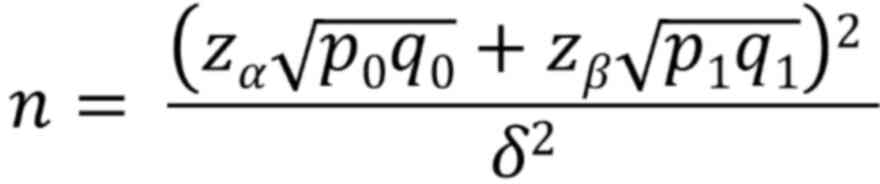Introduction
Prostate cancer (PCa) is the most commonly diagnosed
cancer type among men in the United States, with an estimated
288,300 new cases and 34,700 deaths expected in 2023 (1). Early detection of PCa is crucial, as
it improves survival rates significantly (2). Unfortunately, in the early stages of
PCa, patients may not present with any symptoms.
Serum prostate-specific antigen (PSA) is the most
widely used biomarker for PCa screening. A study that followed
>20,000 middle-aged and older men for 10 years found that the
sensitivity of the PSA test for PCa detection was 46% at a cut-off
of 4.0 ng/ml (3). The issue is that
elevated PSA levels are not exclusively indicative of PCa; indeed,
they can be caused by benign prostatic hyperplasia (BPH), urinary
tract infections, urinary retention and ejaculation (3–6).
However, despite its limitations, PSA remains a key biomarker for
PCa diagnosis and monitoring.
The low specificity of the PSA test results in a
high rate of negative biopsies, particularly in patients with serum
PSA levels between 3.0–10.0 ng/ml, a range known as the ‘PSA gray
zone’. For patients within this PSA gray zone, the positive
predictive value (PPV) of PSA for PCa detection by biopsy is
between 30–42% (7). This implies
that ~60–70% of these patients undergo an unnecessary prostate
biopsy.
MicroRNAs (miRNAs) are small, single-stranded,
non-coding RNA molecules of ~18-22 nucleotides in length that
regulate gene expression at the post-transcriptional level by
degrading or repressing target mRNAs. Genetic alterations in cancer
play significant roles in the specific activity of miRNAs. In PCa,
30% of epigenetic silencing regions contain miRNA loci. DNA
methylation of promoter regions and p53 mutations lead to miRNA
downregulation. As a result, tissue-derived and circulating miRNAs
have been proposed as biomarkers for the diagnosis and prognosis of
PCa (8). Herpes simplex virus
(Hsv)2-miR-H9 is an miRNA derived from the herpes virus, while
hsa-miR-3659 is a human miRNA. The biological functions of
hsv2-miR-H9 and hsa-miR-3659 have remained largely elusive.
However, a small number of reports have demonstrated the
tumor-promoting roles of these miRNAs. Hsv2-miR-H9 may be
associated with metastasis in lung cancer (9). miR-3659 was upregulated in
drug-resistant squamous cell carcinomas of the esophagus (10). A previous study by our group
demonstrated that increased levels of hsv1-miR-H18 and hsv2-miR-H9
may be associated with PCa, and suggested that these two viral
miRNAs may be relevant diagnostic biomarkers for PCa, thereby
reducing the biopsy burden (11–13).
Indeed, miR-3659 shows similar and stable expression levels in
patients with PCa and in BPH controls (14). Recent studies by our group suggest
that hsv2-miR-H9 and the human miRNA hsa-miR-3659 may serve as
potential diagnostic markers for PCa (11–14).
In the present study, a single-center, retrospective,
evaluator-blinded, pilot and pivotal clinical trial was conducted
to assess the clinical performance of the mirCaP kit (Urotech,
Inc.), which detects hsv2-miR-H9/hsa-miR-3659, to assist physicians
in making decisions regarding further tests for patients within the
PSA gray zone.
Patients and methods
Patients and samples
In total, 388 urine samples were collected from the
National Biobank of Korea. The biospecimens and data used in this
study were provided by the Biobank of Chungbuk National University
Hospital (Cheongju, Korea), a member of the KoreaBiobank Network
(project no. 2024ER051000). All patients had undergone prostate
biopsy to determine whether they had PCa or not. A positive
diagnosis of prostate cancer was confirmed by prostate biopsy,
whereas the diagnosis was deemed negative when no tumor was
identified after a transrectal 12-core prostate biopsy. The initial
clinical trial enrolled 70 PCa-positive and 70 biopsy-negative
samples, along with 5 cases each of bladder cancer (BC) and renal
cell carcinoma (RCC). The checklist with the inclusion and
exclusion criteria for the initial clinical trial is provided in
Table SI. The pivotal clinical
trial enrolled 109 PCa-positive and 139 biopsy-negative samples.
The checklist with the inclusion and exclusion criteria used in the
pivotal clinical trial is provided in Table SII. For the initial trial, the
collection periods were as follows: PCa from February 2012 to
December 2020; biopsy-negative from December 2018 to December 2021;
bladder cancer from November 2019 to December 2021; and renal cell
carcinoma from October 2019 to September 2022. For the pivotal
trial, the collection periods were as follows: PCa from September
2010 to October 2023; and biopsy-negative from August 2020 to June
2023. Serum PSA concentrations between 3–10 ng/ml were classified
as the ‘PSA gray zone’. The pivotal clinical trial enrolled only
patients with PSA levels within this gray zone, categorizing them
as positive or negative by biopsy. Urine samples selected from
positive or negative patients were collected during the period from
the date of the PSA test to the date of prostate biopsy or surgery.
In general, voided urine samples were collected prior to surgery;
however, in cases that underwent biopsy, urine samples were
collected immediately before the procedure. In addition, positive
urine samples were obtained from patients who underwent radical
prostatectomy, palliative transurethral resection of the prostate
(TURP) or prostate biopsy, and all had histologically-confirmed
adenocarcinoma. Negative urine samples were obtained from patients
with BPH who underwent TURP or had no tumors reported after
prostate biopsy. Urine samples were obtained from patients with BC
who underwent transurethral resection of bladder tumor with
histologically confirmed urothelial carcinoma, and from patients
with RCC who underwent radical or partial nephrectomy and had
histologically-confirmed clear-cell RCC.
All tissue specimens were examined by an experienced
senior pathologist. The pathologist who was responsible for
diagnosing PCa is highly experienced; they have been diagnosing
urologic tumors for >20 years and have completed a 2-year
urologic oncology fellowship. Gleason grading and TNM 2002 staging
were performed (15). Patients with
urinary tract infections or other diagnosed cancers were also
excluded. The study design is presented in Fig. 1.
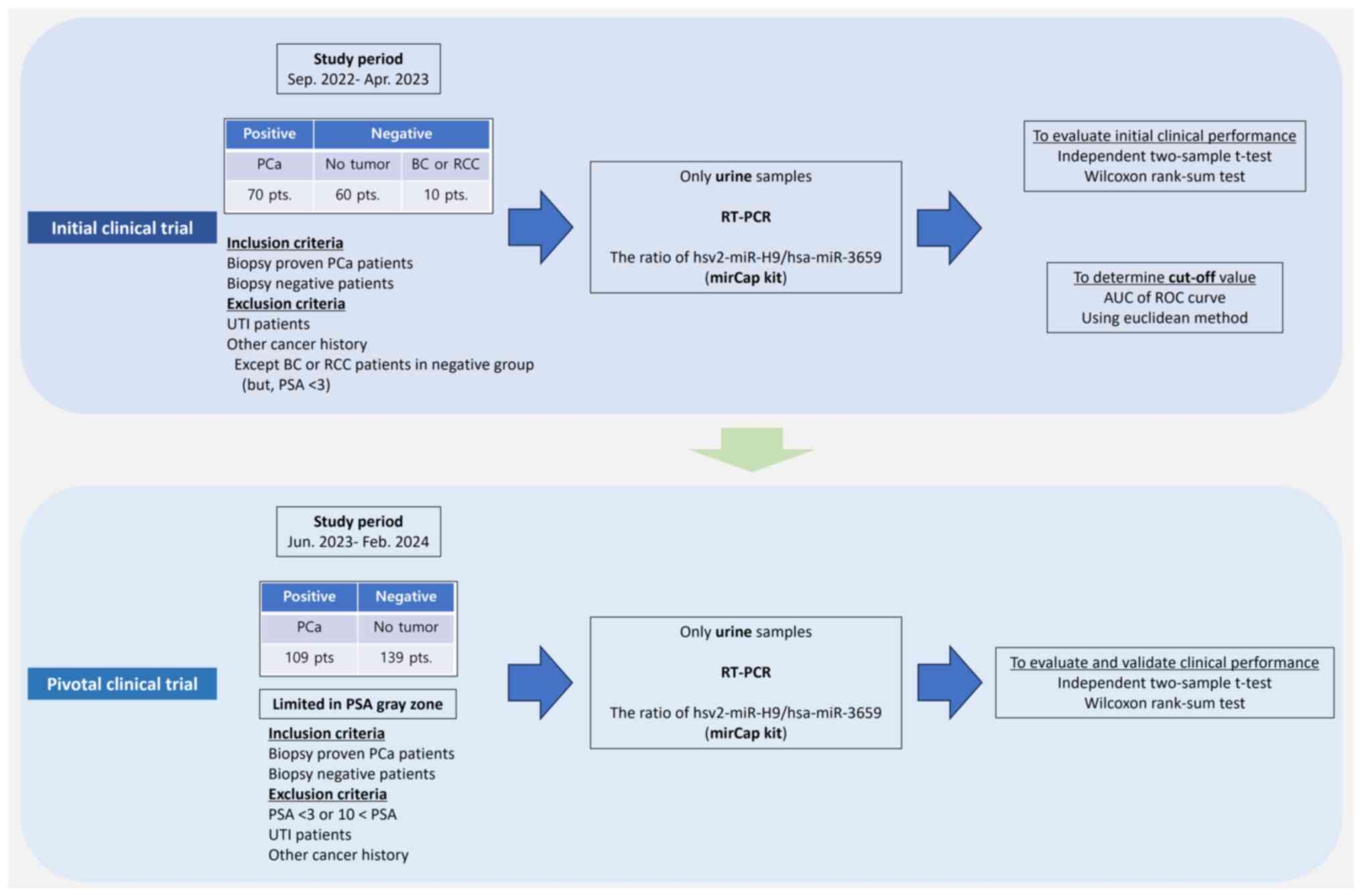 | Figure 1.Study design of the initial and
pivotal clinical trials. PCa, prostate cancer; BC, bladder cancer;
RCC, renal cell carcinoma; UTI, urinary tract infection; PSA,
prostate specific antigen; RT-PCR, real-time polymerase chain
reaction; hsv2-miR, herpes simplex virus 2 microRNA; hsa-miR,
Homo sapiens microRNA; AUC, area under curve; ROC, receiver
operating characteristic. |
Basis for determining sample size in
the initial clinical trial
The sample size for this clinical performance test
was determined based on a literature review, which indicated that
studies using comparable devices typically collected a certain
number of specimens. For instance, the study by Byun et al
(14) collected ~40 positive and 40
negative specimens. Likewise, Kang et al (13) gathered ~60 specimens of both
positive and negative samples. Based on these studies, the present
study collected 140 specimens for the initial clinical trial (70
positive and 70 negative specimens), thereby exceeding the sample
sizes reported in the referenced literature.
Basis for determining the sample size
for the pivotal clinical trial
The number of positive specimens to be used for the
pivotal clinical trial was calculated based on a one-sided
significance level of 0.025 and a power of 0.95. The study aimed to
achieve a minimum sensitivity of 65% and specificity of 60%. The
performance of this medical device for clinical performance testing
is expected to have a sensitivity of 81% and a specificity of 75%.
Therefore, the study required 98 positive specimens. To account for
a potential dropout rate of 10%, this number was adjusted to 109
positive specimens. Similarly, the number of required negative
samples was 125, increasing to 139 after adjusting for a dropout
rate of 10%. The formula used to calculate sample numbers was as
follows:
where Zα is the Z-value for a
significance level of 0.025 (≈1.96), Zβ is the Z-value
for a power of 0.95 (≈1.645), p0 is the minimum
sensitivity (=0.65), q0 is calculated as 1-p0
(1–0.65=0.35), p1 is the expected sensitivity (=0.81),
q1 is calculated as 1-p1 (1–0.81=0.19) and δ is the
effect size. When calculating the difference in sensitivity in this
clinical trial, the difference between the two sensitivities is
δ=p1-p0=0.81–0.65=0.16.
Isolation of miRNA from urine
Urine samples were processed for miRNA purification
using the Genolution Nucleic Acid Extraction Kit (Genolution
Pharmaceuticals, Inc.) For each urine sample, 500 µl of supernatant
was transferred to a tube containing the proprietary miRNA
separation solution from Genolution and then vortexed for 20 sec.
Subsequently, 200 µl of chloroform was added to the mixture,
followed by vortexing for another 10 sec. The samples were then
centrifuged at 15,928 × g for 10 min at 4°C. From the resulting
mixture, 650 µl of the upper aqueous phase was carefully removed
without disturbing the underlying white precipitate and transferred
to a new 1.5-ml tube. To this, 0.8 ml of isopropanol was added and
the mixture was centrifuged again for 20 min at 21,206 × g at 4°C.
The supernatant was carefully decanted so as not to disturb the RNA
pellet. The pellet was washed with 1 ml of 70% ethanol and
centrifuged for an additional 20 min at 21,206 × g at 4°C. After
discarding the residual ethanol, the RNA pellet was resuspended in
40 µl of RNase-free water and stored at −80°C until further
analysis.
Synthesis of cDNA from urinary
cell-free RNA
The concentration and purity of the isolated RNA
were assessed using a spectrophotometer. A cell-free RNA
concentration ≥5 ng/µl was deemed suitable for further processing.
A ratio of the absorbance at 260 nm (A260)/A280 of ≤2.0 for
cell-free RNA was considered to be indicative of acceptable purity.
Following these assessments, cDNA synthesis was performed using the
mirCaP Kit™ cDNA Synthesis Kit (Urotech, Inc.) according to the
manufacturer's protocol. cDNA synthesis is performed using the
maximum allowed template volume of 3.75 µl due to the minimal
amount of miRNA present in the extracted cell-free RNA, 5 µl of
Urotech buffer (Urotech, Inc.) and 1.25 µl of Urotech enzyme
(reverse transcriptase; Urotech, Inc.), resulting in a final volume
of 10 µl. The reverse transcription reaction was carried out at
37°C for 60 min, followed by inactivation of the reverse
transcriptase at 85°C for 5 min. The synthesized cDNA was then
stabilized at 4°C for 20 min. Subsequently, 90 µl of 1X Tris-EDTA
buffer was added to the cDNA solution and the mixture was stored at
2–8°C for a minimum of 2 h prior to use in the experiments.
Real-time (RT) PCR
To measure the levels of urinary hsv2-miR-H9 and
hsa-miR-3659, RT-qPCR was conducted using a Rotor-Gene Q instrument
(Qiagen, Inc.) and UROTECH TB premix (Urotech, Inc.). The reactions
were conducted by mixing 10 µl of Urotech TB premix, 0.4 µl of
Urotech RT primer (Urotech, Inc.), 7.2 µl of distilled water
(buffer), and 0.4 µl of the 20 ng/µl solution of the mirCaP (A or
B) primer to create a master mix, followed by the addition of 2 µl
of cDNA template for a final reaction volume of 20 µl in micro
tubes (Corbett Research). The PCR conditions were as follows: A
single cycle of denaturation at 95°C for 180 sec, followed by the
amplification process consisting of 35 cycles of 95°C for 5 sec and
60°C for 20 sec. Finally, a single cycle of dissociation was
conducted at 95°C for 60 sec, 55°C for 30 sec and 95°C for 30 sec.
The melting program was performed by heating from 65°C to 95°C,
increasing by 1°C increments. A standard curve was established
using the Vector Standard containing the target miRNA, ranging from
2.25×103 to 2.25×106 copies (2,250 to
2,250,000). The mirCaP A and B primers, which are specific for
hsv2-miR-H9 (A gene) and hsa-miR-3659 (B gene), respectively
(Urotech, Inc.), were used for amplification. The following forward
primers were used to amplify the miRNAs: hsv-miR-H9,
5′-CTCGGAGGTGGAGTCGCGGT-3′; and hsa-miR-3659,
5′-TGAGTGTTGTCTACGAGGGCA-3′. The reverse primer used was the
UROTECH RT primer (Urotech Inc.). All samples were analyzed in
triplicate. RT-PCR was performed according to the manufacturer's
instructions. Rotor-Gene Q software 2.3.4.3 (Qiagen, Inc.) was used
to acquire and analyze the spectral data.
Statistical analysis
The expression ratio of the two urinary miRNAs was
calculated by using the upregulated miRNA (hsv2-miR-H9-5p) as the
numerator and the downregulated miRNA (hsa-miR-3659) as the
denominator. Currently, there are no reliable housekeeping genes
for miRNAs in urine samples. Expression ratio analysis does not
require normalization based on housekeeping genes in urine
(11–14,16–18).
An independent two-samples t-test was used to
compare the ages of the two groups. The Wilcoxon rank-sum test was
used to compare PSA levels, expression of hsv2-miR-H9 and
hsa-miR-3659, and the expression ratio of urinary hsv2-miR-H9 to
hsa-miR-3659 between the groups. To determine the cut-off values
for the initial clinical trial, the area under the curve (AUC) of
the receiver operating characteristic curve was calculated using
the Euclidean method, based on the expression ratio of hsv2-miR-H9
and hsa-miR-3659. All statistical analyses were performed using SAS
software (SAS Institute). P<0.05 was considered to indicate
statistical significance.
Results
Baseline characteristics
The clinical characteristics of patients in the
initial and pivotal clinical trials are presented in Table SIII. In the initial clinical trial,
the mean age of the positive group was 67.50±6.19 years, with a
median baseline PSA level of 8.72 [interquartile range (IQR),
5.08–13.57] ng/ml. The mean age of the negative group was
63.97±9.30 years, with a median baseline PSA level of 5.21 (IQR,
3.34–9.13) ng/ml. In the pivotal clinical trial, the mean age of
the positive group was 69.19±6.79 years, with a median baseline PSA
of 5.92 (IQR, 4.63–7.63) ng/ml. The mean age of the negative group
was 64.56±7.92 years, with a median baseline PSA level of 5.80
(IQR, 4.50–7.23) ng/ml. However, when dividing the ages into <60
years, 60–69 years and ≥70 years, no statistically significant
differences in the expression ratio determied with the mirCaP kit
were observed in neither the positive group nor the biopsy-negative
group (P=0.293 and P=0.110, respectively).
Diagnostic value of the mirCaP kit in
the initial clinical trial
The ratio of hsv2-miR-H9 to hsa-miR-3659 was
significantly higher in the positive group than in the negative
group (P<0.0001; Table SIV).
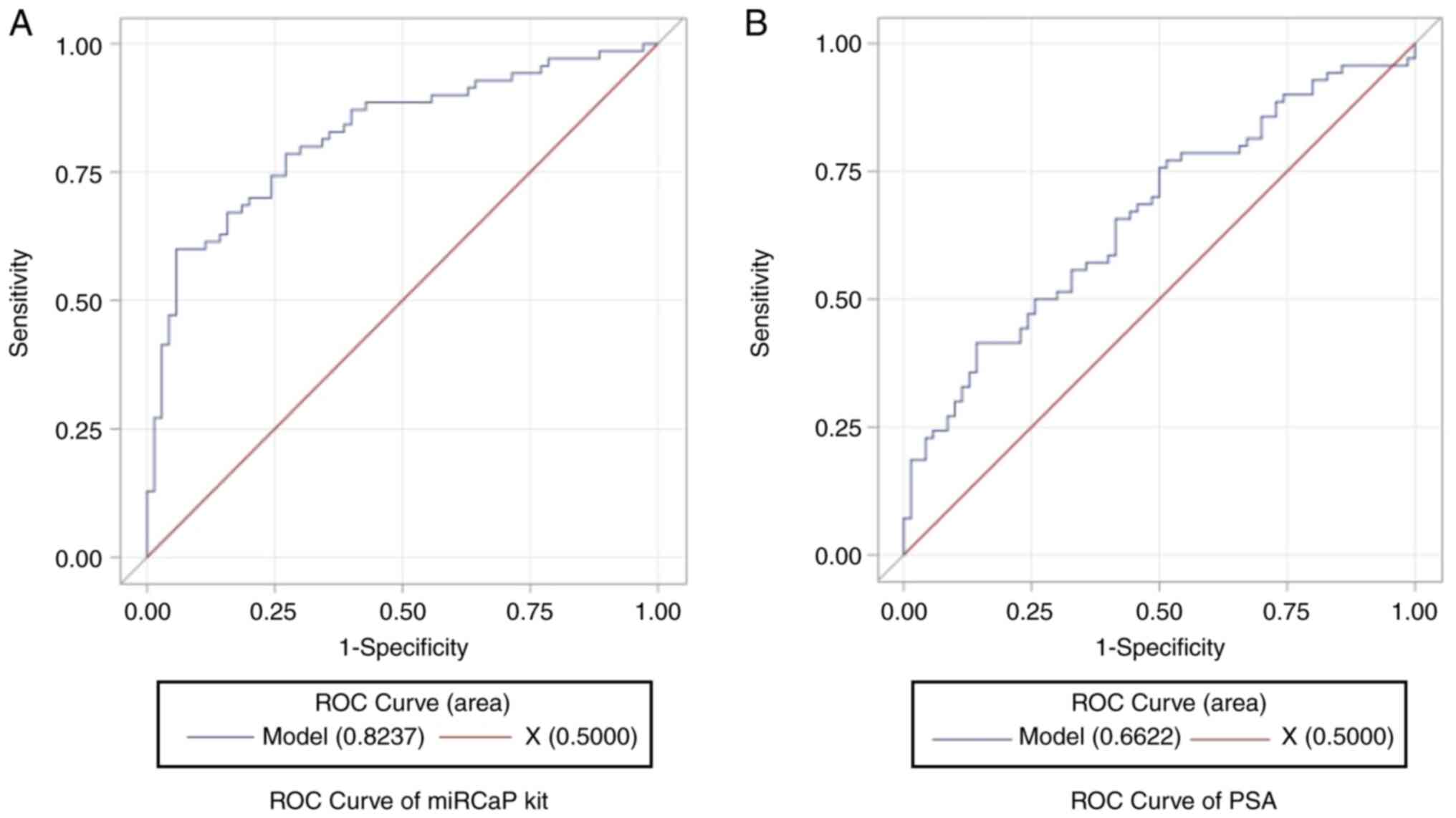
The AUC for the expression ratio of urinary hsv2-miR-H9 to
hsa-miR-3659 was 0.8237 (95% CI, 0.7539–0.8934; Fig. 2A), which was better than that for
PSA (AUC, 0.6622; 95% CI, 0.5723–0.7522; Fig. 2B). The optimal cut-off value for the
miRCaP kit, as determined by the Euclidean method, was 80.64
(Table SV). The sensitivity,
specificity, accuracy, PPV and NPV were 77.14, 72.86, 75.00, 73.97
and 76.12%, respectively (Table I).
Applying this cut-off value increased the diagnostic value in
patients within the PSA gray zone; in these patients, the
sensitivity, specificity, accuracy, PPV and NPV were 94.29, 77.50,
85.33, 78.57 and 93.94%, respectively (Table II).
 | Table I.Diagnostic value of the miRCaP kit in
the initial clinical trial. |
Table I.
Diagnostic value of the miRCaP kit in
the initial clinical trial.
|
| Reference standard
(n=140) |
|---|
|
|
|
|---|
| Item | Positive
(n=70) |
| Negative
(n=70) |
|---|
| Results with the
miRCaP kit (cut-off=80.64) |
|
|
|
|
Positive (n=73) | 54 |
| 19 |
|
Negative (n=67) | 16 |
| 51 |
| Sensitivity, % (95%
CI) |
| 77.14 (65.55,
86.33) |
|
| Specificity, % (95%
CI) |
| 72.86 (60.90,
82.80) |
|
| Accuracy, % (95%
CI) |
| 75.00 (66.98,
81.93) |
|
| Positive predictive
value, % (95% CI) |
| 73.97 (62.83,
83.55) |
|
| Negative predictive
value, % (95% CI) |
| 76.12 (64.14,
85.69) |
|
 | Table II.Diagnostic value of the miRCaP kit
for samples in the PSA Gray zone (3–10 ng/ml) in the initial
clinical trial. |
Table II.
Diagnostic value of the miRCaP kit
for samples in the PSA Gray zone (3–10 ng/ml) in the initial
clinical trial.
|
| Reference standard
(n=75) |
|---|
|
|
|
|---|
| Item | Positive
(n=35) |
| Negative
(n=40) |
|---|
| Results with the
miRCaP kit (cut-off=80.64) |
|
|
|
|
Positive (n=42) | 33 |
| 9 |
|
Negative (n=33) | 2 |
| 31 |
| Sensitivity, % (95%
CI) |
| 94.29 (80.84,
99.30) |
|
| Specificity, % (95%
CI) |
| 77.50 (61.55,
89.16) |
|
| Accuracy, % (95%
CI) |
| 85.33 (75.27,
92.44) |
|
| Positive predictive
value, % (95% CI) |
| 78.57 (63.19,
89.70) |
|
| Negative predictive
value, % (95% CI) |
| 93.94 (79.77,
99.26) |
|
Diagnostic value of the mirCaP kit for
patients within the PSA gray zone in the pivotal clinical
trial
The ratio of hsv2-miR-H9 to hsa-miR-3659 was
significantly higher in the positive group than in the negative
group (P<0.0001; Table SVI).
The sensitivity, specificity, accuracy, PPV and NPV were 94.50,
82.73, 87.90, 81.10 and 95.04%, respectively (Table III). There was no observed
difference in sensitivity according to the Gleason grade (P=0.736;
Table SVII).
 | Table III.Diagnostic value of the miRCaP kit
for samples in the PSA Gray zone (3–10 ng/ml) in the pivotal
clinical trial. |
Table III.
Diagnostic value of the miRCaP kit
for samples in the PSA Gray zone (3–10 ng/ml) in the pivotal
clinical trial.
|
| Reference standard
(n=248) |
|---|
|
|
|
|---|
| Item | Positive
(n=109) |
| Negative
(n=139) |
|---|
| Results with miRCaP
kit (cut-off=80.64) |
|
|
|
| Positive
(n=127) | 103 |
| 24 |
| Negative
(n=121) | 6 |
| 115 |
| Sensitivity, % (95%
CI) |
| 94.50 (88.40,
97.95) |
|
| Specificity, % (95%
CI) |
| 82.73 (75.41,
88.61) |
|
| Accuracy, % (95%
CI) |
| 87.90 (83.18,
91.69) |
|
| Positive predictive
value, % (95% CI) |
| 81.10 (73.20,
87.50) |
|
| Negative predictive
value, % (95% CI) |
| 95.04 (89.52,
98.16) |
|
Discussion
In a previous study, our group showed that the
urinary hsv2-miR-H9 to hsa-miR-3659 expression ratio is a reliable
non-invasive diagnostic marker for PCa (13). In the present study, the diagnostic
utility and cut-off values of the ‘miRCaP’ kit, which measures the
hsv2-miR-H9/hsa-miR-3659 ratio, was evaluated in an initial
clinical trial, and its performance as a non-invasive diagnostic
marker was then validated in a pivotal clinical trial enrolling
only patients within the PSA gray zone.
The PSA gray zone is a well-established concept in
urology. In general, the PSA gray zone is considered to be 4–10
ng/ml. However, Cho (19) reported
that Korean men tend to have slightly lower PSA levels and smaller
prostate volumes compared to Caucasians. The age-specific reference
ranges for serum PSA levels in Korean men were found to be lower.
The predictive factors and characteristics of prostate cancer in
patients with serum PSA levels of 4.0 ng/ml or less are very
similar in Korean men. Therefore, Cho (19) suggested lowering the PSA cutoff for
biopsy recommendations in prostate cancer screening for Korean men.
Following this report, our institution adopted a policy of
performing prostate biopsies in patients with PSA levels >3
ng/ml.
Given the low specificity of PSA, numerous studies
have attempted to overcome these limitations. Adjustments such as
age-adjusted PSA, free PSA, PSA density, PSA velocity and PSA
doubling time have been explored to optimize the accuracy of PSA or
PSA derivatives (20–23). In addition, the prostate health
index (PHI), the 4K score (four-kallikrein panel), PCA gene 3
(PCA3) levels, ExoDX Prostate IntelliScore and SelectMDX have been
used; however, they show a weak clinical performance in real-world
clinical practice (24–28). In a biopsy-naïve population, the PHI
detects aggressive PCa with greater specificity than total and %
free PSA. Although the sensitivity of PHI is 95%, its specificity
was only 36.0% (that for total and % free PSA is 17.2 and 19.4%,
respectively). These data show that the PHI, PSA and % free PSA
have high sensitivity but low specificity (29). In the initial biopsy group, the
diagnostic sensitivity and specificity of PCA3 were 42 and 91%,
respectively, indicating that PCA3 has high specificity but low
sensitivity (26). The performance
of the ExoDx Prostate IntelliScore showed a sensitivity of 91.89%
and specificity of 33.96%, again suggesting high sensitivity but
low specificity (27). SelectMDX, a
tool for diagnosing PCa by measuring the expression of homeobox C6
and distal-less homeobox 1 from mRNA, demonstrated a sensitivity of
91% and a specificity of 36% (28).
The mirCaP kit showed both high sensitivity (94.5%) and high
specificity (82.73%), making it a highly effective diagnostic
marker for PCa. Therefore, in comparison to other tests where high
sensitivity often coincides with low specificity, or high
specificity comes at the cost of low sensitivity, the mirCaP kit,
having both high sensitivity and specificity, is expected to be
superior. This hypothesis should be confirmed through future
head-to-head studies.
Recently, liquid biopsy (LB), which detects
tumor-derived material circulating in blood or urine, has been
developed as a tool for detecting PCa; an obstacle is that, during
early-stage PCa, circulating tumor cells (CTCs) and ctDNA are
either undetectable or detected at rates similar to biopsy-negative
controls (30,31). However, CTC PSA mRNA is a potential
biomarker that could replace serum PSA, and also provide
information about the metastatic stage of PCa (32). The ExoDx™ Prostate
IntelliScore, an established LB diagnostic marker for early PCa,
was shown to prevent 26% of unnecessary biopsies, with a negative
predictive value of 89%, in men aged >50 years with borderline
PSA levels (2–10 ng/ml) (27,33). A
panel that includes miR-141, miR-1290, miR-100 and miR-335 could
serve as urine and/or tissue biomarkers for diagnosing BPH and PCa
(34). The present study
demonstrates the diagnostic performance of urinary miRNAs as a
potential LB. Although RNA is highly unstable, miRNAs are stable in
both serum and urine, and are resistant to RNase activity, extreme
pH, extreme temperatures and multiple freeze-thaw cycles (16,35). A
previous study showed that the urinary miR-H9 to miR-3659 ratio
could be a relevant non-invasive biomarker for PCa diagnosis
(13). In the present study, an
accurate cut-off value was calculated for the mirCaP kit (miR-H9 to
miR-3659) and its clinical performance was validated in a cohort of
patients within the PSA gray zone. The results show that the mirCaP
kit has higher sensitivity and specificity than PSA, suggesting its
potential use as a urinary miRNA diagnostic marker for PCa. Of
note, the NPVs in the two cohorts within the PSA gray zone were
93.94 and 95.04%, respectively, indicating that if patients
received a negative result from the mirCaP Kit, the likelihood of
them being tumor-free was 95%. This suggests that the mirCaP kit
can reduce the number of unnecessary prostate biopsies by
>90%.
This present study has both limitations and
strengths. One limitation is that these two clinical trials were
not multicenter studies. Another limitation is that this is a
retrospective study, which may have introduced bias due to
different enrollment periods between the groups. In order to
achieve the initially planned sample size, it was required to
enroll patients in different groups with samples provided during
different periods. However, a strength of the study is that
sequential evaluator-blinded, pilot and pivotal studies were
conducted. An evaluator-blinded study has several advantages.
First, it minimizes expectation bias, as the researchers do not
know whether a participant has cancer or not. Second,
evaluator-blinded studies increase the credibility of the study
results. Blinding of researchers helps ensure that the findings are
based on the actual effects of the markers. Third, blinding of
researchers helps ensure that the results of the study can be more
easily generalized to a broader population. Finally, the sample
size required for each trial was calculated by statistical experts.
The importance of sample size calculations cannot be
overemphasized. Regardless of the aim of a research study, one can
draw a precise and accurate conclusion only with an appropriate
sample size. The two trails were managed by the Expert Contract
Research Organization (Synex Consulting Ltd.), although prior
approval by the Korean National Institute of Food and Drug Safety
Evaluation was not required.
In conclusion, the present study shows that the
expression ratio of urinary hsv-miR-H9 to hsa-miR-3659 (mirCaP Kit)
could serve as a highly effective non-invasive diagnostic marker
for PCa in patients within the PSA gray zone. Thus, the mirCaP kit
shows promise as a tool for determining the need for prostate
biopsy in these patients.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This research was supported by a Basic Science Research Program
through the National Research Foundation of Korea (NRF) funded by
the Ministry of Education (grant no. 2020R1I1A3062508); the NRF
Regional Innovation Strategy funded by the Ministry of Education
(grant no. 2021RIS-001); and the NRF BK21 FOUR program funded by
the Ministry of Education (grant no. 5199990614277).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
Conception and design: WTK; data analysis and
interpretation: WTK, YSH, JK and IYK; data acquisition: KK, HWK,
YJB, XMP, YJK, SCL and SJY; drafting the manuscript and statistical
analysis: WTK; checking and confirming the authenticity of the raw
data: WTK, KK and HWK. All authors have read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
This study adhered to the applicable laws and
regulations, good clinical practice and the ethical principles
outlined in the Declaration of Helsinki. The study protocol was
approved by the Ethics Committee of Chungbuk National University
Hospital (Cheongju, Korea). The Institutional Review Board (IRB)
approval no. for the initial clinical trial is 2022-09-038, and for
the pivotal clinical trial, it is 2023-06-034. The requirement of
consent was waived prior to enrollment, as the samples were
obtained from the National Biobank of Korea, where patients had
already provided consent at the time of donation. The IRB of
Chungbuk National University Hospital (Cheongju, Korea) approved
all procedures related to sample collection and analysis. This
clinical trial is retrospective and non-invasive, thus it does not
fall under the category requiring medical device clinical trial
plan approval. In such cases, the Ministry of Food and Drug Safety
(MFDS) conducts a pre-review of the protocol. This study was not
subject to prior approval by the Korean MFDS; however, it underwent
a preliminary review. The clinical trial has been submitted to the
Korean MFDS. The registration number for approval is 20240050719,
and it was submitted on March 15, 2024. The data have been
submitted as a ‘Results report’. The review is still in progress
and it will take more time for the results to be finalized.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
AI tools were used to improve the readability and
language of the manuscript.
Glossary
Abbreviations
Abbreviations:
|
PCa
|
prostate cancer
|
|
PPV
|
positive predictive value
|
|
NPV
|
negative predictive value
|
|
PSA
|
prostate-specific antigen
|
|
BPH
|
benign prostatic hyperplasia
|
|
BC
|
bladder cancer
|
|
RCC
|
renal cell carcinoma
|
|
TURP
|
transurethral resection of the
prostate
|
|
AUC
|
area under the curve
|
|
ROC
|
receiver operating characteristic
|
|
IQR
|
interquartile range
|
|
LB
|
liquid biopsy
|
|
CTC
|
circulating tumor cell
|
References
|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Obirieze AC, Moten A, Allen D and Ahaghotu
CA: African-American men with low-risk prostate cancer: Modern
treatment and outcome trends. J Racial Ethn Health Disparities.
2:295–302. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gann PH, Hennekens CH and Stampfer MJ: A
prospective evaluation of plasma prostate-specific antigen for
detection of prostatic cancer. JAMA. 273:289–294. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shahab AA, Soebadi DM, Djatisoesanto W,
Hardjowijoto S, Soetojo S and Hakim L: Prostate-specific antigen
and prostate-specific antigen density cutoff points among
Indonesian population suspected for prostate cancer. Prostate Int.
1:23–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stamey TA, Yang N, Hay AR, McNeal JE,
Freiha FS and Redwine E: Prostate-specific antigen as a serum
marker for adenocarcinoma of the prostate. N Engl J Med.
317:909–916. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kobayashi M, Kijima T, Yashi M and Kamai
T: Prostate-specific antigen kinetics contributes to decision
making for biopsy referral: The predictive implication for PSA
retest in patients with elevated PSA levels. Prostate Int.
11:27–33. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilson S and Crawford ED: Screening for
prostate cancer: Current recommendations. Urol Clin North Am.
31:219–226. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim WT and Kim WJ: MicroRNAs in prostate
cancer. Prostate Int. 1:3–9. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang X, Liu S, Zhou Z, Yan H and Xiao J: A
herpes simplex virus type 2-encoded microRNA promotes tumor cell
metastasis by targeting suppressor of cytokine signaling 2 in lung
cancer. Tumor Biol. 39:10104283177016332017.PubMed/NCBI
|
|
10
|
Choi J, Kwon SM, Moon SU, Yoon S, Shah M,
Lee BG, Yang J, Park YN, Wang HJ and Woo HG: TPRG1-AS1 induces
RBM24 expression and inhibits liver cancer progression by sponging
miR-4691-5p and miR-3659. Liver Int. 41:2788–2800. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yun SJ, Jeong P, Kang HW, Kim YH, Kim EA,
Yan C, Choi YK, Kim D, Kim JM, Kim SK, et al: Urinary MicroRNAs of
prostate cancer: Virus-Encoded hsv1-miRH18 and hsv2-miR-H9-5p could
be valuable diagnostic markers. Int Neurourol J. 19:74–84. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Byun YJ, Kang HW, Piao X, Zheng CM, Moon
SK, Choi YH, Kim WT, Lee SC, Yun SJ and Kim WJ: Expression of
hsv1-miR-H18 and hsv2-miR-H9 as a field defect marker for detecting
prostate cancer. Prostate Int. 10:1–6. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kang HW, Byun YJ, Kim K, Moon SM, Kim K,
Piao XM, Zheng CM, Moon SK, Choi YH, Kim WT, et al: Urinary
hsv2-miR-H9 to hsa-miR-3659 ratio is an effective marker for
discriminating prostate cancer from benign prostate hyperplasia in
patients within the prostate-specific antigen grey zone. Invest
Clin Urol. 63:238–244. 2022. View Article : Google Scholar
|
|
14
|
Byun YJ, Piao XM, Jeong P, Kang HW, Seo
SP, Moon SK, Lee JY, Choi YH, Lee HY, Kim WT, et al: Urinary
microRNA-1913 to microRNA-3659 expression ratio as a non-invasive
diagnostic biomarker for prostate cancer. Invest Clin Urol.
62:340–348. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Borley N and Feneley MR: Prostate cacner:
Diagnosis and staging. Asian J Androl. 11:74–80. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yun SJ, Jeong P, Kim W, Kim TH, Lee YS,
Song PH, Choi YH, Kim IY, Moon SK and Kim WJ: Cell-free microRNAs
in urine as diagnostic and prognostic biomarkers of bladder cancer.
Int J Oncol. 41:1871–1878. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim WT, Jeong P, Yan C, Kim YH, Lee IS,
Kang HW, Kim YJ, Lee SC, Kim SJ, Kim YT, et al: UBE2C cell-free RNA
in urine can discriminate between bladder cancer and hematuria.
Oncotarget. 7:58193–58202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Piao XM, Jeong P, Kim YH, Byun YJ, Xu Y,
Kang HW, Ha YS, Kim WT, Lee JY, Woo SH, et al: Urinary cell-free
microRNA biomarker could discriminate bladder cancer from benign
hematuria. Int J Cancer. 144:380–388. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cho JS: Optimal prostate-specific antigen
cutoff value in Korean. Korean J Urol Oncol. 5:1–5. 2007.(In
Korean).
|
|
20
|
Oesterling JE, Jacobsen SJ and Cooner WH:
The use of age-specific reference ranges for serum prostate
specific antigen in men 60 years old or older. J Urol.
153:1160–1163. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee R, Localio AR, Armstrong K, Malkowicz
SB and Schwartz JS; Free PSA Study Group, : A meta-analysis of the
performance characteristics of the free prostate-specific antigen
test. Urology. 67:762–768. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang P, Du W, Xie K, Deng X, Fu J, Chen H
and Yang W: Transition zone PSA density improves the prostate
cancer detection rate both in PSA 4.0–10.0 and 10.1–20.0 ng/ml in
Chinese men. Urol Oncol. 31:744–748. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Carter HB, Ferrucci L, Kettermann A,
Landis P, Wright EJ, Epstein JI, Trock BJ and Metter EJ: Detection
of life-threatening prostate cancer with prostate-specific antigen
velocity during a window of curability. J Natl Cancer Inst.
98:1521–1527. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lepor A, Catalona WJ and Loeb S: The
prostate health index: Its utility in prostate cancer detection.
Urol Clin North Am. 43:1–6. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Parekh DJ, Punnen S, Sjoberg D, Asroff SW,
Bailen JL, Cochran JS, Concepcion R, David RD, Deck KB, Dumbadze I,
et al: A multi-institutional prospective trial in the USA confirms
that the 4Kscore accurately identifies men with high-grade prostate
cancer. Eur Urol. 68:464–470. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wei JT, Feng Z, Partin AW, Brown E,
Thompson I, Sokoll L, Chan DW, Lotan Y, Kibel AS, Busby JE, et al:
Can urinary PCA3 supplement PSA in the early detection of prostate
cancer? J Clin Oncol. 32:4066–4072. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
McKiernan J, Donovan MJ, O'Neill V,
Bentink S, Noerholm M, Belzer S, Skog J, Kattan MW, Partin A,
Andriole G, et al: A novel urine exosome gene expression assay to
predict high-grade prostate cancer at initial biopsy. JAMA Oncol.
2:882–889. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Van Neste L, Hendriks RJ, Dijkstra S,
Trooskens G, Cornel EB, Jannink SA, de Jong H, Hessels D, Smit FP,
Melchers WJ, et al: Detection of high-grade prostate cancer using a
urinary molecular biomarker-based risk score. Eur Urol. 70:740–748.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
de la Calle C, Patil D, Wei JT, Scherr DS,
Sokoll L, Chan DW, Siddiqui J, Mosquera JM, Rubin MA and Sanda MG:
Multicenter evaluation of the prostate health index to detect
aggressive prostate cancer in biopsy Naïve men. J Urol. 194:65–72.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Davis JW, Nakanishi H, Kumar VS,
Bhadkamkar VA, McCormack R, Fritsche HA, Handy B, Gornet T and
Babaian RJ: Circulating tumor cells in peripheral blood samples
from patients with increased serum prostate specific antigen:
Initial results in early prostate cancer. J Urol. 179:2187–2191;
discussion 2191. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lau E, McCoy P, Reeves F, Chow K, Clarkson
M, Kwan EM, Packwood K, Northen H, He M, Kingsbury Z, et al:
Detection of ctDNA in plasma of patients with clinically localised
prostate cancer is associated with rapid disease progression.
Genome Med. 12:722020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cho H, Oh CK, Cha J, Chung JI, Byun SS,
Hong SK, Chung JS and Han KH: Association of serum
prostate-specific antigen (PSA) level and circulating tumor
cell-based PSA mRNA in prostate cancer. Prostate Int. 10:14–20.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
McKiernan J, Donovan MJ, Margolis E,
Partin A, Carter B, Brown G, Torkler P, Noerholm M, Skog J, Shore
N, et al: A prospective adaptive utility trial to validate
performance of a novel urine exosome gene expression assay to
predict high-grade prostate cancer in patients with
prostate-specific antigen 2–10 ng/ml at initial biopsy. Eur Urol.
74:731–738. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Manoj A, Ahmad MK, Prasad G, Kumar D,
Mahdi A and Kumar M: Screening and validation of novel serum panel
of microRNA in stratification of prostate cancer. Prostate Int.
11:150–158. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Glinge C, Clauss S, Boddum K, Jabbari R,
Jabbari J, Risgaard B, Tomsits P, Hildebrand B, Kääb S, Wakili R,
et al: Stability of circulating blood-based MicroRNAs-pre-analytic
methodological considerations. PLoS One. 12:e01679692017.
View Article : Google Scholar : PubMed/NCBI
|