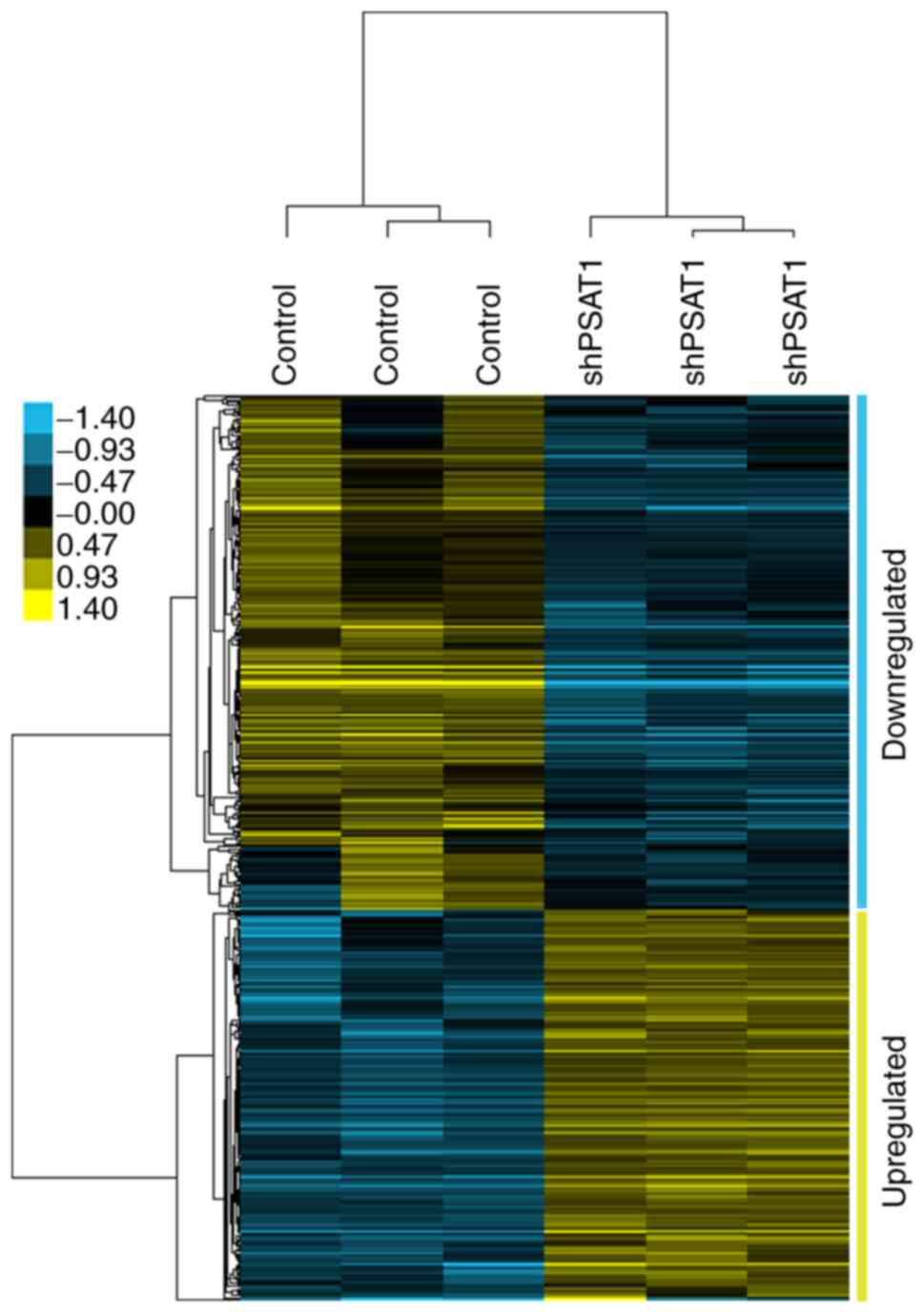
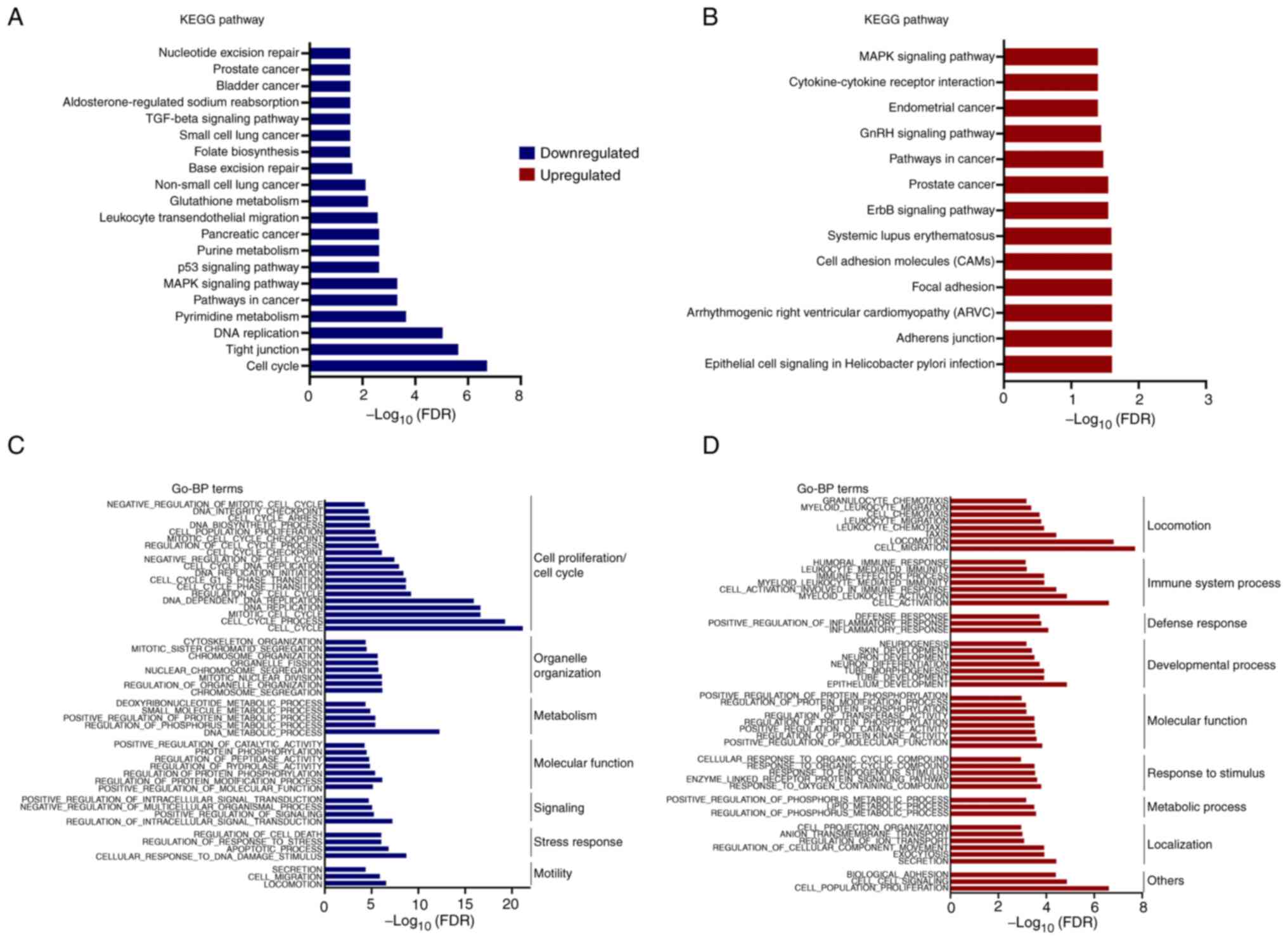
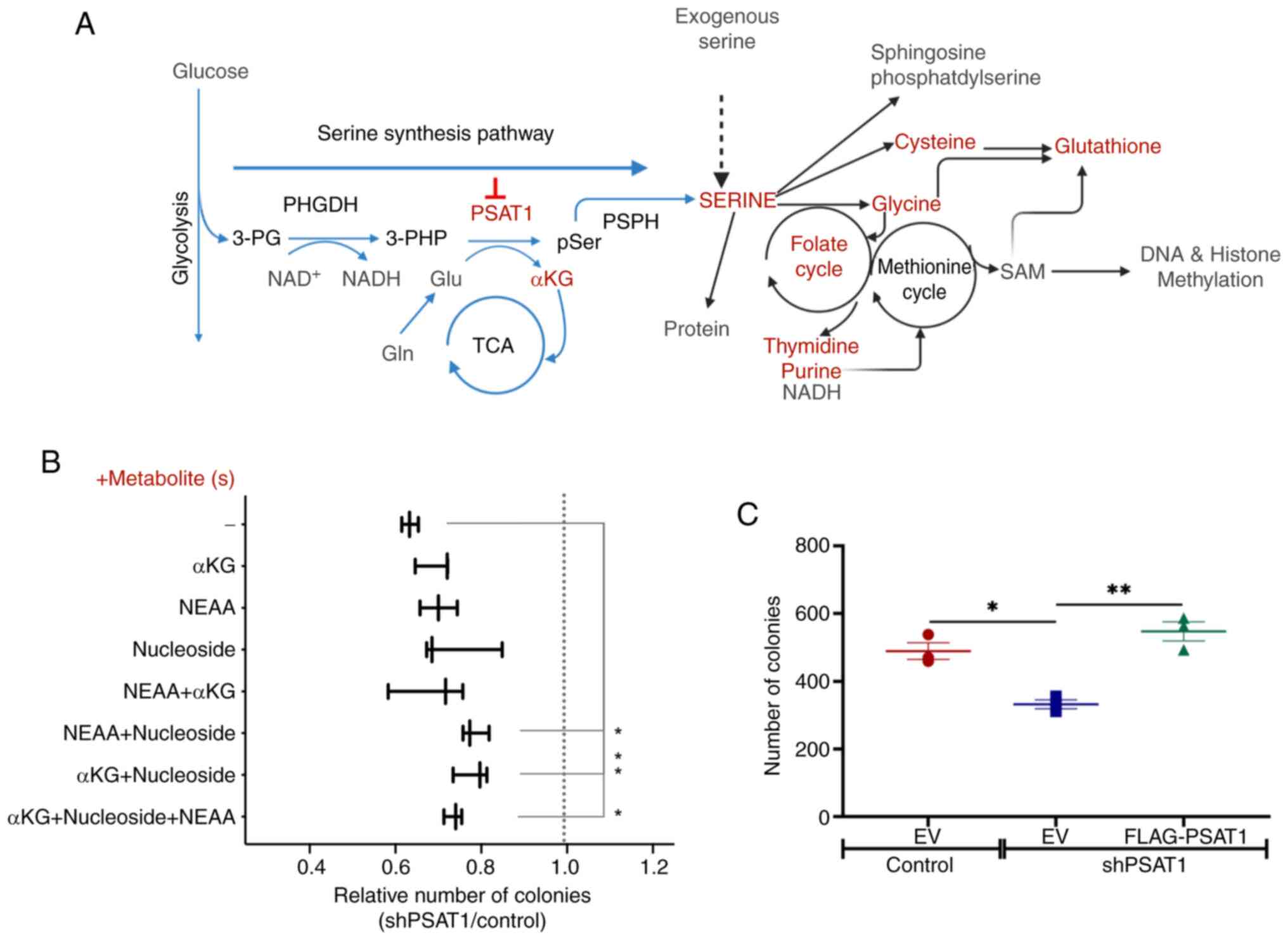
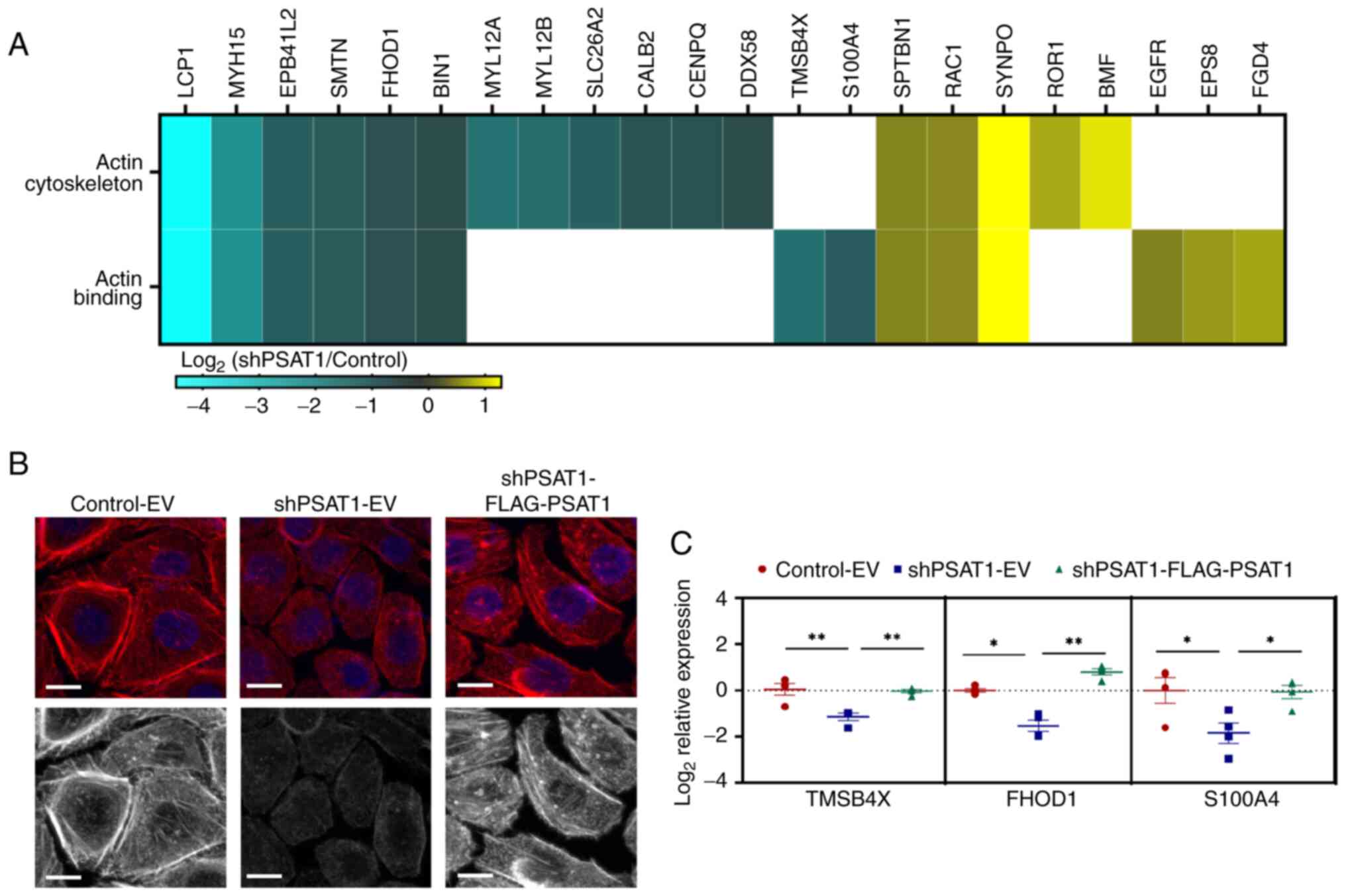
|
1
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou X, Tian C, Cao Y, Zhao M and Wang K:
The role of serine metabolism in lung cancer: From oncogenesis to
tumor treatment. Front Genet. 13:10846092023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim SK, Jung WH and Koo JS: Differential
expression of enzymes associated with serine/glycine metabolism in
different breast cancer subtypes. PLoS One. 9:e1010042014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun WY, Kim HM, Jung WH and Koo JS:
Expression of serine/glycine metabolism-related proteins is
different according to the thyroid cancer subtype. J Transl Med.
14:1682016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mullarky E, Lucki NC, Beheshti Zavareh R,
Anglin JL, Gomes AP, Nicolay BN, Wong JC, Christen S, Takahashi H,
Singh PK, et al: Identification of a small molecule inhibitor of
3-phosphoglycerate dehydrogenase to target serine biosynthesis in
cancers. Proc Natl Acad Sci USA. 113:1778–1783. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pacold ME, Brimacombe KR, Chan SH, Rohde
JM, Lewis CA, Swier LJ, Possemato R, Chen WW, Sullivan LB, Fiske
BP, et al: A PHGDH inhibitor reveals coordination of serine
synthesis and one-carbon unit fate. Nat Chem Biol. 12:452–458.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu S, Wang X, Liu L and Ren G:
Stabilization of Notch1 and β-catenin in response to ER-breast
cancer-specific up-regulation of PSAT1 mediates distant metastasis.
Transl Oncol. 20:1013992022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Y, Li J, Dong X, Meng D, Zhi X, Yuan
L and Yao L: PSAT1 regulated oxidation-reduction balance affects
the growth and prognosis of epithelial ovarian cancer. Onco Targets
Ther. 13:5443–5453. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fang Y, Liang X, Xu J and Cai X: miR-424
targets AKT3 and PSAT1 and has a tumor-suppressive role in human
colorectal cancer. Cancer Manag Res. 10:6537–6547. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang H, Cui L, Li D, Fan M, Liu Z, Liu C,
Pan S, Zhang L, Zhang H and Zhao Y: Overexpression of PSAT1
regulated by G9A sustains cell proliferation in colorectal cancer.
Signal Transduct Target Ther. 5:472020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Biyik-Sit R, Kruer T, Dougherty S, Bradley
JA, Wilkey DW, Merchant ML, Trent JO and Clem BF: Nuclear pyruvate
kinase M2 (PKM2) contributes to phosphoserine aminotransferase 1
(PSAT1)-mediated cell migration in EGFR-activated lung cancer
cells. Cancers (Basel). 13:39382021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo MY, Zhou Y, Gu WM, Wang C, Shen NX,
Dong JK, Lei HM, Tang YB, Liang Q, Zou JH, et al: Metabolic and
nonmetabolic functions of PSAT1 coordinate signaling cascades to
confer EGFR inhibitor resistance and drive progression in lung
adenocarcinoma. Cancer Res. 82:3516–3531. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Duan W and Liu X: PSAT1 upregulation
contributes to cell growth and cisplatin resistance in cervical
cancer cells via regulating PI3K/AKT signaling pathway. Ann Clin
Lab Sci. 50:512–518. 2020.PubMed/NCBI
|
|
14
|
Gao S, Ge A, Xu S, You Z, Ning S, Zhao Y
and Pang D: PSAT1 is regulated by ATF4 and enhances cell
proliferation via the GSK3β/β-catenin/cyclin D1 signaling pathway
in ER-negative breast cancer. J Exp Clin Cancer Res. 36:1792017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang Y, Wu J, Cai J, He Z, Yuan J, Zhu X,
Li Y, Li M and Guan H: PSAT1 regulates cyclin D1 degradation and
sustains proliferation of non-small cell lung cancer cells. Int J
Cancer. 136:E39–E50. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu S and Le H: Dual roles of PKM2 in
cancer metabolism. Acta Biochim Biophys Sin (Shanghai). 45:27–35.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sutendra G, Kinnaird A, Dromparis P,
Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E and
Michelakis ED: A nuclear pyruvate dehydrogenase complex is
important for the generation of acetyl-CoA and histone acetylation.
Cell. 158:84–97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
O'Cathail SM, Wu CH, Lewis A, Holmes C,
Hawkins MA and Maughan T: NRF2 metagene signature is a novel
prognostic biomarker in colorectal cancer. Cancer Genet. 248–249.
1–10. 2020.
|
|
19
|
Wang X, Yu Q, Ghareeb WM, Zhang Y, Lu X,
Huang Y, Huang S, Sun Y, Lin J, Liu J and Chi P: Downregulated
SPINK4 is associated with poor survival in colorectal cancer. BMC
Cancer. 19:12582019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
You GR, Cheng AJ, Lee LY, Huang YC, Liu H,
Chen YJ and Chang JT: Prognostic signature associated with
radioresistance in head and neck cancer via transcriptomic and
bioinformatic analyses. BMC Cancer. 19:642019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
de Hoon MJL, Imoto S, Nolan J and Miyano
S: Open source clustering software. Bioinformatics. 20:1453–1454.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Saldanha AJ: Java Treeview-extensible
visualization of microarray data. Bioinformatics. 20:3246–3248.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Simon R, Lam A, Li MC, Ngan M, Menenzes S
and Zhao Y: Analysis of gene expression data using BRB-ArrayTools.
Cancer Inform. 3:11–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Simon RM, Subramanian J, Li MC and Menezes
S: Using cross-validation to evaluate predictive accuracy of
survival risk classifiers based on high-dimensional data. Brief
Bioinform. 12:203–214. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nguyen DX, Chiang AC, Zhang XHF, Kim JY,
Kris MG, Ladanyi M, Gerald WL and Massagué J: WNT/TCF signaling
through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis.
Cell. 138:51–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bair E and Tibshirani R: Semi-supervised
methods to predict patient survival from gene expression data. PLoS
Biol. 2:E1082004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang W, Xia Y, Ji H, Zheng Y, Liang J,
Huang W, Gao X, Aldape K and Lu Z: Nuclear PKM2 regulates β-catenin
transactivation upon EGFR activation. Nature. 480:118–122. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Snaebjornsson MT and Schulze A:
Non-canonical functions of enzymes facilitate cross-talk between
cell metabolic and regulatory pathways. Exp Mol Med. 50:1–16. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liberzon A, Subramanian A, Pinchback R,
Thorvaldsdóttir H, Tamayo P and Mesirov JP: Molecular signatures
database (MSigDB) 3.0. Bioinformatics. 27:1739–1740. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gross SR: Actin binding proteins: Their
ups and downs in metastatic life. Cell Adh Migr. 7:199–213. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Beurel E, Grieco SF and Jope RS: Glycogen
synthase kinase-3 (GSK3): Regulation, actions, and diseases.
Pharmacol Ther. 148:114–131. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nakata A, Yoshida R, Yamaguchi R, Yamauchi
M, Tamada Y, Fujita A, Shimamura T, Imoto S, Higuchi T, Nomura M,
et al: Elevated β-catenin pathway as a novel target for patients
with resistance to EGF receptor targeting drugs. Sci Rep.
5:130762015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nakayama S, Sng N, Carretero J, Welner R,
Hayashi Y, Yamamoto M, Tan AJ, Yamaguchi N, Yasuda H, Li D, et al:
β-catenin contributes to lung tumor development induced by EGFR
mutations. Cancer Res. 74:5891–5902. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang F, Li Y, Liu B, You J and Zhou Q:
Cancer stem cell-like population is preferentially suppressed by
EGFR-TKIs in EGFR-mutated PC-9 tumor models. Exp Cell Res.
362:195–202. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang F, Xu J, Li H, Tan M, Xiong X and Sun
Y: FBXW2 suppresses migration and invasion of lung cancer cells via
promoting β-catenin ubiquitylation and degradation. Nat Commun.
10:13822019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fan FT, Shen CS, Tao L, Tian C, Liu ZG,
Zhu ZJ, Liu YP, Pei CS, Wu HY, Zhang L, et al: PKM2 regulates
hepatocellular carcinoma cell epithelial-mesenchymal transition and
migration upon EGFR activation. Asian Pac J Cancer Prev.
15:1961–1970. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Aktary Z, Bertrand JU and Larue L: The
WNT-less wonder: WNT-independent β-catenin signaling. Pigment Cell
Melanoma Res. 29:524–540. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Arce L, Yokoyama NN and Waterman ML:
Diversity of LEF/TCF action in development and disease. Oncogene.
25:7492–7504. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Robertson H, Hayes JD and Sutherland C: A
partnership with the proteasome; the destructive nature of GSK3.
Biochem Pharmacol. 147:77–92. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Valenta T, Hausmann G and Basler K: The
many faces and functions of β-catenin. EMBO J. 31:2714–2736. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Arao T, Fukumoto H, Takeda M, Tamura T,
Saijo N and Nishio K: Small in-frame deletion in the epidermal
growth factor receptor as a target for ZD6474. Cancer Res.
64:9101–9104. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou W, Han L and Altman RB: Imputing gene
expression to maximize platform compatibility. Bioinformatics.
33:522–528. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Amelio I, Markert EK, Rufini A, Antonov
AV, Sayan BS, Tucci P, Agostini M, Mineo TC, Levine AJ and Melino
G: p73 regulates serine biosynthesis in cancer. Oncogene.
33:5039–5046. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chan YC, Chang YC, Chuang HH, Yang YC, Lin
YF, Huang MS, Hsiao M, Yang CJ and Hua KT: Overexpression of PSAT1
promotes metastasis of lung adenocarcinoma by suppressing the
IRF1-IFNγ axis. Oncogene. 39:2509–2522. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mattaini KR, Sullivan MR and Vander Heiden
MG: The importance of serine metabolism in cancer. J Cell Biol.
214:249–257. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
DeNicola GM, Chen PH, Mullarky E, Sudderth
JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, et al: NRF2
regulates serine biosynthesis in non-small cell lung cancer. Nat
Genet. 47:1475–1481. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu B, Jia Y, Cao Y, Wu S, Jiang H, Sun X,
Ma J, Yin X, Mao A and Shang M: Overexpression of phosphoserine
aminotransferase 1 (PSAT1) predicts poor prognosis and associates
with tumor progression in human esophageal squamous cell carcinoma.
Cell Physiol Biochem. 39:395–406. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Morita T, Mayanagi T and Sobue K:
Reorganization of the actin cytoskeleton via transcriptional
regulation of cytoskeletal/focal adhesion genes by
myocardin-related transcription factors (MRTFs/MAL/MKLs). Exp Cell
Res. 313:3432–3445. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gau D and Roy P: SRF'ing and SAP'ing-the
role of MRTF proteins in cell migration. J Cell Sci.
131:jcs2182222018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shi X, Zhao S, Cai J, Wong G and Jiu Y:
Active FHOD1 promotes the formation of functional actin stress
fibers. Biochem J. 476:2953–2963. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Schönichen A, Mannherz HG, Behrmann E,
Mazur AJ, Kühn S, Silván U, Schoenenberger CA, Fackler OT, Raunser
S, Dehmelt L and Geyer M: FHOD1 is a combined actin filament
capping and bundling factor that selectively associates with actin
arcs and stress fibers. J Cell Sci. 126:1891–1901. 2013.PubMed/NCBI
|
|
56
|
Heuser VD, Mansuri N, Mogg J, Kurki S,
Repo H, Kronqvist P, Carpén O and Gardberg M: Formin proteins FHOD1
and INF2 in triple-negative breast cancer: Association with basal
markers and functional activities. Breast Cancer (Auckl).
12:11782234187922472018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gardberg M, Kaipio K, Lehtinen L, Mikkonen
P, Heuser VD, Talvinen K, Iljin K, Kampf C, Uhlen M, Grénman R, et
al: FHOD1, a formin upregulated in epithelial-mesenchymal
transition, participates in cancer cell migration and invasion.
PLoS One. 8:e749232013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Koka S, Neudauer CL, Li X, Lewis RE,
McCarthy JB and Westendorf JJ: The
formin-homology-domain-containing protein FHOD1 enhances cell
migration. J Cell Sci. 116:1745–1755. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Rottner K, Faix J, Bogdan S, Linder S and
Kerkhoff E: Actin assembly mechanisms at a glance. J Cell Sci.
130:3427–3435. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lee CW, Vitriol EA, Shim S, Wise AL,
Velayutham RP and Zheng JQ: Dynamic localization of G-actin during
membrane protrusion in neuronal motility. Curr Biol. 23:1046–1056.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Fei F, Qu J, Zhang M, Li Y and Zhang S:
S100A4 in cancer progression and metastasis: A systematic review.
Oncotarget. 8:73219–73239. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Peippo M, Gardberg M, Lamminen T, Kaipio
K, Carpén O and Heuser VD: FHOD1 formin is upregulated in melanomas
and modifies proliferation and tumor growth. Exp Cell Res.
350:267–278. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Morita T and Hayashi K: Tumor progression
is mediated by thymosin-β4 through a TGFβ/MRTF signaling axis. Mol
Cancer Res. 16:880–893. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Morita T and Hayashi K: G-actin
sequestering protein thymosin-β4 regulates the activity of
myocardin-related transcription factor. Biochem Biophys Res Commun.
437:331–335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
da Cunha BR, Domingos C, Stefanini ACB,
Henrique T, Polachini GM, Castelo-Branco P and Tajara EH: Cellular
interactions in the tumor microenvironment: The role of secretome.
J Cancer. 10:4574–4587. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Karagiannis GS, Pavlou MP and Diamandis
EP: Cancer secretomics reveal pathophysiological pathways in cancer
molecular oncology. Mol Oncol. 4:496–510. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lin A, Wei T, Meng H, Luo P and Zhang J:
Role of the dynamic tumor microenvironment in controversies
regarding immune checkpoint inhibitors for the treatment of
non-small cell lung cancer (NSCLC) with EGFR mutations. Mol Cancer.
18:1392019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li H, Wu C, Chang W, Zhong L, Gao W, Zeng
M, Wen Z, Mai S and Chen Y: Overexpression of PSAT1 is correlated
with poor prognosis and immune infiltration in non-small cell lung
cancer. Front Biosci (Landmark Ed). 28:2432023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sivanand S, Rhoades S, Jiang Q, Lee JV,
Benci J, Zhang J, Yuan S, Viney I, Zhao S, Carrer A, et al: Nuclear
Acetyl-CoA production by ACLY promotes homologous recombination.
Mol Cell. 67:252–265.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang Y, Guo YR, Liu K, Yin Z, Liu R, Xia
Y, Tan L, Yang P, Lee JH, Li XJ, et al: KAT2A coupled with the
α-KGDH complex acts as a histone H3 succinyltransferase. Nature.
552:273–277. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kottakis F, Nicolay BN, Roumane A, Karnik
R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Chen T, et al:
LKB1 loss links serine metabolism to DNA methylation and
tumorigenesis. Nature. 539:390–395. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hwang IY, Kwak S, Lee S, Kim H, Lee SE,
Kim JH, Kim YA, Jeon YK, Chung DH, Jin X, et al: Psat1-dependent
fluctuations in α-ketoglutarate affect the timing of ESC
differentiation. Cell Metab. 24:494–501. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Harmston N and Lenhard B: Chromatin and
epigenetic features of long-range gene regulation. Nucleic Acids
Res. 41:7185–7199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Clark SJ: Action at a distance: Epigenetic
silencing of large chromosomal regions in carcinogenesis. Hum Mol
Genet. 16:Spec No 1. R88–R95. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ahn MJ, Won HH, Lee J, Lee ST, Sun JM,
Park YH, Ahn JS, Kwon OJ, Kim H, Shim YM, et al: The 18p11.22 locus
is associated with never smoker non-small cell lung cancer
susceptibility in Korean populations. Hum Genet. 131:365–372. 2012.
View Article : Google Scholar : PubMed/NCBI
|