilms' tumor cell
sensitivity to doxorubicin, independent of the cell cycle
Building upon findings from our previous study
(11), which indicated the
involvement of UNC13B in tumor cell resistance to chemotherapy, the
present study further assessed whether UNC13B is associated with
drug resistance and sensitivity in Wilms' tumor cells. In clinical
treatment for Wilms' tumor, chemotherapy drugs such as vincristine,
doxorubicin and actinomycin-D are primarily used for preoperative
treatment (21). Therefore, the
altered sensitivity of UNC13B knockdown 17.94 cells to these drugs
was analyzed. The results demonstrated that UNC13B knockdown
notably increased the sensitivity of 17.94 cells to doxorubicin and
actinomycin-D, although the IC50 value for vincristine was lower in
the UNC13B knockdown group (88.45±12.16 vs. 59.06±10.80 nM;
Fig. 2A) compared to the scramble
group, this difference was not statistically significant.
Significantly lowering the IC50 values for actinomycin-D
(1.544±0.09 vs. 1.005±0.07 nM; Fig.
2B) and doxorubicin (1882±124.3 vs. 697.2±46.29 nM; Fig. 2C) compared to the scramble group.
The changes in sensitivity to the aforementioned drugs were also
tested in G401 cells after UNC13B knockdown. Following UNC13B
knockdown, there was a significant decrease in the IC50
for all three drugs: Vincristine (77.74±6.82 vs. 33.17±2.83 nM;
Fig. 2D), actinomycin-D
(1.524±0.085 vs. 1.17±0.056 nM; Fig.
2E) and doxorubicin (1,018±65.23 vs. 239.0±30.14 nM; Fig. 2F) compare to the scramble group.
This demonstrates that reducing UNC13B levels enhances the
sensitivity of G401 cells to these drugs. Subsequently, the cell
cycle changes upon doxorubicin treatment were evaluated to
elucidate the role of UNC13B in drug sensitivity and resistance.
The results demonstrated that UNC13B had no notable impact on cell
cycle alterations (Fig. 2G and H),
suggesting the involvement of other mechanisms in UNC13B-mediated
drug resistance in Wilms' tumor cells. Furthermore, apoptosis in
17.94 cells following UNC13B knockdown was analyzed. The results
indicated that knockdown of UNC13B significantly increased
apoptosis levels at 0.5 µM doxorubicin (Fig. 2I and J) compared to the scramble
group; however, at 2 µM doxorubicin, the increase in apoptosis
levels was significant but less pronounced. This suggests that
UNC13B may negatively regulate the drug sensitivity of 17.94 cells
to doxorubicin-induced apoptosis.
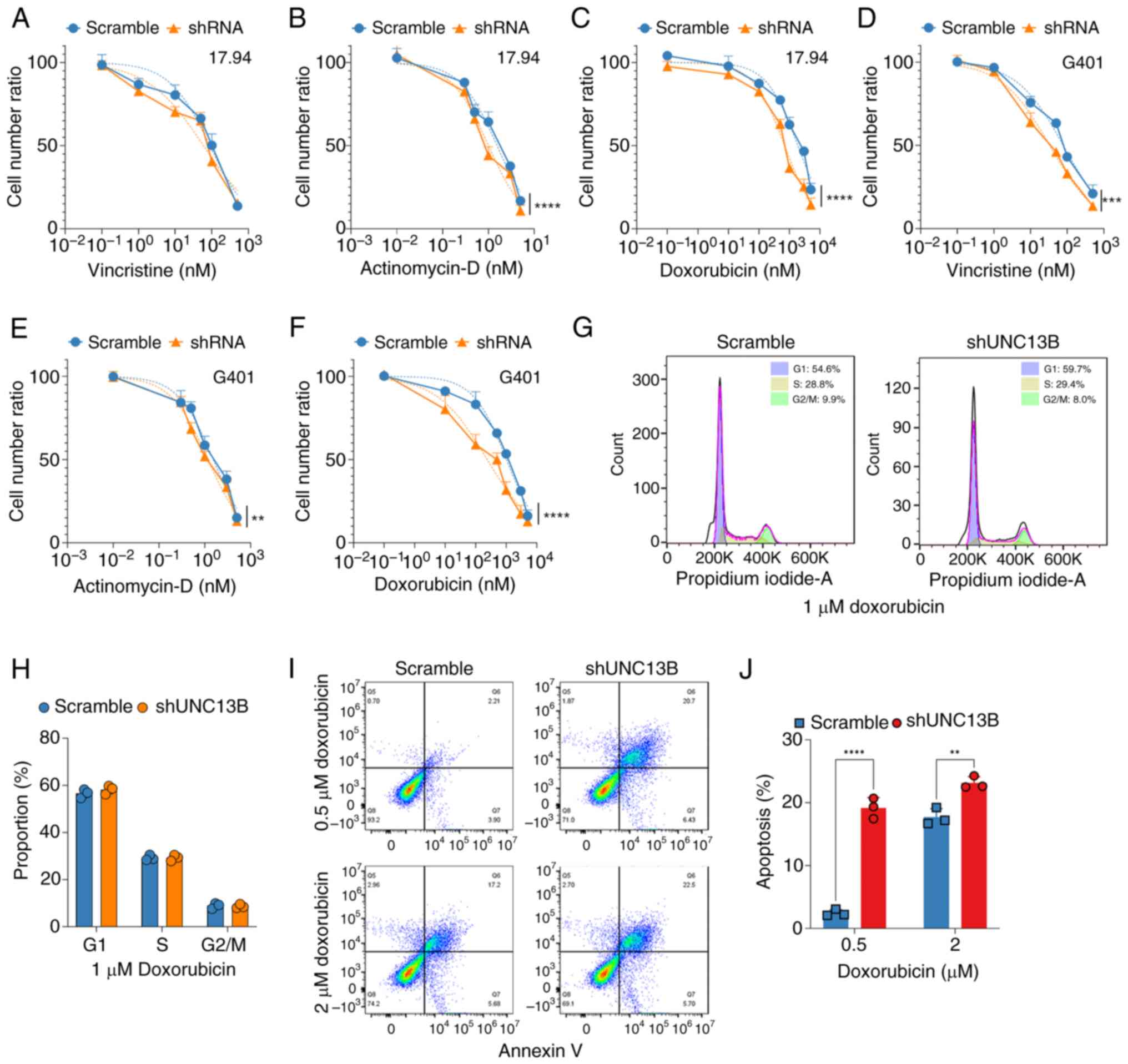 | Figure 2.UNC13B influences Wilms’ tumor
sensitivity to chemotherapy drugs independent of the cell cycle.
Evaluation of cell proliferation post-shRNA-mediated UNC13B
knockdown after treatment with varying concentrations of (A)
vincristine, (B) actinomycin-D and (C) doxorubicin for 48 h in
17.94 cells, and (D) vincristine, (E) actinomycin-D and (F)
doxorubicin in the G401 cell line, assessed using Cell Counting
Kit-8 assays. Changes in drug sensitivity were analyzed, with
dashed lines representing fitted curves for half-maximal inhibitory
concentration calculated using GraphPad software, and the cell
number ratio indicating the relative number of viable cells
compared between initial cell number and different time points. (G)
Cell cycle analysis of 1 µM doxorubicin treatment on control and
shUNC13B knockdown cells, detected after 48 h post-drug treatment.
(H) Quantification of the G1, S and G2 phases of the scramble and
shUNC13B groups. (I) Typical pseudocolor scatter plots of the
apoptosis analysis of UNC13B-knockdown 17.94 cells after 48 h
treatment with 0.5 and 2 µM doxorubicin, and (J) statistical
results. **P<0.01; ***P<0.001; ****P<0.0001. UNC13B,
unc-13 homolog B; sh, short hairpin. |
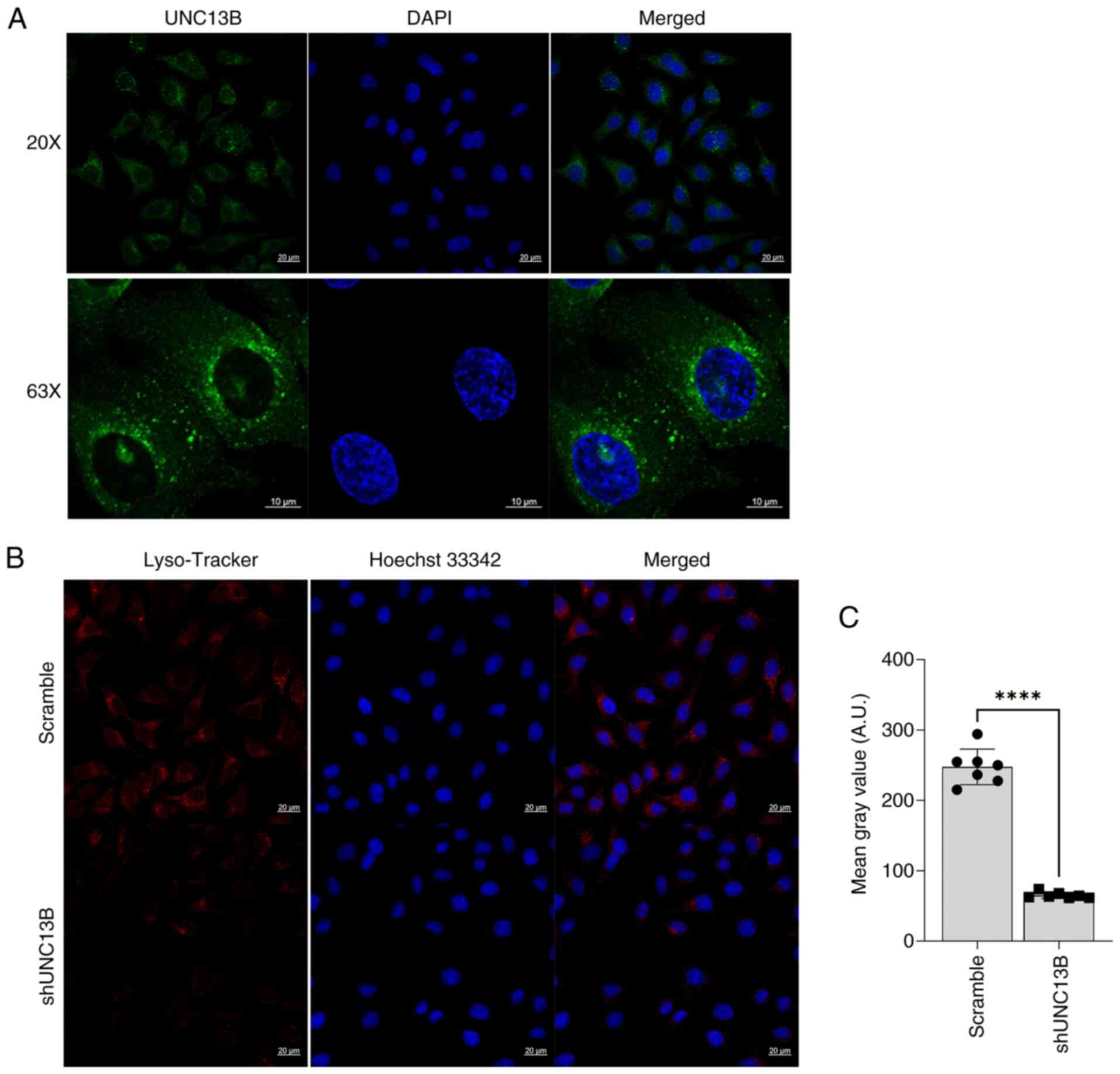
Involvement of UNC13B in Wilms' tumor
lysosome formation
A previous report suggested an association between
doxorubicin drug sensitivity and lysosomes (22). Additionally, several studies have
highlighted the notable role of UNC13B in regulating synaptic
vesicles (23,24). Hence, we hypothesized that UNC13B
may modulate drug sensitivity by participating in vesicle
regulation within cells and localization of UNC13B within the
cellular vesicles was detected using an indirect immunofluorescence
method. The results demonstrated the presence of UNC13B within the
cellular vesicles (Fig. 3A),
indicating its potential involvement in vesicle-related functions.
Furthermore, the lysosome status post-UNC13B knockdown was
analyzed, revealing a significant decrease in lysosome level
compared to the scramble group (Fig. 3B
and C). This suggests that UNC13B may regulate lysosome
formation through certain pathways.
UNC13B-induced lysosomal changes
affect cell sensitivity to doxorubicin
The transfection efficiency of the 17.94 cell line
was first evaluated using pCDNA3.1 as the expression vector. The
results revealed a significant increase in the transcription level
of UNC13B after transfection compared with the negative control,
indicating that overexpression of UNC13B using the transient
transfection system was effective in the 17.94 cell line (Fig. 4A). To further assess the influence
of UNC13B on lysosomes, the endogenous UNC13B was deleted, creating
17.94-delUNC13B. Subsequently, exogenous UNC13B was overexpressed
in this cell line. The results demonstrated that the expression
levels of LAMP1, a lysosome-associated protein, significantly
increased in the 17.94 OE group compared to the 17.94 cell NC
group, and decreased significantly in the 17.94 cell KO group
compared to the 17.94 cell NC group (Fig. 4B and C). As UNC13B expression
increased, the levels of LAMP1 expression also rose proportionally.
Simultaneously, using Lyso-Tracker, a significant increase in
lysosomal levels was observed in the 17.94 cell OE group compared
to the 17.94 cell NC group, while a significant decrease was
observed in the 17.94 cell KO group compared to the 17.94 cell NC
group (Fig. 4D and E). These
findings collectively indicate a clear positive association between
lysosomal levels and UNC13B expression. The significant results
compared the doxorubicin sensitivity between the 17.94 cell KO
group and the 17.94 cell NC group, showing significantly increased
sensitivity in the KO strain (1782±101.3 vs. 702.9±50.49 nM;
Fig. 4F). Conversely, following
UNC13B overexpression, there was a significant but minimal
reduction in 17.94 sensitivity to doxorubicin compared to the 17.94
cell NC group (1782±101.3 vs. 1928±84.38 nM).
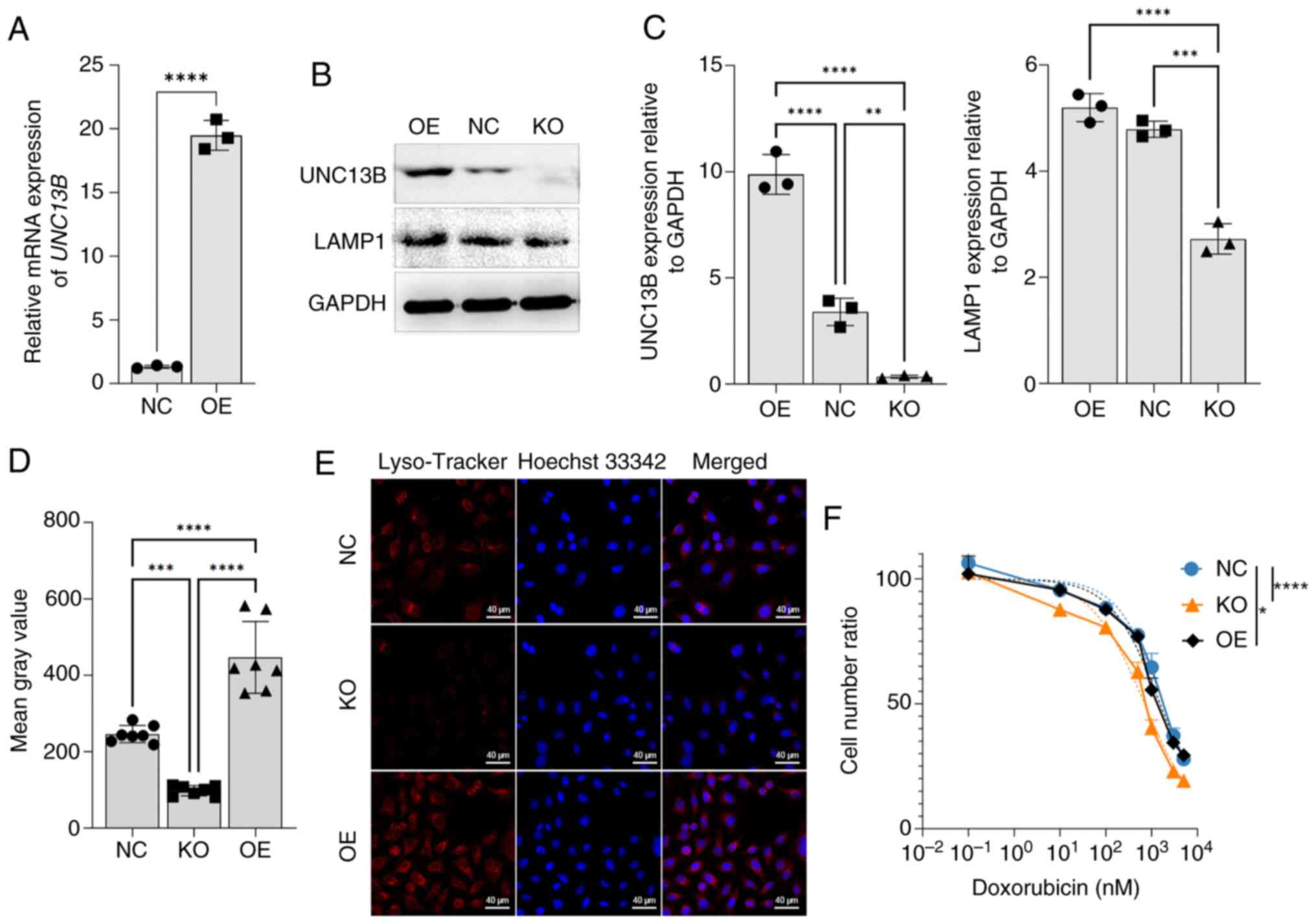 | Figure 4.UNC13B modulates cell drug
sensitivity by affecting lysosome formation. (A) 17.94 cell line
was transiently transfected with the UNC13B-pCDNA3.1 overexpression
vector using Lipofectamine 3,000. Reverse
transcription-quantitative PCR validation was performed 24 h
post-transfection. NC was the transfection with an empty pcDNA3.1
vector; n=3. (B) UNC13B and LAMP1 expression levels in 17.94 NC
cells, UNC13B 17.94 OE cells and UNC13B 17.94 KO cells, using GAPDH
as a reference, and (C) the associated semi-quantitative results.
(D) Analysis of Mean Gray Value of Lyso-Tracker in 6 random fields
of 17.94 NC, UNC13B 17.94 OE cells and UNC13B 17.94 KO cells, and
(E) representative images of 17.94 NC cells, UNC13B 17.94 OE cells
and UNC13B 17.94 KO cells. Lyso-Tracker indicated lysosomes, as the
dye is highly selective for acidic environments, with an excitation
wavelength of 577 nm and an emission wavelength of 590 nm. (F)
Assessment of doxorubicin sensitivity changes in 17.94 NC, UNC13B
17.94 OE cells and UNC13B 17.94 KO cells; n=3. The cell number
ratio indicates the relative number of viable cells compared
between initial cell number and different time points. *P<0.05;
**P<0.01; ***P<0.001; ****P<0.0001. UNC13B, unc-13 homolog
B; NC, negative control; OE, over-expressed; KO, knock-out; LAMP1,
lysosomal-associated membrane protein 1. |
Discussion
Doxorubicin, also known as Adriamycin, is an
anthracycline antibiotic derived from Streptomyces
peucetius, exerting its effect through several molecular
mechanisms that induce cell death or growth arrest, including the
inhibition of topoisomerase II, DNA intercalation and free radical
production (25). Resistance of
tumor cells to doxorubicin poses a significant challenge in its
clinical use. Moreover, clinical studies have revealed late effects
associated with doxorubicin, particularly cardiac system effects
such as arrhythmias (26,27). Therefore, reducing drug dosage and
lowering the doxorubicin's IC50 has become a critical
need in clinical application. Mechanisms influencing doxorubicin
drug sensitivity and resistance primarily involve
epithelial-to-mesenchymal transition (28,29),
alterations in topoisomerase II activity (30) and excessive activation of the ERK1/2
pathway (31). In addition to
molecular changes, cells develop resistance by enhancing their
detoxification capabilities, including reported overexpression of
glutathione S-transferase (32).
Notably, one key factor in doxorubicin resistance is lysosomes
(22). Research has reported
doxorubicin accumulation in the lysosomes of resistant strains
(33,34). In lymphoma cell lines, doxorubicin
has been reported to be sequestered within lysosomes (22). In the present study, post-UNC13B
knockdown, there was a decrease in the IC50 of
doxorubicin in the Wilms' tumor 17.94 cell line, from 1882±124.3 to
697.2±46.29 nM (Fig. 2C),
suggesting the involvement of UNC13B in the sensitivity of the
tumor to doxorubicin. Furthermore, the results of the present study
indicated that as lysosome quantity decreases in Wilms' tumor,
17.94 cells exhibited increased sensitivity to doxorubicin
(Fig. 4D-F). After UNC13B
knockdown, varying degrees of sensitivity changes to the other two
drugs, vincristine and actinomycin-D, were also observed in 17.94
cells. However, these changes were not significant for vincristine
(P=0.0726; Fig. 2A) or were
significant but minor for actinomycin-D (1.544±0.09 vs 1.005±0.07
nM; P<0.0001; IC50 reduced by 50%; Fig. 2B). Therefore, further exploration of
the mechanisms of sensitivity changes to vincristine and
actinomycin-D in 17.94 cells after UNC13B knockdown was not pursued
in the current study.
Lysosomes, a type of intracellular vesicle, are
crucial for maintaining cellular homeostasis, digestion and
breakdown of aging cellular components and ingested nutrients
(35,36). They also participate in cellular
signaling regulation (37,38) and programmed cell death (39). Given the multifaceted functions of
lysosomes, it is plausible that they serve a role in regulating
cellular sensitivity to drugs. Extensive research has reported
lysosome involvement in cellular drug sensitivity and resistance.
Drugs captured by lysosomes via cationic chelation or sequestration
led to drug isolation within the lysosomal acidic compartment,
thereby isolating the drug's action on its target (40,41).
Several chemotherapy drugs, including sunitinib (42), doxorubicin (43), rotenone (22) and vincristine (44), exhibited lysosomal capture,
contributing to cell resistance. Additionally, lysosomes regulate
cellular resistance through lysosome-mediated exocytosis (45). Due to the broad and critical role
lysosomes serve in cellular sensitivity and resistance to drugs,
researchers have targeted lysosomes to develop new strategies
against resistance. These include altering drug structures
(46), inhibiting key enzymes
involved in lysosomal acidification (47,48)
and interfering with intravesicular acidity (49). Several lysosomal proteins, including
ion channels (50) and PINK-1
(51), could serve as specific
targets to impair lysosomal activity, thereby selectively altering
the drug resistance characteristics of cancer cells. Changes in
lysosomal function have been reported in several types of tumors,
such as colorectal cancer (52) and
gliomas (53). In cancer, tumor
cells promote proliferation, migration and potentially resistance
to chemotherapy by altering the number of lysosomes, the protein
levels and activity of lysosomal hydrolases (54). In addition to the role in drug
resistance mentioned in the present study, lysosomes also
participate in the regulation of programmed cell death in tumor
cells (55). Moreover, lysosomes
can promote angiogenesis by secreting tissue proteases through
exocytosis, activating Matrix metalloproteinases (56). Therefore, a wide range of candidate
antitumor drugs has been developed targeting lysosomal organelle
membranes (57), several Cathepsins
contained within lysosomes (58),
lysosomal pH (59) and key cellular
processes involving lysosomes such as autophagy (60).
Most drugs targeting lysosomes in cancer research
are still in the laboratory or preclinical stages. Among
lysosome-mediated drugs in clinical research, hydroxychloroquine
(HCQ) is a clinically approved and widely studied lysosome-related
inhibitor. HCQ can inhibit autophagic degradation by blocking
lysosomal acidification (61). Vogl
et al (62) demonstrated in
a Phase I trial that combining bortezomib (a proteasome inhibitor)
with HCQ improved the efficacy of proteasome inhibition in multiple
myeloma. A total of 45% of the 25 patients treated with the
combination of HCQ and bortezomib demonstrated stable disease as
their best response (62).
Furthermore, in a clinical trial combining HCQ with the histone
deacetylase inhibitor vorinostat, 24/27 treated patients were
considered fully evaluable for study assessments and toxicity,
including one patient with renal cell carcinoma showing durable
partial response and two patients with colorectal cancer showing
long-term stable disease (63).
However, in a clinical trial for patients with malignant gliomas,
the overall survival of patients with malignant gliomas was not
notably improved when HCQ was used in combination with low-dose
continuous temozolomide at a dose below the maximum tolerated dose
(600 mg/day) (64). Additionally,
low-dose, short-term use of HCQ has few side effects (65); however, at higher doses and longer
durations, certain serious side effects have been observed, such as
retinal toxicity (66). Therefore,
improving the efficacy of HCQ should also be a focus of future
research. We hypothesize that, as a gene involved in lysosome
regulation, UNC13B will synergize with HCQ to achieve better
efficacy.
In the present study, a significant positive
association between UNC13B and lysosome formation was demonstrated
(Fig. 4B-E). UNC13B participates in
regulating vesicle precursor formation within neurons, essential
for neurotransmitter storage and release (67). UNC13B, in collaboration with other
proteins such as SNAP receptor proteins, facilitates vesicle fusion
with the cell membrane (68). These
findings suggest that in tumor cells, UNC13B may serve a role in
vesicle maturation and regulation. The specific role of UNC13B in
lysosome-mediated drug resistance involves several potential
mechanisms based on its function in vesicular transport and its
interactions with cellular organelles. UNC13B may influence the
trafficking and fusion of lysosomes with autophagosomes or other
vesicles. If UNC13B enhances the efficiency of lysosome fusion with
autophagosomes, this may lead to more effective sequestration and
degradation of drugs, thereby contributing to drug resistance. As
autophagy serves a role in drug resistance by degrading damaged
organelles and proteins that could be targeted by chemotherapeutic
agents, lysosomes may serve a central role in cellular autophagy
(69). Furthermore, UNC13B may be
involved in regulating the fusion of autophagy-related vesicles, a
mechanism that could contribute to drug resistance. A UNC13 family
protein, UNC13D has been reported to regulate the autophagy process
(70). UNC13B may also influence
the autophagy process by regulating the transport and fusion of
vesicles, especially during the fusion step between autophagosomes
and lysosomes. The results of the current study demonstrated a
clear positive association between UNC13B levels and lysosomal
levels, but the specific interactions and biological significance
of UNC13B with lysosomes still need to be validated and clarified
through further experimental research.
The findings of the present study should be
validated using more Wilms' tumor cell lines; to the best of our
knowledge, however, the only commercially available Wilms' tumor
cell line is 17.94. Previous cell lines such as SK-NEP-1 and G401,
which have been widely used for investigating Wilms' tumor
mechanisms, are actually Ewing sarcoma and rhabdoid tumor cell
lines, respectively (19,20). The use of only one Wilms' tumor cell
line is acknowledged as a limitation of the present study, and it
is suggested that additional Wilms' tumor cell lines would be used
in future experiments to validate the findings. Although we have
demonstrated a positive association between UNC13B levels and
lysosome formation, the specific signaling pathways through which
UNC13B regulates lysosome formation remain to be elucidated.
Additionally, it is necessary to verify whether the regulatory role
of UNC13B on lysosome formation is universal. Further studies are
needed to determine if UNC13B can positively regulate lysosome
formation in other cell lines as well.
In conclusion, the results of the present study
demonstrated elevated expression of UNC13B in the Wilms' tumor cell
line, 17.94, whilst its expression in normal cells (HK-2) remained
low. This indicates that UNC13B could serve as a specific
regulatory target for combined therapy with doxorubicin.
Additionally, the results preliminarily demonstrated increased
sensitivity to doxorubicin in 17.94 cells when UNC13B was
inhibited. Furthermore, considering previous reports on
lysosome-mediated resistance with drugs such as vincristine and
rotenone, UNC13B holds promise as a target for combined therapy
with such drugs. Lysosomes have become increasingly significant due
to their role in cancer progression and resistance. However,
lysosomal activity is crucial for almost all types of cells, hence,
targeting them is generally not a cancer-specific strategy and may
lead to severe adverse effects. Cancer-specific or cancer-enriched
targets in this regard are still uncommon. The results of the
present study suggest that UNC13B is likely an enriched target
involved in lysosomal regulation in Wilms' tumor, providing a new
intervention target for optimizing chemotherapy approaches in
Wilms' tumor and other cancer types with high UNC13B
expression.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Key R&D
Program of China (grant no. 2022YFC2703500) and the Natural Science
Foundation of Guangdong Province (grant no. 2019A1515110703).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JZ and XW contributed to the study conception and
design. Material preparation, data collection and analysis were
performed by XC, YB and GS. XC and XW confirm the authenticity of
all the raw data. The first draft of the manuscript was written by
XC and XW. All authors commented on previous versions of the
manuscript. All the authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pastore G, Znaor A, Spreafico F, Graf N,
Pritchard-Jones K and Steliarova-Foucher E: Malignant renal tumours
incidence and survival in European children (1978–1997): Report
from the automated childhood cancer information system project. Eur
J Cancer. 42:2103–2114. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Spreafico F, Fernandez CV, Brok J, Nakata
K, Vujanic G, Geller JI, Gessler M, Maschietto M, Behjati S,
Polanco A, et al: Wilms tumour. Nat Rev Dis Primers. 7:752021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dome JS, Graf N, Geller JI, Fernandez CV,
Mullen EA, Spreafico F, Van den Heuvel-Eibrink M and
Pritchard-Jones K: Advances in wilms tumor treatment and biology:
Progress through international collaboration. J Clin Oncol.
33:2999–3007. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Termuhlen AM, Tersak JM, Liu Q, Yasui Y,
Stovall M, Weathers R, Deutsch M, Sklar CA, Oeffinger KC, Armstrong
G, et al: Twenty-five year follow-up of childhood Wilms tumor: A
report from the childhood cancer survivor study. Pediatr Blood
Cancer. 57:1210–1216. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dome JS, Fernandez CV, Mullen EA,
Kalapurakal JA, Geller JI, Huff V, Gratias EJ, Dix DB, Ehrlich PF,
Khanna G, et al: Children's oncology group's 2013 blueprint for
research: Renal tumors. Pediatr Blood Cancer. 60:994–1000. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vujanic GM, Gessler M, Ooms A, Collini P,
Coulomb-l'Hermine A, D'Hooghe E, de Krijger RR, Perotti D,
Pritchard-Jones K, Vokuhl C, et al: The UMBRELLA SIOP-RTSG 2016
Wilms tumour pathology and molecular biology protocol. Nat Rev
Urol. 15:693–701. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pritchard-Jones K, Bergeron C, de Camargo
B, van den Heuvel-Eibrink MM, Acha T, Godzinski J, Oldenburger F,
Boccon-Gibod L, Leuschner I, Vujanic G, et al: Omission of
doxorubicin from the treatment of stage II–III, intermediate-risk
Wilms' tumour (SIOP WT 2001): An open-label, non-inferiority,
randomised controlled trial. Lancet. 386:1156–1164. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Israels T, Moreira C, Scanlan T, Molyneux
L, Kampondeni S, Hesseling P, Heij H, Borgstein E, Vujanic G,
Pritchard-Jones K and Hadley L: SIOP PODC: Clinical guidelines for
the management of children with Wilms tumour in a low income
setting. Pediatr Blood Cancer. 60:5–11. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van den Heuvel-Eibrink MM, Hol JA,
Pritchard-Jones K, van Tinteren H, Furtwängler R, Verschuur AC,
Vujanic GM, Leuschner I, Brok J, Rübe C, et al: Position paper:
Rationale for the treatment of Wilms tumour in the UMBRELLA
SIOP-RTSG 2016 protocol. Nat Rev Urol. 14:743–752. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kaste SC, Dome JS, Babyn PS, Graf NM,
Grundy P, Godzinski J, Levitt GA and Jenkinson H: Wilms tumour:
Prognostic factors, staging, therapy and late effects. Pediatr
Radiol. 38:2–17. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang XB, Yuan LH, Yan LP, Ye YB, Lu B and
Xu X: UNC13B promote arsenic trioxide resistance in chronic
lymphoid leukemia through mitochondria quality control. Front
Oncol. 12:9209992022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mansoori B, Mohammadi A, Davudian S,
Shirjang S and Baradaran B: The different mechanisms of cancer drug
resistance: A brief review. Adv Pharm Bull. 7:339–348. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pooryasin A, Maglione M, Schubert M,
Matkovic-Rachid T, Hasheminasab SM, Pech U, Fiala A, Mielke T and
Sigrist SJ: Unc13A and Unc13B contribute to the decoding of
distinct sensory information in Drosophila. Nat Commun.
12:19322021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Green TE, Scheffer IE, Berkovic SF and
Hildebrand MS: UNC13B and focal epilepsy. Brain. 145:e10–e12. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Corrigendum to: UNC13B variants associated
with partial epilepsy with favourable outcome. Brain. 145:e52022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Warr MR, Binnewies M, Flach J, Reynaud D,
Garg T, Malhotra R, Debnath J and Passegué E: FOXO3A directs a
protective autophagy program in haematopoietic stem cells. Nature.
494:323–327. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brown KW, Charles A, Dallosso A, White G,
Charlet J, Standen GR and Malik K: Characterization of 17.94, a
novel anaplastic Wilms' tumor cell line. Cancer Gene. 205:319–326.
2012. View Article : Google Scholar
|
|
19
|
Smith MA, Morton CL, Phelps D, Girtman K,
Neale G and Houghton PJ: SK-NEP-1 and Rh1 are Ewing family tumor
lines. Pediatr Blood Cancer. 50:703–706. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Garvin AJ, Re GG, Tarnowski BI,
Hazen-Martin DJ and Sens DA: The G401 cell line, utilized for
studies of chromosomal changes in Wilms' tumor, is derived from a
rhabdoid tumor of the kidney. Am J Pathol. 142:375–380.
1993.PubMed/NCBI
|
|
21
|
Oostveen RM and Pritchard-Jones K:
Pharmacotherapeutic management of wilms tumor: An update. Paediatr
Drugs. 21:1–13. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hurwitz SJ, Terashima M, Mizunuma N and
Slapak CA: Vesicular anthracycline accumulation in
doxorubicin-selected U-937 cells: Participation of lysosomes.
Blood. 89:3745–3754. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Qiao JD, Liu XR, Liu DT, Chen YH,
Wu Y, Sun Y, Yu J, Ren RN, Mei Z, et al: UNC13B variants associated
with partial epilepsy with favourable outcome. Brain.
144:3050–3060. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bohme MA, Beis C, Reddy-Alla S, Reynolds
E, Mampell MM, Grasskamp AT, Lützkendorf J, Bergeron DD, Driller
JH, Babikir H, et al: Active zone scaffolds differentially
accumulate Unc13 isoforms to tune Ca(2+) channel-vesicle coupling.
Nat Neurosci. 19:1311–1320. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Meredith AM and Dass CR: Increasing role
of the cancer chemotherapeutic doxorubicin in cellular metabolism.
J Pharm Pharmacol. 68:729–741. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Murtagh G, Lyons T, O'Connell E, Ballot J,
Geraghty L, Fennelly D, Gullo G, Ledwidge M, Crown J, Gallagher J,
et al: Late cardiac effects of chemotherapy in breast cancer
survivors treated with adjuvant doxorubicin: 10-year follow-up.
Breast Cancer Res Treat. 156:501–506. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kilickap S, Barista I, Akgul E, Aytemir K,
Aksoy S and Tekuzman G: Early and late arrhythmogenic effects of
doxorubicin. South Med J. 100:262–265. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Saxena M, Stephens MA, Pathak H and
Rangarajan A: Transcription factors that mediate
epithelial-mesenchymal transition lead to multidrug resistance by
upregulating ABC transporters. Cell Death Dis. 2:e1792011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu T, Zhang J, Chen W, Pan S, Zhi X, Wen
L, Zhou Y, Chen BW, Qiu J, Zhang Y, et al: ARK5 promotes
doxorubicin resistance in hepatocellular carcinoma via
epithelial-mesenchymal transition. Cancer Lett. 377:140–148. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Withoff S, De Jong S, De Vries EG and
Mulder NH: Human DNA topoisomerase II: Biochemistry and role in
chemotherapy resistance (review). Anticancer Res. 16:1867–1880.
1996.PubMed/NCBI
|
|
31
|
Shukla A, Hillegass JM, MacPherson MB,
Beuschel SL, Vacek PM, Pass HI, Carbone M, Testa JR and Mossman BT:
Blocking of ERK1 and ERK2 sensitizes human mesothelioma cells to
doxorubicin. Mol Cancer. 9:3142010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Singh SV, Nair S, Ahmad H, Awasthi YC and
Krishan A: Glutathione S-transferases and glutathione peroxidases
in doxorubicin-resistant murine leukemic P388 cells. Biochem
Pharmacol. 38:3505–3510. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo B, Tam A, Santi SA and Parissenti AM:
Role of autophagy and lysosomal drug sequestration in acquired
resistance to doxorubicin in MCF-7 cells. BMC Cancer. 16:7622016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seebacher NA, Richardson DR and Jansson
PJ: A mechanism for overcoming P-glycoprotein-mediated drug
resistance: Novel combination therapy that releases stored
doxorubicin from lysosomes via lysosomal permeabilization using
Dp44mT or DpC. Cell Death Dis. 7:e25102016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lamming DW and Bar-Peled L: Lysosome: The
metabolic signaling hub. Traffic. 20:27–38. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mahapatra KK, Mishra SR, Behera BP, Patil
S, Gewirtz DA and Bhutia SK: The lysosome as an imperative
regulator of autophagy and cell death. Cell Mol Life Sci.
78:7435–7449. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang C and Wang X: Lysosome biogenesis:
Regulation and functions. J Cell Biol. 220:e2021020012021.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Perera RM and Zoncu R: The lysosome as a
regulatory hub. Annu Rev Cell Dev Biol. 32:223–253. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Patra S, Patil S, Klionsky DJ and Bhutia
SK: Lysosome signaling in cell survival and programmed cell death
for cellular homeostasis. J Cell Physiol. 238:287–305. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Adar Y, Stark M, Bram EE, Nowak-Sliwinska
P, van den Bergh H, Szewczyk G, Sarna T, Skladanowski A, Griffioen
AW and Assaraf YG: Imidazoacridinone-dependent lysosomal
photodestruction: A pharmacological Trojan horse approach to
eradicate multidrug-resistant cancers. Cell Death Disease.
3:e2932012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kaufmann AM and Krise JP: Lysosomal
sequestration of amine-containing drugs: Analysis and therapeutic
implications. J Pharma Sci. 96:729–746. 2007. View Article : Google Scholar
|
|
42
|
Gotink KJ, Broxterman HJ, Labots M, de
Haas RR, Dekker H, Honeywell RJ, Rudek MA, Beerepoot LV, Musters
RJ, Jansen G, et al: Lysosomal sequestration of sunitinib: A novel
mechanism of drug resistance. Clin Cancer Res. 17:7337–7346. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Herlevsen M, Oxford G, Owens CR, Conaway M
and Theodorescu D: Depletion of major vault protein increases
doxorubicin sensitivity and nuclear accumulation and disrupts its
sequestration in lysosomes. Mol Cancer Ther. 6:1804–1813. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Groth-Pedersen L, Ostenfeld MS,
Hoyer-Hansen M, Nylandsted J and Jäättelä M: Vincristine induces
dramatic lysosomal changes and sensitizes cancer cells to
lysosome-destabilizing siramesine. Cancer Res. 67:2217–2225. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yanes RE, Tarn D, Hwang AA, Ferris DP,
Sherman SP, Thomas CR, Lu J, Pyle AD, Zink JI and Tamanoi F:
Involvement of lysosomal exocytosis in the excretion of mesoporous
silica nanoparticles and enhancement of the drug delivery effect by
exocytosis inhibition. Small. 9:697–704. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Duvvuri M, Konkar S, Funk RS, Krise JM and
Krise JP: A chemical strategy to manipulate the intracellular
localization of drugs in resistant cancer cells. Biochemistry.
44:15743–15749. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ouar Z, Bens M, Vignes C, Paulais M,
Pringel C, Fleury J, Cluzeaud F, Lacave R and Vandewalle A:
Inhibitors of vacuolar H+-ATPase impair the preferential
accumulation of daunomycin in lysosomes and reverse the resistance
to anthracyclines in drug-resistant renal epithelial cells. Biochem
J. 370:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hrabeta J, Groh T, Khalil MA, Poljakova J,
Adam V, Kizek R, Uhlik J, Doktorova H, Cerna T, Frei E, et al:
Vacuolar-ATPase-mediated intracellular sequestration of ellipticine
contributes to drug resistance in neuroblastoma cells. Int J Oncol.
47:971–980. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kazmi F, Hensley T, Pope C, Funk RS,
Loewen GJ, Buckley DB and Parkinson A: Lysosomal sequestration
(trapping) of lipophilic amine (cationic amphiphilic) drugs in
immortalized human hepatocytes (Fa2N-4 cells). Drug Metab Dispos.
41:897–905. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Geisslinger F, Muller M, Vollmar AM and
Bartel K: Targeting lysosomes in cancer as promising strategy to
overcome chemoresistance-a mini review. Front Oncol. 10:11562020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dykstra KM, Fay HRS, Massey AC, Yang N,
Johnson M, Portwood S, Guzman ML and Wang ES: Inhibiting autophagy
targets human leukemic stem cells and hypoxic AML blasts by
disrupting mitochondrial homeostasis. Blood Adv. 5:2087–2100. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Abdulla MH, Valli-Mohammed MA, Al-Khayal
K, Al Shkieh A, Zubaidi A, Ahmad R, Al-Saleh K, Al-Obeed O and
McKerrow J: Cathepsin B expression in colorectal cancer in a Middle
East population: Potential value as a tumor biomarker for late
disease stages. Oncol Rep. 37:3175–3180. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fukuda ME, Iwadate Y, Machida T, Hiwasa T,
Nimura Y, Nagai Y, Takiguchi M, Tanzawa H, Yamaura A and Seki N:
Cathepsin D is a potential serum marker for poor prognosis in
glioma patients. Cancer Res. 65:5190–5194. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Davidson SM and Heiden MG: Critical
functions of the lysosome in cancer biology. Ann Rev Pharmacol
Toxicol. 57:481–507. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Iulianna T, Kuldeep N and Eric F: The
Achilles' heel of cancer: Targeting tumors via lysosome-induced
immunogenic cell death. Cell Death Dis. 13:5092022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kallunki T, Olsen OD and Jäättelä M:
Cancer-associated lysosomal changes: Friends or foes? Oncogene.
32:1995–2004. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Boya P and Kroemer G: Lysosomal membrane
permeabilization in cell death. Oncogene. 27:6434–6451. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Groth-Pedersen L and Jäättelä M: Combating
apoptosis and multidrug resistant cancers by targeting lysosomes.
Cancer Lett. 332:265–274. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Manic G, Obrist F, Kroemer G, Vitale I and
Galluzzi L: Chloroquine and hydroxychloroquine for cancer therapy.
Mol Cell Oncol. 1:e299112014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dufour M, Dormond-Meuwly A, Demartines N
and Dormond O: Targeting the mammalian target of rapamycin (mTOR)
in cancer therapy: Lessons from past and future perspectives.
Cancers (Basel). 3:2478–2500. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Pellegrini P, Strambi A, Zipoli C,
Hägg-Olofsson M, Buoncervello M, Linder S and De Milito A: Acidic
extracellular pH neutralizes the autophagy-inhibiting activity of
chloroquine: Implications for cancer therapies. Autophagy.
10:562–571. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Vogl DT, Stadtmauer EA, Tan KS, Heitjan
DF, Davis LE, Pontiggia L, Rangwala R, Piao S, Chang YC, Scott EC,
et al: Combined autophagy and proteasome inhibition: A phase 1
trial of hydroxychloroquine and bortezomib in patients with
relapsed/refractory myeloma. Autophagy. 10:1380–1390. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Mahalingam D, Mita M, Sarantopoulos J,
Wood L, Amaravadi RK, Davis LE, Mita AC, Curiel TJ, Espitia CM,
Nawrocki ST, et al: Combined autophagy and HDAC inhibition: A phase
I safety, tolerability, pharmacokinetic, and pharmacodynamic
analysis of hydroxychloroquine in combination with the HDAC
inhibitor vorinostat in patients with advanced solid tumors.
Autophagy. 10:1403–1414. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Rosenfeld MR, Ye X, Supko JG, Desideri S,
Grossman SA, Brem S, Mikkelson T, Wang D, Chang YC, Hu J, et al: A
phase I/II trial of hydroxychloroquine in conjunction with
radiation therapy and concurrent and adjuvant temozolomide in
patients with newly diagnosed glioblastoma multiforme. Autophagy.
10:1359–1368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Amaravadi RK, Lippincott-Schwartz J, Yin
XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT and White E:
Principles and current strategies for targeting autophagy for
cancer treatment. Clin Cancer Res. 17:654–666. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Rangwala R, Leone R, Chang YC, Fecher LA,
Schuchter LM, Kramer A, Tan KS, Heitjan DF, Rodgers G, Gallagher M,
et al: Phase I trial of hydroxychloroquine with dose-intense
temozolomide in patients with advanced solid tumors and melanoma.
Autophagy. 10:1369–1379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Rossner S, Fuchsbrunner K, Lange-Dohna C,
Hartlage-Rübsamen M, Bigl V, Betz A, Reim K and Brose N:
Munc13-1-mediated vesicle priming contributes to secretory amyloid
precursor protein processing. J Biol Chem. 279:27841–27844. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Dittman JS: Unc13: A multifunctional
synaptic marvel. Curr Opin Neurobiol. 57:17–25. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yamamoto H, Zhang S and Mizushima N:
Autophagy genes in biology and disease. Nat Rev Gene. 24:382–400.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang J, He J, Johnson JL, Napolitano G,
Ramadass M, Rahman F and Catz SD: Cross-regulation of defective
endolysosome trafficking and enhanced autophagy through TFEB in
UNC13D deficiency. Autophagy. 15:1738–1756. 2019. View Article : Google Scholar : PubMed/NCBI
|