Introduction
The four and a half LIM domain protein 1 (FHL1)
belongs to the FHL family and is located in Xq26.3. It is also
known as SLIM-1 or KYO-T (1,2).
As a cytoskeletal protein, FHL1 is widely expressed
in humans. Relevant studies have shown that it is mainly expressed
in the skeletal muscle and myocardium. FHL1 regulates cell
proliferation, differentiation, apoptosis, adhesion, migration,
transcription and other cellular processes, and plays an important
role in cell growth (3).
FHL1 is composed of four and a half LIM domains and
three subtypes have been identified. The LIM domain of FHL1 is
responsible for mediating interactions between proteins. Each
domain of FHL1 has a different regulatory role in protein function.
Mutations in LIM2 and LIM4 cause a loss of function in FHL1, and
the function of FHL1 depends on the integrity of its various
domains (4).
FHL1 also plays an important regulatory role in the
growth of numerous tumors, in addition to affecting the occurrence
and development of skeletal muscle and myocardial diseases. In
different tumors, the expression of FHL1 is upregulated or
downregulated and plays a role in promoting or inhibiting tumor
development. Owing to the wide expression of FHL1 in numerous
tumors and its different regulatory roles in tumor development,
FHL1 has become a hot topic in the field of tumor research. The
present article reviewed the role of FHL1 in tumors.
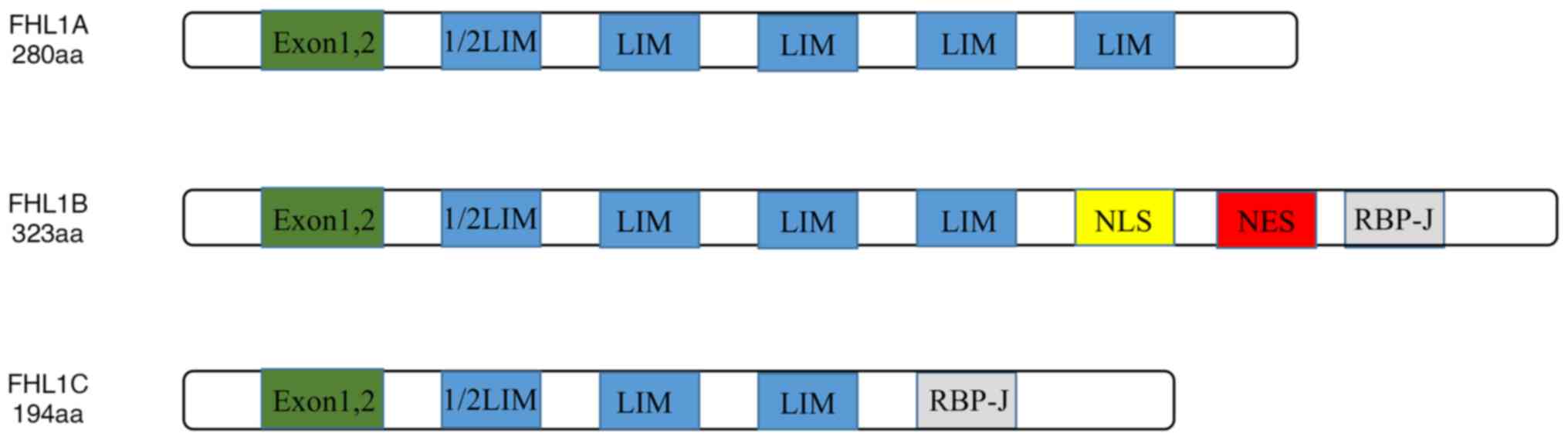
Domain architectures of FHL1
A total of 8 exons were found in FHL1 gene and exons
1 and 2 are non-coding. The structure of FHL1 is characterized by
the structure of the LIM domain. The N-terminus is a half LIM
domain, followed by four complete LIM domains. The LIM domain was
first identified during the isolation and identification of the
LIN1 gene in Caenorhabditis elegans, the ISL-1 gene in rats
and the MEC-1 gene in C. elegans (5). The consensus amino acid sequence of
the LIM domains has been defined as
Cys-X2-Cys-X16-23-His-X2-Cys-X2-Cys-X2-Cys-X16-21-Cys-X2-Cys/His/Asp,
where X represents any amino acid.
The LIM domain is an important sequence mediating
interactions between proteins. The structure of the LIM domain is
highly conserved. It mediates the interaction between FHL1 and
transcriptional regulatory factors, kinases and structural proteins
such as receptor-interacting protein140 (RIP140), cytosolic
tyrosine kinase Src and α-tubulin.
The alternative splicing results in the existence of
three subtypes of FHL1: FHL1A is a complete FHL1 protein; FHL1B
lacks the last LIM domain, which includes nuclear localization,
output signal and RBP-J binding domain; and FHL1C lacks the last
two LIM domains, which includes the RBP-J binding domain, and has
no nuclear location signal. The RBP-J binding domain binds to RBP-J
and thus inhibits the binding of RBP-J to DNA and further inhibits
its transcriptional activity. The structures of FHL1A, FHL1B and
FHL1C are shown in Fig. 1. Studies
have indicated that FHL1A is mainly expressed in the skeletal
muscle, myocardium and fibroblasts, and FHL1B/FHL1C are mainly
expressed in the muscle, brain and testis (6).
The LIM domain, which is composed of ~55 amino acid
residues, binds to zinc and is rich in cysteine. It also plays an
important role in protein transcription and signal transduction
(7). Its cysteine-rich double zinc
fingers contain the consensus sequence
X2:CX2CX3I–X11-15WHX2CFXCX2CX3(I/L)X4(F/Y)X8CX2C
(8). The consensus sequence
facilitates protein-protein interactions. The highly conserved
cysteine and histidine residues mediate Zn2+ binding,
thereby stabilizing the folding and structure of the LIM domain
(9,10). LIM domain is able to form homo- or
heterodimers via interacting with other domains. It also binds to
motifs such as PDZ domains and helix-loop-helix domains and
performs its function. Multiple mutations in FHL1 affect its
function and are associated with various skeletal and cardiac
diseases. Relevant studies have shown that FHL1 mutations mostly
occur in the second and fourth LIM domains (11).
The expression of FHL1 is also affected by a variety
of post-transcriptional modulations; for example, methylation
silences the expression of FHL1. The deubiquitinating enzyme 15
(USP15) stabilizes FHL1 expression by regulating the ubiquitin
proteasome (12).
As an important cytoskeletal protein, the structure
of FHL1 and the physiological functions of each domain require
further study.
FHL1 in normal physiology
FHL1 performs its physiological functions by
interacting with a variety of molecules. The proteins that interact
with FHL1 include structural proteins, signal transduction
proteins, transcription regulators, receptors and channels, such as
ERK and non-structural protein 3 (Nsp3) (13).
FHL1 plays a role in the regulation of gene
transcription, viral infection, thyroid function, blood glucose
levels, myoblast differentiation and other physiological processes.
Its abnormal expression is closely related to numerous diseases of
the skeletal muscle and myocardium (14–17).
FHL1 is closely associated with gene transcription.
For instance, FHL1A interacts with Gβγ- and Gαs-dependent guanine
nucleotide exchange factor pleckstrin homology and RhoGEF domain
containing G2 to enhance its induced serum response element
(Ser)-dependent gene transcription (14).
FHL1 interacts with Pol1 transcription factor HMO1
to regulate ribosome synthesis. Interacts with Forkhead 1 (IFH1) is
a weak multicopy suppressor with FHL1 deletion, and there is an
interaction between FHL1 and the coactivator IFH1. TOR promotes
this interaction and inhibits the interaction between FHL1 and the
corepressor CRF1 to regulate the expression of ribosomal protein
genes (18).
The expression of FHL1 is also associated with viral
infections. For instance, FHL1 promotes the development of
chikungunya and cashmere virus infection (19). The interaction between Nsp3 (HVD)
and FHL1 is important for the proviral function of FHL1 (20). In addition, FHL1 interacts with the
viral protein Nsp3 to regulate viral RNA replication. It is also
highly expressed in chikungunya virus (CHIKV) target muscle cells
and fibroblasts, and plays an important role in CHIKV gene
expansion. FHL1 knockdown confers resistance to viral infections
(21).
Mutations in FHL1 are associated with skeletal
muscle and myocardial diseases. For instance, c.370–375del leads to
reductive myopathy and c.763T>c,p.Cys255Arg is associated with
hypertrophic cardiomyopathy (22,23).
Role of FHL1 in tumors
FHL1 is abnormally expressed in tumors and the
expression of FHL1 is not consistent among different tumors. It
promotes or inhibits the tumor development by upregulating or
downregulating its expression. Simultaneously, the expression of
FHL1 in tumors is regulated by epigenetic modifications. Therefore,
FHL1 is a potential target in cancer research.
Dysregulation of FHL1 in tumors
Low expression of FHL1 in tumors
The expression of FHL1 is downregulated in cancer
types including lung, prostate, breast, ovarian, colon, thyroid,
brain, kidney, liver and oral cancers, as well as melanoma
(24,25).
In non-small cell lung cancer, a decrease in long
intergenic noncoding RNA (LINC)00261, which has a tumor suppressor
function, leads to an increased expression of miR-105, which leads
to a decrease in FHL1 expression (26,27).
The expression level of FHL1 correlates with that of numerous
immune cells. For instance, its expression level is positively
correlated with the levels of monocytes, eosinophils and
neutrophils, and negatively correlated with the level of M0
macrophages (28). Increased FHL1
expression in lung adenocarcinoma predicts a good prognosis
(29). In lung adenocarcinoma, the
expression of FHL1 is lower than that in normal lung tissue, and
patients with relatively high expression have longer overall
survival (30). In papillary
thyroid microcarcinoma (PTMC), the expression of FHL1 in PTMC is
lower than that in normal tissues, and the expression of FHL1 is
significantly lower in PTC than in PTMC (31). In gastrointestinal tumors, the FHL1
promoter region is methylated, which silences the expression of
FHL1. The expression of FHL1 in gastric cancer tissues is decreased
compared to that in normal mucosa. In addition, more invasion and
metastasis are found in these patients (24). In liver and colorectal tumors,
miR-410 is highly expressed and downregulates the expression of
FHL1 to promote tumor growth (27,32,33).
In breast cancer, miR-183-5p downregulates the expression of FHL1,
resulting in enhanced activity of breast cancer cells (32). In head and neck and esophageal
squamous cell carcinomas, FHL1 expression is downregulated, and the
prognosis of these patients is poor (2,34). In
pediatric acute myelocytic leukemia (AML) with FLT3-ITD mutations,
high expression of FHL1 predicts poor prognosis (35).
High expression of FHL1 in tumors
The expression of FHL1 is increased in laryngeal
carcinomas (36). Increased FHL1
expression has been associated with gastric signet ring cell
carcinoma (37). In addition, the
expression of FHL1 in patients with acute promyelocytic leukemia
and AML without FLT3 mutations or PML/RAR fusion is positively
correlated with the expression of Rho-related BTB domain 2, which
is an indicator of poor prognosis (38). High FHL1 expression promotes
glioblastoma development (39).
FHL1 as a prognostic marker
Significant differences were observed in the
expressions of FHL1 between different tumors. Its expression is
closely related to the prognosis of patients with various tumors.
Via causing the activation of TGF-β and other signaling pathways,
promoting or inhibiting the expression of tumor
proliferation-related proteins such as putative specific protein 1
(SP1), FHL1 influences the proliferation and migration of tumor
cells, thus resembling a prognostic marker in tumors. FHL1 is also
regulated by posttranslational modifications, such as
phosphorylation and methylation, thus influencing the prognosis of
bladder and other tumors (40).
FHL1 acts through different pathways in different tumors, thus
predicting different prognoses in different tumors. Increased FHL1
expression in lung adenocarcinoma, gastric cancer, colorectal
tumors, head and neck squamous cell carcinoma, glioma, liver,
breast and papillary thyroid cancers (PTCs), and esophageal
squamous cell carcinoma, predicts a good prognosis. A high FHL1
expression in glioblastoma and pediatric AML with FLT3-ITD
mutations predicts a poor prognosis. To conclude, the expression of
FHL1 is closely related to the prognoses and clinical indicators of
different tumors and the role of FHL1 in different tumors is
summarized in Table I.
 | Table I.FHL1 as a prognostic marker in
tumors. |
Table I.
FHL1 as a prognostic marker in
tumors.
| Tumor type | FHL1
expression | Target | Mechanism | Prognosis | (Refs.) |
|---|
| Glioblastoma | Upregulated | SP1 | Promoting cell
proliferation | Poor | (46) |
| Acute myelocytic
leukemia | Upregulated | CD14 CD11b | Promoting cell
proliferation | Poor | (39) |
| Gastric cancer | Downregulated | CCDC43 | Inhibiting cell
proliferation | Good | (51) |
| Colorectal
tumor | Downregulated | GSK3β | Inhibiting cell
proliferation | Good | (3) |
| Glioma | Downregulated | AKT | Inhibiting cell
proliferation | Good | (5) |
| Head and neck
squamous cell carcinoma | Downregulated | Cyclin D1 Cyclin E
p27 | Inhibiting cell
proliferation | Good | (2) |
| Liver cancer | Downregulated | Smad2 | Inhibiting cell
proliferation | Good | (58) |
| Breast cancer | Downregulated | VEGF N-cadherin
vimentin | Inhibiting
migration and invasion | Good | (32) |
| Lung cancer | Downregulated | RhoGDIβ | Inhibiting
migration and invasion | Good | (48) |
| Papillary thyroid
cancer | Downregulated | Wnt/β-catenin | Inhibiting cell
proliferation | Good | (59) |
Regulation of FHL1 signaling
The expression of FHL1 is influenced by several
factors. Its expression is regulated by miRNA, Src and other
proteins, as well as by methylation, ubiquitination and other
post-translational modifications.
Interaction with RIP140
In breast cancer, FHL1 interacts with RIP140, in
which process all of the domains of FHL1 are required.
Overexpression of FHL1 has a synergistic effect on the inhibition
of RIP140 in estrogen signal transduction (41).
Mediation by
immortalization-upregulated protein (IMUP)
IMUP is highly expressed in pancreatic ductal
adenocarcinoma and knockdown of IMUP prevents tumor cell growth.
IMUP and nucleophosmin (NPMI) are co-localized in the cell nucleus.
IMUP inhibits FHL1 transcription via NPMI-induced promoter
methylation. Downregulation of IMUP decreases the expression of
CyclinA2, cyclinE1 and cyclin-dependent kinase (CDK)2 by
upregulating FHL1 expression. FHL1 regulates CyclinA2, cyclinE1 and
CDK2 by promoting the degradation of CDC25A through phosphorylation
(40).
Mediation by Src
Src is a membrane-bound tyrosine kinase. Src
phosphorylates Crk-associated substrate and thus reduces FHL1
expression. In breast, kidney and prostate cancers, the expression
of FHL1 is inhibited, possibly owing to gene methylation. FHL1
induces the expression of serum deprivation response factor in
Src-transformed cells, whereas the expression of both molecules is
inhibited in tumors (2,42).
Silencing by miR-410, enhancer of
zeste homolog 2 (EZH2) and methylation
FHL1 may be silenced by miR-410 or EZH2 (30). Methylation of the FHL1 promoter
region has been observed in gastrointestinal tumors and oral
squamous cell carcinoma-derived cells, which silences the
expression of FHL1 (43,44).
FHL1 signaling in cell proliferation
and cell cycle
The role of FHL1 in tumor growth is complex and this
effect varies among different tumors (Fig. 2). For instance, FHL1 can inhibit the
progression of lung, prostate, ovarian, colon, thyroid, brain,
kidney, liver and stomach cancers and promote the progression of
glioblastoma. High expression of FHL1 is a risk factor for acute
glioma and patients with breast cancer undergoing radiotherapy;
however, it can inhibit the progression of colorectal cancer
through the Wnt/β-catenin signaling pathway (45).
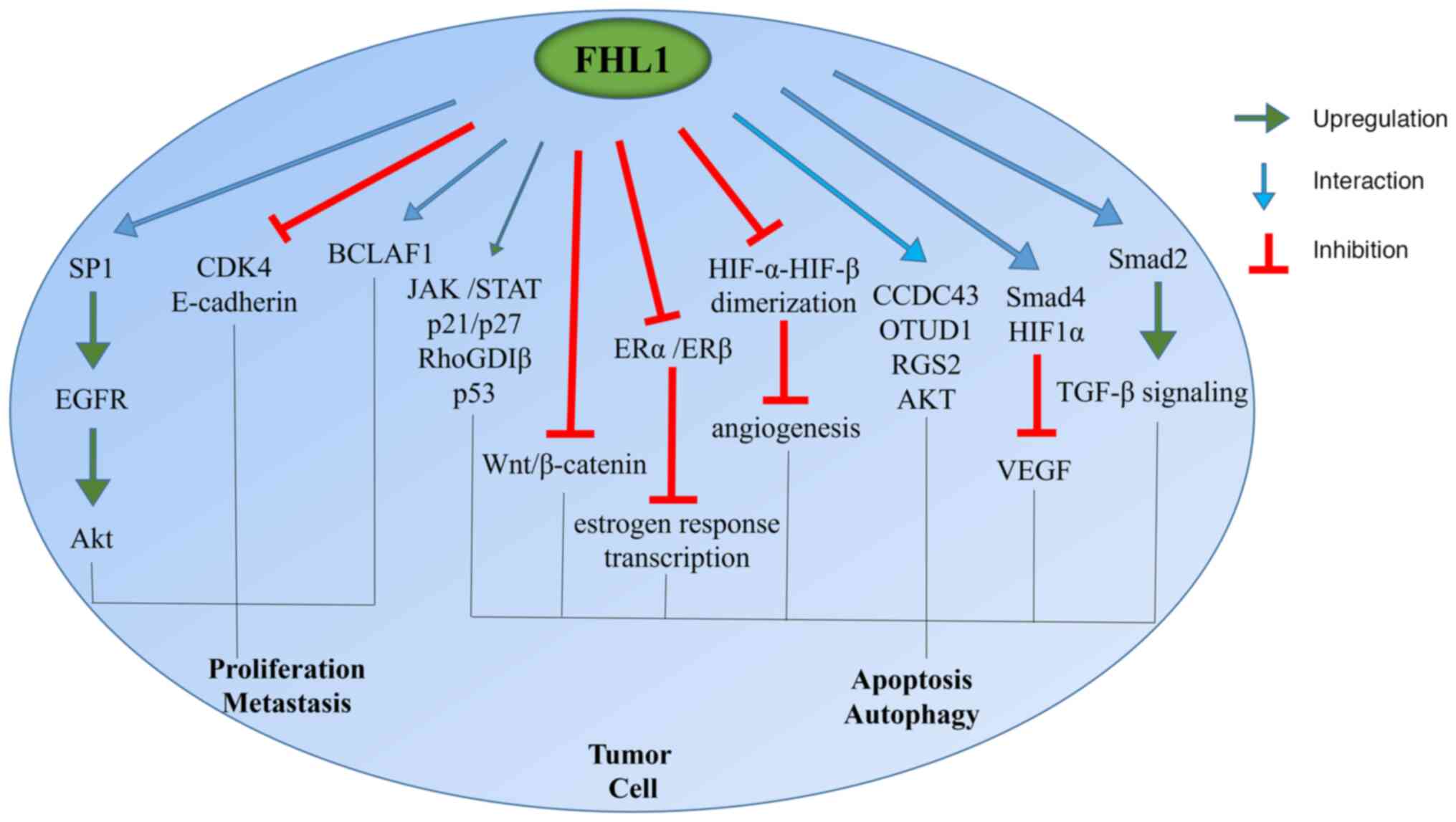 | Figure 2.Role of FHL1 in tumor progression.
FHL1, four and a half LIM domain protein 1; SP1, putative specific
protein 1; EGFR, epidermal growth factor receptor; CDK4,
cyclin-dependent kinase 4; BCLAF1, Bcl2-associated transcription
factor 1; RhoGDIβ, Rho GDP-dissociation inhibitor β; CCDC43,
coiled-coil domain-containing protein 43; OTUD1, OTU domain
containing 1; RGS2, G protein signal regulatory factor 2; HIF1,
hypoxia-inducible factor 1. |
Promotion of tumor proliferation
FHL1 promotes the proliferation of tumor cells via
interacting with SP1, Src and other proteins.
Interacting with the transcription
factor SP1
FHL1 and SP1 are co-located in the cell and they
form a complex. FHL1 interacts with the transcription factor SP1 to
upregulate EGFR expression and activate the downstream signaling
pathways, including Src, Akt, ERK1/2 and STAT3, thereby causing
glioblastoma. When FHL1 is phosphorylated by Src at Y149 and Y272,
it is transformed into an oncogene (46).
Mediated by Src and Kindlin-2
Src interacts with FHL1 through its kinase domain
and phosphorylates FHL1 via Y149 and Y272 of FHL1. The LIM4 domain
is also involved in this interaction. This process makes it a
pro-cancer factor that promotes tumor proliferation. Phosphorylated
FHL1 is more likely to enter the nucleus and bind to the
transcription factor Bcl2 associated transcription factor 1
(BCLAF1) to promote tumor growth. Kindlin2 also interacts with FHL1
via its FERM domain to inhibit cell growth. The last LIM domain is
involved in this interaction. Kindlin2 competes with Src to bind
FHL1 and inhibits the SRC-mediated phosphorylation of FHL1
(46–48).
Inhibition of tumor proliferation
FHL1 inhibits the growth of colon, bladder and liver
tumor cells via mechanisms including blocking hypoxia-inducible
factor (HIF)-α-HIF-β dimerization, interacting with coiled-coil
domain-containing protein 43 (CCDC43) and regulating the
Wnt/β-catenin signaling pathway. For instance, FHL1 knockdown
promoted the growth of HeLa and HepG2 cells. Increased FHL1
methylation in bladder and gastrointestinal tumors was observed to
lead to decreased FHL1 expression, which promoted tumor cell growth
and metastasis (49).
Mediation of HIF-α-HIF-β
dimerization
Kruppel-like factor 17 increases the expression of
FHL1 by binding to FHL1 promoter and promoting its transcription.
This process inhibits the proliferation, invasion and migration of
colon tumor cells via influencing the expression of E-cadherin and
N-cadherin. In colon cancer, the expression of FHL1 is
downregulated, which leads to the proliferation, invasion and
migration of tumor cells. As a tumor suppressor gene, FHL1
regulates tumor angiogenesis by blocking HIF-α-HIF-β dimerization
and reducing VEGF expression (50).
Interaction with CCDC43
In gastric cancer, CCDC43 and FHL1 are co-localized
in perinuclear and nuclear regions and the expression of CCDC43 is
negatively correlated with the expression of FHL1. CCDC43 is highly
expressed, whereas FHL1 is weakly expressed. There is an
interaction between the two molecules. Knocking down CCDC43
improves the stability of FHL1, inhibits the growth and metastasis
of gastric cancer cells and promotes apoptosis (51).
Interaction with Smad2, Smad3 and
Smad4
FHL1 interacts with Smad2, Smad3 and Smad4 to
activate the transcription of p21, inhibit c-Myc transcription and
inhibit the growth of liver cancer cells. FHL1 and the tumor
suppressor gene Smad4 synergistically inhibit VEGF promoter
activity. Furthermore, a reduction in Smad4 abolishes the
inhibitory effect of FHL1 on VEGF promoter activity, suggesting
that FHL1 inhibits VEGF promoter activity in a Smad4-dependent way
(52,53).
Interaction with estrogen receptor
(ER)α and ERβ
FHL1 inhibits the growth of breast cancer cells.
FHL1 interacts with ERα and ERβ in a 17β-estradiol-independent
manner to reduce the transcriptional activity of ERα and ERβ, which
is necessary to inhibit estrogen response transcription, resulting
in lower expressions of estrogen response genes PS2 and cathepsin
D. LIM1, LIM2 and LIM3 of FHL1 are required for this interaction
(44,54).
Interaction with OTU domain containing
1 (OTUD1)
The ATR inhibitor VE-822 upregulates the expression
level of the deubiquitinating enzyme OTUD1 to stabilize FHL1
expression through its OUT domain and inhibits the progression of
lung adenocarcinoma. In this process, the deubiquitinase activity
of OTUD1 is necessary. The expression of OTUD1 is downregulated in
lung cancer. Similarly, the expression of FHL1 is downregulated and
OTUD1 has lost its ability to inhibit lung cancer (55)
Interaction with G protein signal
regulatory factor 2 (RGS2)
In tumors, the expression of RGS2 is lower in cancer
than in paracancer cells. High expression of RGS2 can inhibit the
invasion and proliferation of tumors. FHL1 competes with
damage-specific DNA binding protein 1 to bind to RGS2, thereby
reducing its ubiquitination level, stabilizing its expression and
playing a role in inhibiting tumor growth.
Inhibition of VEGF promoter
activity
FHL1 inhibits the VEGF promoter activity, thereby
inhibiting VEGF expression. FHL1 interacts with HIF1α vis its LIM
domain. The interaction between FHL1 and HIF1α can inhibit the
interaction between HIF1α and HIF1β, thereby inhibiting the
dimerization of HIF1α and HIF1β. This can further inhibit the
binding of HIF1α and VEGF promoter, thereby inhibiting the
expression of HIF1α-dependent VEGF (56).
Taking part in the transcription
process
Phosphorylated FHL1 binds to the nuclear
transcription factor BCLAF1 and takes part in the DNA-binding
transcriptional activator activity and transcription factor
binding. FHL1 is involved in an IL-15-mediated signaling pathway
that activates JAK and STAT family proteins. Its expression is
positively correlated with the levels of B, CD8 and CD4 cells and
macrophages, and is also correlated with the expression of
immune-related genes, such as CD48 and CD80 (57).
Mediation of the Wnt/β-catenin
signaling pathway
The expression of FHL1 is downregulated in
colorectal tumors and is associated with poor prognosis. FHL1
reduces the phosphorylation of glycogen synthase kinase 3β and
activation of β-catenin, thereby negatively regulating the
Wnt/β-catenin signaling pathway, causing the downregulation of
cyclin D1 and CDK4 and thus inhibiting the proliferation of colon
tumor cells (3).
Mediation by miR-410
MiR-410 is highly expressed in liver and colorectal
tumors. MiR-410 targets FHL1 via the 3′-UTR of FHL1 and promotes
FHL1 methylation, thus negatively regulating FHL1 to promote tumor
growth (27,32).
Mediation of Cyclin D1, Cyclin E and
p27
In head and neck squamous cell carcinoma, FHL1
expression is downregulated and it promotes tumor cell growth
through dysregulation of Cyclin D1, Cyclin E and p27. Decreased
FHL1 expression is closely related to poor differentiation.
Downregulation of FHL1 expression in head and neck squamous cell
carcinoma is usually caused by hypermethylation of its DNA promoter
region and EZH2-mediated regulation of histone methylation
(2).
Activation of the TGF-β signaling
pathway
FHL1 and Smad2 interact in liver cancer cells by
phosphorylating Smad2 at Ser465 and Ser467. FHL1 activates the
TGF-β signaling pathway in a TGF-β-independent manner, thereby
inhibiting tumor growth (58).
Mediation of the cell circle
FHL1 mediates cell cycle in AML and other tumors to
influence the development of tumor. The expression of cell
cycle-related proteins such as p21 and p27 is regulated in this
process.
AML
In AML, FHL1 is highly expressed and promotes the
proliferation of AML cells. High FHL1 expression can lead to a
decrease in the proportion of cells in G1 phase and a decrease in
CDK4. FHL1 knockdown leads to an increase in the proportion of
cells in G1 phase. Loss of FHL1 also leads to increased expression
of CD11b and CD14. Overexpression of FHL1 prevents all-trans
retinoic acid-induced differentiation of HL-60 cells. Consumption
of FHL1 inhibits cell proliferation and induces cell
differentiation (39).
Lung cancer
The expression of FHL1 in lung tumor tissues is
lower than that in normal tissues. FHL1 inhibits the growth of
tumor cells by inducing G1 and G2/M phase cell cycle arrest and
decreasing the number of cells in S-phase. Overexpression of FHL1
increases the expression of p21 to increase the proportion of cells
in G1 and G2/M phases, increases the expression of p27 to promote
G1 phase arrest, and decreases the expression of CyclinD1, CyclinB1
and CyclinA (53).
Glioma
FHL1 inhibits the growth of glioma cells and its
expression level is negatively associated with prognosis. FHL1
maintains cells in the G0/G1 phase and inhibits tumor growth. When
FHL1 is knocked down, cells enter the S phase, which promotes cell
growth. FHL1 interacts with AKT and inhibits the binding of AKT and
PI3K, thereby inhibiting glioma growth (5).
In conclusion, FHL1 mediates various cellular
pathways in tumor progression and mediates tumor growth. FHL1
mainly functions through interacting with proteins, particularly
transcription factors such as SP1, to activate downstream signaling
pathways. It also affects the transcriptional activity of proteins
such as p21 to regulate the growth of tumor cells. Cyclins are also
regulated by FHL1 to mediate tumor growth.
FHL1 signaling in tumor invasion and
migration
In bladder, breast, lung, papillary thyroid and
other cancers, FHL1 inhibits tumor invasion and metastasis by
inhibiting the Wnt/β-catenin signaling pathway, and is a good
potential therapeutic target. In bladder tumors, the expression of
FHL1 is decreased and its methylation level is increased, leading
to tumor invasion and migration. Given the complexity of its
regulatory mechanisms, the different mechanisms by which FHL1
mediates tumor invasion and migration need to be further explored
(49).
Mediation by miR-183-5p
In breast cancer, miR-183-5p downregulates the
expression of FHL1 and mediates the upregulation of VEGF,
N-cadherin and vimentin, and the downregulation of p53 and
E-cadherin, resulting in enhanced activity of breast cancer cells
and enhanced invasion and migration abilities (32).
Increase in the expression of
RhoGDIβ
The expression of FHL1 is inhibited in lung cancer,
thus reducing the expression of RhoGDIβ and leading to tumor
invasion (48).
Mediation by miR-96-5p
MiR-96-5p inhibits the expression of FHL1 and
promotes the proliferation, invasion and migration of lung
adenocarcinomas, resulting in a decrease in the overall survival of
patients (27).
Inhibition of the Wnt/β-catenin
signaling pathway
FHL1 expression is downregulated in PTC and its
expression is negatively associated with T/N staging. Its
expression is also related to the progression-free and disease-free
intervals. FHL1 inhibits the Wnt/β-catenin signaling pathway,
thereby inhibiting the proliferation, invasion and migration of PTC
cells (59).
FHL1 signaling in tumor therapy
resistance
The comprehensive application of various treatment
methods can effectively improve the curative effects in patients
with tumors. However, treatment resistance remains an important
reason for poor prognosis. FHL1 is abnormally expressed in various
tumors and is closely associated with drug resistance during tumor
therapy. The mechanism by which it mediates tumor resistance also
differs among different tumors.
Induction of radiation resistance
In breast cancer, ionizing radiation can induce the
expression of FHL1, thereby inhibiting the activity of CDC25C
through its interaction with checkpoint kinase 2 and CDC25, and
thus causes radiation resistance (46,60).
Increase of sensitivity to
paclitaxel
In hepatocellular carcinoma, knockdown of FHL1 can
increase the sensitivity to paclitaxel by inducing the activation
of caspase-3 and caspase-9 (46).
Induction of resistance to
cytarabin-based induction chemotherapy
High expression of FHL1 is associated with poor
prognosis in AML and is an independent prognostic factor. Src
phosphorylation of FHL1 promotes cell proliferation. High
expression of FHL1 is associated with drug resistance in AML. In
patients with AML receiving cytarabin-based induction chemotherapy,
high FHL1 predicts a poor prognosis. High FHL1 levels lead to
increased expression of ATP-binding cassette, sub-family C
(CFTR/MRP), member 1 (ABCC1) and ABCC4, which are associated with
the transport of chemotherapy drugs, and which are subsequently
excreted into the extracellular system. Simultaneously, the
expression of SLC29A1 decreases and the entry of cytarabine into
the cell, resulting in drug resistance (61).
Increasing sensitivity to
olaparib
In patients with ovarian cancer, tabersonine
increases the sensitivity of ovarian cancer cells to the poly
ADP-ribose polymerase inhibitor olaparib. In addition, glycyrrhizin
increases FHL expression. They act synergistically to decrease
epithelial-mesenchymal transformation, thereby inhibiting the
invasion and migration of ovarian cancer cells. The combination of
tabersonine and olaparib increases the expression of E-cadherin,
decreases the expression of N-cadherin and vimentin, and
effectively inhibits tumor growth (62).
Inducing resistance to platinum and
paclitaxel
In patients with ovarian cancer, low FHL1 expression
predicts higher sensitivity to platinum and paclitaxel and a better
prognosis. Relevant studies have shown that FHL1 is co-expressed
with Filamin C, γ (FLNC), Caveolin-1 and FLNA, and is related to
the abundance of macrophages, thus affecting the sensitivity of
patients to chemotherapy (45).
In brief, the expression of FHL1 in tumors is
complex and the role of FHL1 varies in different tumors, which
deserves further study. At the same time, FHL1 has become a
potential target in tumor treatment. In different tumors, tumor
growth could be inhibited by inducing or inhibiting the expression
level of FHL1 or by affecting its post-translational modifications,
such as phosphorylation, ubiquitination and methylation (40,44).
The expression level of FHL1 could also be used as a reference for
estimating the sensitivity to chemoradiotherapy treatment in
certain tumors.
However, research on the role of FHL1 in tumor
progression and treatment resistance is still limited. The
mechanisms of FHL1 affecting tumor cell proliferation and migration
have mainly been obtained through the study of cell lines and need
to be further verified in clinical samples. There is also a lack of
multicenter clinical trials to further increase the reliability of
the findings. Furthermore, research methods such as bioinformatics
have been rarely used in this field.
Conclusions
FHL1 is an important cytoskeletal protein that is
expressed in various cells (63–66).
It plays a regulatory role in the development of myocardial and
skeletal muscle diseases, such as reducing body myopathy and
cardiac hypertrophy (67–72). It is also widely expressed in tumor
cells, including lung, ovarian and liver cancers. In different
tumors, FHL1 functions differently to influence the fate of the
tumor (73,74). In numerous tumors such as colon
cancer and esophageal cancer, FHL1 functions as a tumor suppressor
gene that inhibits tumor growth, while it is also reported that
FHL1 promotes the development of glioblastoma and other tumors
(75–77). In the process of tumor occurrence
and development, it is regulated by numerous signaling pathways,
and it promotes or inhibits tumor occurrence and development by
regulating the EGFR and Wnt/β-catenin signaling pathways.
Post-transcriptional modifications also affect the function of
FHL1. Its expression can be silenced via methylation and it can
also be stabilized by deubiquitinating enzymes such as USP15. The
regulatory role of FHL1 in tumors is complex and relevant studies
suggest that the phosphorylation of FHL1 by Src may play an
important regulatory role in the function of FHL1 in tumor growth.
When phosphorylated, FHL1 is more likely to be located in the
nucleus and facilitates tumor growth.
However, owing to the complexity of its regulatory
mechanisms, the role of FHL1 in different tumors has remained to be
fully elucidated (78–80). The results of the present study
suggest that FHL1 can regulate tumor growth by regulating the cell
cycle of tumor cells, as well as other signaling pathways such as
Wnt/β-catenin and TGF-β. The different roles of FHL1 in different
tumors may be caused by factors including regional differences,
tumor differences and small sample size. At the same time, the
location of FHL1 and its regulatory mechanism is rarely reported.
FHL1 was mainly located in the cytoplasm and whether FHL1 has
different functions in the nucleus and cytoplasm, and its
regulatory mechanism, still warrant further study. The relationship
between the location of FHL1 and its role in tumor progression are
also a potential research field.
In addition, the subtype of FHL1 expressed in
various tumors deserves further study, and the relationship between
the expression of various subtypes and tumor occurrence and
development has been rarely reported. This could also be a
potential target for treatments against FHL1.
Of note, the abnormal expression of FHL1 is also
closely related to tumor resistance to radiotherapy and
chemotherapy. Therefore, FHL1 is a promising target for cancer
research and may serve as an effective therapeutic target. However,
in view of its complex roles in various tumors, its mechanism of
action in different tumors requires further study. The relationship
between the expression level of FHL1 and sensitivity to
chemoradiotherapy has only been studied in a small number of
tumors. Whether there is a relationship between FHL1 expression and
therapeutic resistance in other tumors deserves further
investigation.
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science Foundation of
Nantong (grant no. JC2023038).
Availability of data and materials
Not applicable.
Authors' contributions
XM and SH contributed to the conceptualization of
this study and provided constructive guidance. YT and YW collected
relevant studies and wrote the manuscript. RS participated in the
study design and completion. All authors read and approved the
final version of the manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Han S, Cui C, He H, Shen X, Chen Y, Wang
Y, Li D, Zhu Q and Yin H: FHL1 regulates myoblast differentiation
and autophagy through its interaction with LC3. J Cell Physiol.
235:4667–4678. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cao W, Liu J, Xia R, Lin L, Wang X, Xiao
M, Zhang C, Li J, Ji T and Chen W: X-linked FHL1 as a novel
therapeutic target for head and neck squamous cell carcinoma.
Oncotarget. 7:14537–14550. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu Y, Wang C, Cheng P, Zhang S, Zhou W,
Xu Y, Xu H and Ji G: FHL1 inhibits the progression of colorectal
cancer by regulating the Wnt/β-catenin signaling pathway. J Cancer.
12:5345–5354. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Keßler M, Kieltsch A, Kayvanpour E, Katus
HA, Schoser B, Schessl J, Just S and Rottbauer W: A zebrafish model
for FHL1-opathy reveals Loss-of-function effects of human FHL1
mutations. Neuromuscul Disord. 28:521–531. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li SZ, Hu YY, Zhao JL, Zang J, Fei Z, Han
H and Qin HY: Downregulation of FHL1 protein in glioma inhibits
tumor growth through PI3K/AKT signaling. Oncol Lett. 19:3781–3788.
2020.PubMed/NCBI
|
|
6
|
Ding J, Cong Y, Li F, Liu B, Wu D, Miao J
and Wang L: Muscle death participates in myofibrillar abnormalities
in FHL1 knockout mice. Biochem Biophys Res Commun. 523:105–111.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Weng J, Liao M, Zou S, Bao J, Zhou J, Qu
L, Feng R, Feng X, Zhao Z and Jing Z: Downregulation of FHL1
expression in thoracic aortic dissection: Implications in aortic
wall remodeling and pathogenesis of thoracic aortic dissection. Ann
Vasc Surg. 25:240–247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee SMY, Tsui SKW, Chan KK, Garcia-Barcelo
M, Waye MM, Fung KP, Liew CC and Lee CY: Chromosomal mapping,
tissue distribution and cDNA sequence of Four-and-a-half LIM domain
protein 1 (FHL1). Gene. 216:163–170. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wilding BR, McGrath MJ, Bonne G and
Mitchell CA: FHL1 mutations that cause clinically distinct human
myopathies form protein aggregates and impair myoblast
differentiation. J Cell Sci. 127:2269–2281. 2014.PubMed/NCBI
|
|
10
|
Wei X and Zhang H: Four and a half LIM
domains protein 1 can be as a double-edged sword in cancer
progression. Cancer Biol Med. 17:270–281. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Selcen D, Bromberg MB, Chin SS and Engel
AG: Reducing bodies and myofibrillar myopathy features in FHL1
muscular dystrophy. Neurology. 77:1951–1959. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Isumi Y, Hirata T, Saitoh H, Miyakawa T,
Murakami K, Kudoh G, Doi H, Ishibashi K and Nakajima H: Transgenic
overexpression of USP15 in the heart induces cardiac remodeling in
mice. Biochem Biophys Res Commun. 405:216–221. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shathasivam T, Kislinger T and Gramolini
AO: Genes, proteins and complexes: The multifaceted nature of FHL
family proteins in diverse tissues. J Cell Mol Med. 14:2702–2720.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nishikawa M, Sato K, Nakano S, Yamakawa H,
Nagase T and Ueda H: Specific activation of PLEKHG2-induced serum
response element-dependent gene transcription by four-and-a-half
LIM domains (FHL) 1, but not FHL2 or FHL3. Small GTPases.
10:361–366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zabalegui F, Castañeda SL, Amin G, Belli
C, Miriuka SG and Moro LN: Derivation of two human induced
pluripotent stem cell lines carrying a missense mutation in FHL1
(c.377G > A, p.C126Y) linked to familial muscular dystrophy.
Stem Cell Res. 75:1033072024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
López Blázquez M, Fernández Ávila AI,
Álvarez García-Rovés R, Centeno Jiménez M, Gómez González C and
Espinosa Castro MÁ: Description of a novel variant in the FHL1 gene
associated with hypertrophic cardiomyopathy with early and
aggressive presentation. Rev Esp Cardiol (Engl Ed). 75:968–970.
2022.PubMed/NCBI
|
|
17
|
Albrecht I, Wick C, Hallgren Å, Tjärnlund
A, Nagaraju K, Andrade F, Thompson K, Coley W, Phadke A, Diaz-Gallo
LM, et al: Development of autoantibodies against muscle-specific
FHL1 in severe inflammatory myopathies. J Clin Invest.
125:4612–4624. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martin DE, Soulard A and Hall MN: TOR
regulates ribosomal protein gene expression via PKA and the
forkhead transcription factor FHL1. Cell. 119:969–979. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ng WH, Liu X, Ling ZL, Santos CNO,
Magalhães LS, Kueh AJ, Herold MJ, Taylor A, Freitas JR, Koit S, et
al: FHL1 promotes chikungunya and o'nyong-nyong virus infection and
pathogenesis with implications for alphavirus vaccine design. Nat
Commun. 14:66052023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meertens L, Hafirassou ML, Couderc T,
Bonnet-Madin L, Kril V, Kümmerer BM, Labeau A, Brugier A,
Simon-Loriere E, Burlaud-Gaillard J, et al: FHL1 is a major host
factor for chikungunya virus infection. Nature. 574:259–263. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meertens L, Hafirassou ML, Couderc T,
Bonnet-Madin L, Kril V, Kümmerer BM, Labeau A, Brugier A,
Simon-Loriere E, Burlaud-Gaillard J, et al: FHL1 is a key player of
chikungunya virus tropism and pathogenesis. C R Biol. 343:79–89.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mota IA, Correia CdC, Fontana PN and
Carvalho AAS: Reducing body myopathy-A new pathogenic FHL1 variant
and literature review. Neuromuscul Disord. 31:847–853. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Binder MS, Brown E, Aversano T, Wagner KR,
Calkins H and Barth AS: Novel FHL1 mutation associated with
hypertrophic cardiomyopathy, sudden cardiac death, and myopathy.
JACC Case Rep. 2:372–377. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sakashita K, Mimori K, Tanaka F, Kamohara
Y, Inoue H, Sawada T, Hirakawa K and Mori M: Clinical significance
of loss of Fhl1 expression in human gastric cancer. Ann Surg Oncol.
15:2293–2300. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pereira CM, de Carvalho AC, da Silva FR,
Melendez ME, Lessa RC, Andrade VCC, Kowalski LP, Vettore AL and
Carvalho AL: In vitro and in silico validation of CA3 and FHL1
downregulation in oral cancer. BMC Cancer. 18:1932018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Z, Zhang J, Yang B, Li R, Jin L, Wang
Z, Yu H, Liu C, Mao Y and You Q: Long intergenic noncoding RNA
00261 acts as a tumor suppressor in non-small cell lung cancer via
regulating miR-105/FHL1 axis. J Cancer. 10:6414–6421. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou F, Qian C, Chen T, Zang X and Huang
T: MiR-96-5p facilitates lung adenocarcinoma cell phenotypes by
inhibiting FHL1. Comput Math Methods Med. 2022:78912222022.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang F, Su Q and Li C: Identidication of
novel biomarkers in Non-small cell lung cancer using machine
learning. Sci Rep. 12:166932022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen H, Lai X, Zhu Y, Huang H, Zeng L and
Zhang L: Quantitative proteomics identified circulating biomarkers
in lung adenocarcinoma diagnosis. Clin Proteomics. 19:442022.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Song J, Liang K, Wei T, Li L, Huang Z,
Chen G, Mao N and Yang J: Expression and predictive significance of
FHL1 and SLIT3 in surgically resected lung adenocarcinoma. Comb
Chem High Throughput Screen. 26:2226–2237. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang F, Lian M, Ma H, Feng L, Shen X, Chen
J and Fang J: Identification of key genes associated with papillary
thyroid microcarcinoma characteristics by integrating transcriptome
sequencing and weighted gene co-expression network analysis. Gene.
811:1460862022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Zeng Qa, Qiu J, Pang T, Ye F, Huang
L and Zhang X: MiR-183-5p promotes proliferation, metastasis and
angiogenesis in breast cancer cells through negatively regulating
four and a Half LIM protein 1. J Breast Cancer. 23:355–372. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Y, Fu J, Jiang M, Zhang X, Cheng L,
Xu X, Fan Z, Zhang J, Ye Q and Song H: MiR-410 is overexpressed in
liver and colorectal tumors and enhances tumor cell growth by
silencing FHL1 via a Direct/Indirect mechanism. PLoS One.
9:e1087082014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ji C, Liu H, Xiang M, Liu J, Yue F, Wang W
and Chu X: Deregulation of decorin and FHL1 are associated with
esophageal squamous cell carcinoma progression and poor prognosis.
Int J Clin Exp Med. 8:20965–20970. 2015.PubMed/NCBI
|
|
35
|
Eshibona N, Giwa A, Rossouw SC, Gamieldien
J, Christoffels A and Bendou H: Upregulation of FHL1, SPNS3, and
MPZL2 predicts poor prognosis in pediatric acute myeloid leukemia
patients with FLT3-ITD mutation. Leuk Lymphoma. 63:1897–1906. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang J, Hu X, Liu S, He Y, Lyu L and
Jiang L: Clinical value screening, prognostic significance, and key
gene identification of TrkB in laryngeal carcinoma. Dis Markers.
2022:13540052022.PubMed/NCBI
|
|
37
|
Das K, Chan XB, Epstein D, Te Teh B, Kim
KM, Kim ST, Park SH, Kang WK, Rozen S, Lee J and Tan P: NanoString
expression profiling identifies candidate biomarkers of RAD001
response in metastatic gastric cancer. ESMO Open. 1:e0000092016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu1 P, Ma Q, Chen H, Zhang L and Zhang X:
Identification of RHOBTB2 aberration as an independent prognostic
indicator in acute myeloid leukemia. Aging (Albany NY).
13:15269–15284. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu Y, Li H, Zhao Y, Li D, Zhang Q, Fu J
and Fan S: Targeting FHL1 impairs cell proliferation and
differentiation of acute myeloid leukemia cells. Biochem Cell Biol.
100:301–308. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Luo Q, Pan Y, Fu Q, Zhang X, Zhou S, Yu P,
Tian H, Liu P, Chen S, Zhang H and Qin T:
Immortalization-upregulated protein promotes pancreatic cancer
progression by regulating NPM1/FHL1-mediated cell-cycle-checkpoint
protein activity. Cell Biol Toxicol. 39:2069–2087. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin J, Ding L, Jin R, Zhang H, Cheng L,
Qin X, Chai J and Ye Q: Four and a half LIM domains 1 (FHL1) and
receptor interacting protein of 140 kDa (RIP140) interact and
cooperate in estrogen signaling. Int J Biochem Cell Biol.
41:1613–1618. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li X, Jia Z, Shen Y, Ichikawa H, Jarvik J,
Nagele RG and Goldberg GS: Coordinate suppression of Sdpr and Fhl1
expression in tumors of the breast, kidney, and prostate. Cancer
Sci. 99:1326–1333. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Koike K, Kasamatsu A, Iyoda M, Saito Y,
Kouzu Y, Koike H, Sakamoto Y, Ogawara K, Tanzawa H and Uzawa K:
High prevalence of epigenetic inactivation of the human four and a
half LIM domains 1 gene in human oral cancer. Int J Oncol.
42:141–150. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Asada K, Ando T, Niwa T, Nanjo S, Watanabe
N, Okochi-Takada E, Yoshida T, Miyamoto K, Enomoto S, Ichinose M,
et al: FHL1 on chromosome X is a single-hit gastrointestinal
tumor-suppressor gene and contributes to the formation of an
epigenetic field defect. Oncogene. 32:2140–2149. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen X, Yu Y, Su Y, Shi L, Xie S, Hong Y,
Liu X and Yin F: Low FHL1 expression indicates a good prognosis and
drug sensitivity in ovarian cancer. Funct Integr Genomics.
24:252024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sun L, Chen L, Zhu H, Li Y, Chen CC and Li
M: FHL1 promotes glioblastoma aggressiveness through regulating
EGFR expression. FEBS Lett. 595:85–98. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang X, Wei X, Yuan Y, Sun Q, Zhan J,
Zhang J, Tang Y, Li F, Ding L, Ye Q and Zhang H: Src-mediated
phosphorylation converts FHL1 from tumor suppressor to tumor
promoter. J Cell Biol. 217:1335–1351. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shi MK, Xuan YL and He XF: FHL1
overexpression as a inhibitor of lung cancer cell invasion via
increasing RhoGDIß mRNA expression. Cell J. 24:239–244.
2022.PubMed/NCBI
|
|
49
|
Matsumoto M, Kawakami K, Enokida H, Toki
K, Matsuda R, Chiyomaru T, Nishiyama K, Kawahara K, Seki N and
Nakagawa M: CpG hypermethylation of human four-and-a-half LIM
domains 1 contributes to migration and invasion activity of human
bladder cancer. Int J Mol Med. 26:241–247. 2010.PubMed/NCBI
|
|
50
|
Yi S, Luo M, Peng Y, Chen Y and Yu D:
Anti-oncogenic mechanism of KLF17 in colon cancer by repressing
cell migration and invasion via FHL1 upregulation. Chi J Physiol.
66:534–545. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen Y, Pei M, Li J, Wang Z, Liu S, Xiang
L, Zhang J, Hong L, Lin J, Dai W, et al: Disruption of the
CCDC43-FHL1 interaction triggers apoptosis in gastric cancer cells.
Exp Cell Res. 415:1131072022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhou Z, Lu J, Dou J, Lv Z, Qin XI and Lin
J: FHL1 and Smad4 synergistically inhibit vascular endothelial
growth factor expression. Mol Med Rep. 7:649–653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Niu C, Liang C, Guo J, Cheng L, Zhang H,
Qin X, Zhang Q, Ding L, Yuan B, Xu X, et al: Downregulation and
growth inhibitory role of FHL1 in lung cancer. Int J Cancer.
130:2549–2556. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ding L, Niu C, Zheng Y, Xiong Z, Liu Y,
Lin J, Sun H, Huang K, Yang W, Li X and Ye Q: FHL1 interacts with
oestrogen receptors and regulates breast cancer cell growth. J Cell
Mol Med. 15:72–85. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang Q, Li J, Chen Z, Jiang K, Yang K,
Huang F, Huang A, Zhang X, Zhang J and Wang H: VE-822 upregulates
the deubiquitinase OTUD1 to stabilize FHL1 to inhibit the
progression of lung adenocarcinoma. Cell Oncol. 46:1001–1014. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lin J, Qin X, Zhu Z, Mu J, Zhu L, Wu K,
Jiao H, Xu X and Ye Q: FHL family members suppress vascular
endothelial growth factor expression through blockade of
dimerization of HIF1α and HIF1β. IUBMB Life. 64:921–930. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang J, Li H, Guo M, Zhang J, Zhang G,
Sun N, Feng Y, Cui W and Xu F: FHL1 as a novel prognostic biomarker
and correlation with immune infiltration levels in lung
adenocarcinoma. Immunotherapy. 15:235–252. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ding L, Wang Z, Yan J, Yang X, Liu A, Qiu
W, Zhu J, Han J, Zhang H, Lin J, et al: Human four-and-a-half LIM
family members suppress tumor cell growth through a TGF-β-like
signaling pathway. J Clin Invest. 119:349–361. 2009.PubMed/NCBI
|
|
59
|
Chen J, Zeng C, Jin J, Zhang P, Zhang Y,
Zhang H, Li Y and Guan H: Overexpression of FHL1 suppresses
papillary thyroid cancer proliferation and progression via
inhibiting Wnt/β-catenin pathway. Endocrine. 85:238–249. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xu X, Fan Z, Liang C, Li L, Wang L, Liang
Y, Wu J, Chang S, Yan Z, Lv Z, et al: A signature motif in LIM
proteins mediates binding to checkpoint proteins and increases
tumour radiosensitivity. Nat Commun. 8:140592017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fu Y, Xu M, Cui Z, Yang Z, Zhang Z, Yin X,
Huang X, Zhou M, Wang X and Chen C: Genome-wide identification of
FHL1 as a powerful prognostic candidate and potential therapeutic
target in acute myeloid leukaemia. EBioMedicine. 52:1026642020.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen X, Yan Y, Liu Y, Yi Q and Xu Z:
Tabersonine enhances olaparib sensitivity through FHL1-Mediated
Epithelial-mesenchymal transition in an ovarian tumor. J Nat Prod.
87:837–848. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu M, Meng J, Chen X, Wang F and Han Z:
Long non-coding RNA Small Nucleolar RNA Host Gene 4 ameliorates
cigarette Smoke-induced proliferation, apoptosis, inflammation, and
airway remodeling in alveolar epithelial cells through the
modulation of the mitogen-activated protein kinase signaling
pathway via the microRNA-409-3p/Four and a Half LIM Domains 1 axis.
Eur J Med Res. 29:3092024. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ding J, Cong YF and Liu B: Aberrant
protein turn-over associated with myofibrillar disorganization in
FHL1 knockout mice. Front Genet. 9:2732018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wong CH, Fung YW, Ng EK, Lee SM, Waye MM
and Tsui SK: LIM domain protein FHL1B interacts with PP2A catalytic
β subunit-A novel cell cycle regulatory pathway. FEBS Lett.
584:4511–4516. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wu J, Zhao K, Du Z, Chen Y, Zhang F, Jiang
W, Zheng J, Wu X, Shen C and Xiao X: Systemic effect of FHL1 on
neuromuscular junction and myotube formation via insulin-like
growth factor and myostatin signaling pathways. Biochem Biophys Res
Commun. 537:125–131. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Schreckenbach T, Henn W, Kress W, Roos A,
Maschke M, Feiden W, Dillmann U, Schulz JB, Weis J and Claeys KG:
Novel FHL1 mutation in a family with reducing body myopathy. Muscle
Nerve. 47:127–134. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Peters S: Electrocardiographic analysis in
unclassifiable arrhythmic cardiomyopathy associated with
Emery-Dreifuss caused by a mutation in FHL1. Int J Cardiol.
214:1362016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Luo YB, Peng Y, Lu Y, Li Q, Duan H, Bi F
and Yang H: Expanding the Clinico-genetic spectrum of myofibrillar
myopathy: Experience from a Chinese neuromuscular center. Front
Neurol. 11:10142020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hu Z, Zhu Y, Liu X, Zhang W, Liu J, Wu S,
Xiao J, Yuan Y and Wang Z: FHL1-related clinical, muscle MRI and
genetic features in six Chinese patients with reducing body
myopathy. J Hum Genet. 64:919–926. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sepp R, Hategan L, Csányi B, Borbás J,
Tringer A, Pálinkás ED, Nagy V, Takács H, Latinovics D, Nyolczas N,
et al: The genetic architecture of hypertrophic cardiomyopathy in
hungary: Analysis of 242 patients with a panel of 98 genes.
Diagnostics (Basel). 12:11322022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lin Y, Ban R, Qiao L, Chen J, Liu M, Liu J
and Shi Q: Identification of novel FHL1 mutations associated with
X-linked scapuloperoneal myopathy in unrelated Chinese patients. J
Hum Genet. 68:477–484. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Liu E, Jelinek J, Pastore YD, Guan Y,
Prchal JF and Prchal JT: Discrimination of polycythemias and
thrombocytoses by novel, simple, accurate clonality assays and
comparison with PRV-1 expression and BFU-E response to
erythropoietin. Blood. 101:3294–3301. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang F, Feng F, Yang P, Li Z, You J, Xie
W, Gao X and Yang J: Four-and-a-half-LIM protein 1 down-regulates
estrogen receptor α activity through repression of AKT
phosphorylation in human breast cancer cell. Int J Biochem Cell
Biol. 44:320–326. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zeng Y, Zeng D, Qi X, Wang H, Wang X, Dai
X and Qu L: FHL1: A novel diagnostic marker for papillary thyroid
carcinoma. Pathol Int. 74:520–529. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Balraj A, Muthamilselvan S, Raja R and
Palaniappan A: PRADclass: Hybrid Gleason grade-informed
computational strategy identifies consensus biomarker features
predictive of aggressive prostate adenocarcinoma. Technol Cancer
Res Treat. 23:153303382312223892024. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Cai Y, Xia L, Zhu H, Cheng H, Tian Y, Sun
L, Wang J, Lu N, Wang J and Chen Y: MiR-3682-3p promotes esophageal
cancer progression by targeting FHL1 and activating the
Wnt/β-catenin signaling pathway. Cell Signal. 119:1111552024.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhao JL, Liang SQ, Fu W, Zhu BK, Li SZ,
Han H and Qin HY: The LIM domain protein FHL1C interacts with tight
junction protein ZO-1 contributing to the epithelial-mesenchymal
transition (EMT) of a breast adenocarcinoma cell line. Gene.
542:182–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Dai J, Yu X, Han Y, Chai L, Liao Y, Zhong
P, Xie R, Sun X, Huang Q and Wang J: TMT-labeling proteomics of
papillary thyroid carcinoma reveal invasive biomarkers. J Cancer.
11:6122–6132. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Fu W, Wang K, Zhao JL, Yu HC, Li SZ, Lin
Y, Liang L, Huang SY, Liang YM, Han H and Qin HY: FHL1C induces
apoptosis in notch1-dependent T-ALL cells through an interaction
with RBP-J. BMC Cancer. 14:4632014. View Article : Google Scholar : PubMed/NCBI
|