Introduction
Breast cancer, a heterogeneous disease associated
with impaired cell proliferation, is the second most prevalent
cause of cancer-related deaths globally and the most common cancer
in women (1,2). It is a heterogeneous disease with
several morphological and molecular features, as well as clinical
outcomes. The majority of breast cancer cases are
hormone-dependent, and a positive relationship has been reported
between long-term exposure to high concentrations of estrogen and
breast cancer incidence (3).
However, there are also types of breast cancer that occur and
develop independently of hormones. Treatment approaches also differ
in breast cancers that differ in terms of formation and development
mechanisms (4).
Currently, the parameters used to determine the
classification of breast carcinomas are as follows: estrogen
receptor (ER) and progesterone receptor (PR), human epidermal
growth factor receptor 2 (HER2/cErbB2) overexpression and/or HER2
gene amplification and Ki-67 proliferation index (5). Accordingly, invasive breast carcinoma
can be classified as hormone receptor-positive, (HER2)-positive and
triple-negative, and these subtypes have their own specific
treatment approaches (6–11).
Luminal A is the most common subgroup and
constitutes the majority of all breast cancers. Whilst these tumors
are ER and/or PR positive, they are HER2 negative. Ki-67 level is
also generally <20%. The Luminal A subgroup has the best
prognosis of all breast cancers and is generally characterized by
low-grade, slow-growing tumors with a high survival rate.
Furthermore, relapse rates are lower compared with that of other
subgroups (12). In the treatment
of hormone receptor-positive breast cancer, selective estrogen
receptor modulators (such as tamoxifen and its derivatives) that
affect the effect of endogenous estrogens via the receptor, and
selective estrogen enzyme modulators (such as formestane and
letrozole) that affect the activity of enzymes involved in the
synthesis of estrogens, are used (13,14).
HER2-positive tumors constitute 13–20% of invasive
breast cancers, and ~50% of them are hormone receptor-negative
(11). Whilst the presence of HER2
was used as an indicator of poor prognosis in the past, this view
has changed with the use of recombinant humanized anti-HER2
antibody (transtuzumab) (7).
In the triple negative subgroup, all three hormone
receptors (ER, PR and HER2) are detected as negative. A
distinguishing feature from other subgroups is that it is more
common in women aged <40 years and constitutes ~20% of all
breast cancers (15).
The majority of cancer drugs used today are tyrosine
kinase (TK) inhibitors. This is a family of enzymes involved in
signal transduction in human cells. TKs are necessary for normal
physiology and regulate signal transduction mechanisms that serve a
role in cell homeostasis, cell proliferation, growth arrest and
apoptosis. The disruption of these functions causes abnormal cell
activity and immunological, neurological, metabolic and infectious
diseases, especially cancer. TKIs stop the cell cycle by preventing
protein phosphorylation catalyzed by TKs and cause tumor cell
apoptosis; it is used in cancer treatment with these effects
(16,17). The MET gene (c-MET) encodes a
receptor tyrosine kinase (RTK) known as MET (18). A significant factor in the onset and
spread of many cancers, including breast cancer, is abnormal MET
activation (19,20). Crizotinib acts as a MET inhibitor
and crizotinib activity has been reported in patients with MET
amplification (21).
Sodium butyrate (SB), as the sodium salt of
butyrate, a short-chain fatty acid, causes the activation of genes
related to apoptosis due to its histone deacetylase (HDAC)
inhibitor (HDACi) activity (22).
The HDACi feature, one of the functions of SB, is a subject that
has been studied extensively in terms of its effects on tumor
formation and development. Acetylated histones form a loose
chromatin structure suitable for the transcription initiation
complex. In this respect, deacetylation inhibited by SB causes a
global increase in transcription (23). It has been reported that SB triggers
apoptosis when applied in combination with different molecules such
as quercetin (24,25).
The present study aimed to compare the use of
Crizotinib as monotherapy and in combination with butyric acid
under in vitro conditions using cell lines from different
breast cancer types and different cell kinetic parameters,
assessing whether the combined use is more effective than its use
alone.
Materials and methods
Cell culture
MCF-7, MDA-MB-231 and SKBR-3 were used as different
breast cancer models. All cells were adherent cell lines growing in
a single layer. The MCF-7 and SKBR3 cell lines were cultured in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) medium containing
2 mM L-glutamine and the MDA-MB-231 cell line was cultured in high
glucose DMEM (Gibco; Thermo Fisher Scientific, Inc.). In order for
the cells to proliferate, 10% (v/v) fetal calf serum (Gibco; Thermo
Fisher Scientific, Inc.) was added to all media. A total of 100
µg/ml streptomycin (streptomycin sulphate; IE Ulugay Ilac Sanayi
TAS), 100 IU/ml penicillin (Pronapen; Pfizer, Inc.), amphotericin B
(Merck KGaA) were added to the media. All cells were grown at 37°C
in a humidified 5% CO2 atmosphere.
Cell index
For all three cell lines, 100 µl medium was added to
each well of the 16-well e-plates (special cell culture containers
containing microelectrodes). All cells were grown at 37°C in a
humidified 5% CO2 atmosphere. Subsequently, cell
counting was performed. After the plate was placed in the station,
measurements were taken by the device every 15 min and continued
until the end of the experiment. A total of 100 µl cells per well
were seeded on e-plates, with 10×103 cells for MCF-7,
8×103 cells for SKBR3 and 5×103 cells for MDA
MB-231 in each well. After 24 h, the cells were treated with
appropriate concentrations of substances (crizotinib: 5, 10 and 15
µM; sodium butyrate: 300, 400 and 500 µM; combined concentrations:
10 µM crizotinib + 50 µM SB, 10 µM crizotinib + 100 µM SB, 10 µM
crizotinib + 200 µM SB, 400 µM SB + 2.5 µM crizotinib, 400 µM SB +
5 µM crizotinib and 400 µM SB + 7.5 µM crizotinib) and changes in
cell proliferation were observed using the xCELLigence RTCA DP
System (Agilent Technologies, Inc.) in the incubator in special
cell culture dishes containing microelectrodes.
Bromodeoxyuridine (BrdU) incorporation
assay
The Colorimetric BrdU Cell Proliferation Assay Kit
(cat. no. 2750; Millipore Sigma) was used to determine the
proliferation levels in the MCF-7, MDA-MB-231 and SKBR-3 cells.
This method was based on the principle of marking BrdU, which is
incorporated into genomic DNA as a thymidine analogue in
proliferating cells, using antibody probes and detecting cells in S
phase. The BrdU test was performed following the instructions for
use provided by the manufacturer. BrdU-labeled cells were evaluated
at 370 nm in a microplate reader spectrophotometer.
Mitotic index
Mitotic index values, which demonstrate mitotic
activity (the division rate of cells), were evaluated using the
Mitotic Assay Kit (cat. no. 18021; Active Motif, Inc.). The mitotic
activity assay was performed following the instructions for use
provided by the manufacturer. Mitotic cells were evaluated at 450
nm in a microplate reader spectrophotometer within 5 min.
Caspase activity
The CaspaTag Caspase 3,7 In Situ Assay Kit
(cat. no. APT403; MilliporeSigma) was used to determine active
caspase-3 or −7 in cells undergoing apoptosis. The methodology was
based on Fluorochrome Inhibitors of Caspases (26). Caspase activity was performed
following the instructions for use provided by the manufacturer,
and the activity was assessed at 490 nm excitation wavelength and
520 nm emission wavelength (λex 490, λem 520) using a BioTek
Agilent FLx-800 Fluorescent Plate Reader (Agilent Technologies,
Inc.).
Statistical analysis
The experiments were performed in three replicates.
The data in the present study is presented as the mean ± standard
deviation. Comparisons between groups were performed using one-way
ANOVA and Dunnett's tests. Statistical evaluations of the cell
index were made by the xCelligence device. Dunnett's post hoc test
was used for all other parameters. Statistical analyses were
reviewed twice in line with referee opinions. Experiments were
performed in triplicate. The statistical analyses were performed
using SPSS statistics software (v22.0; IBM). P<0.05 was
considered to indicate a statistically significant difference.
Results
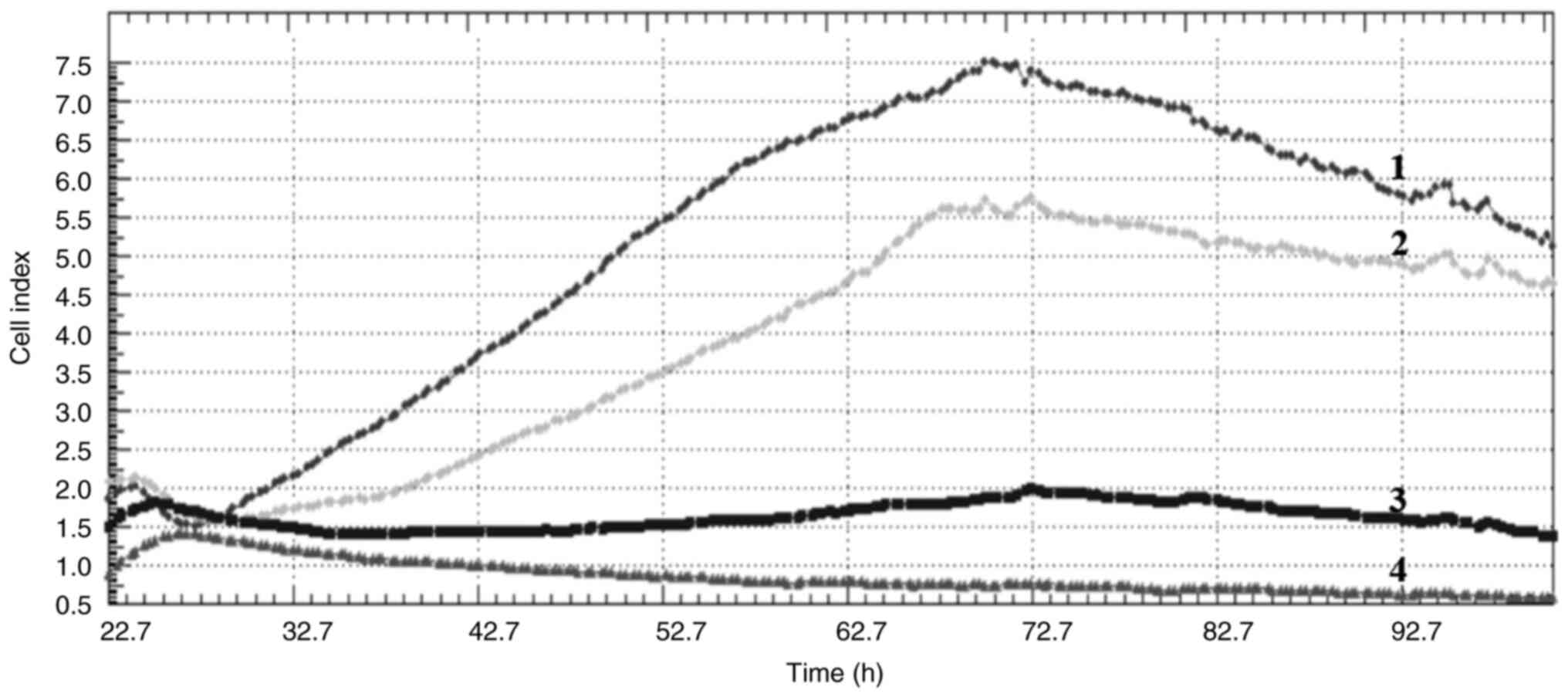
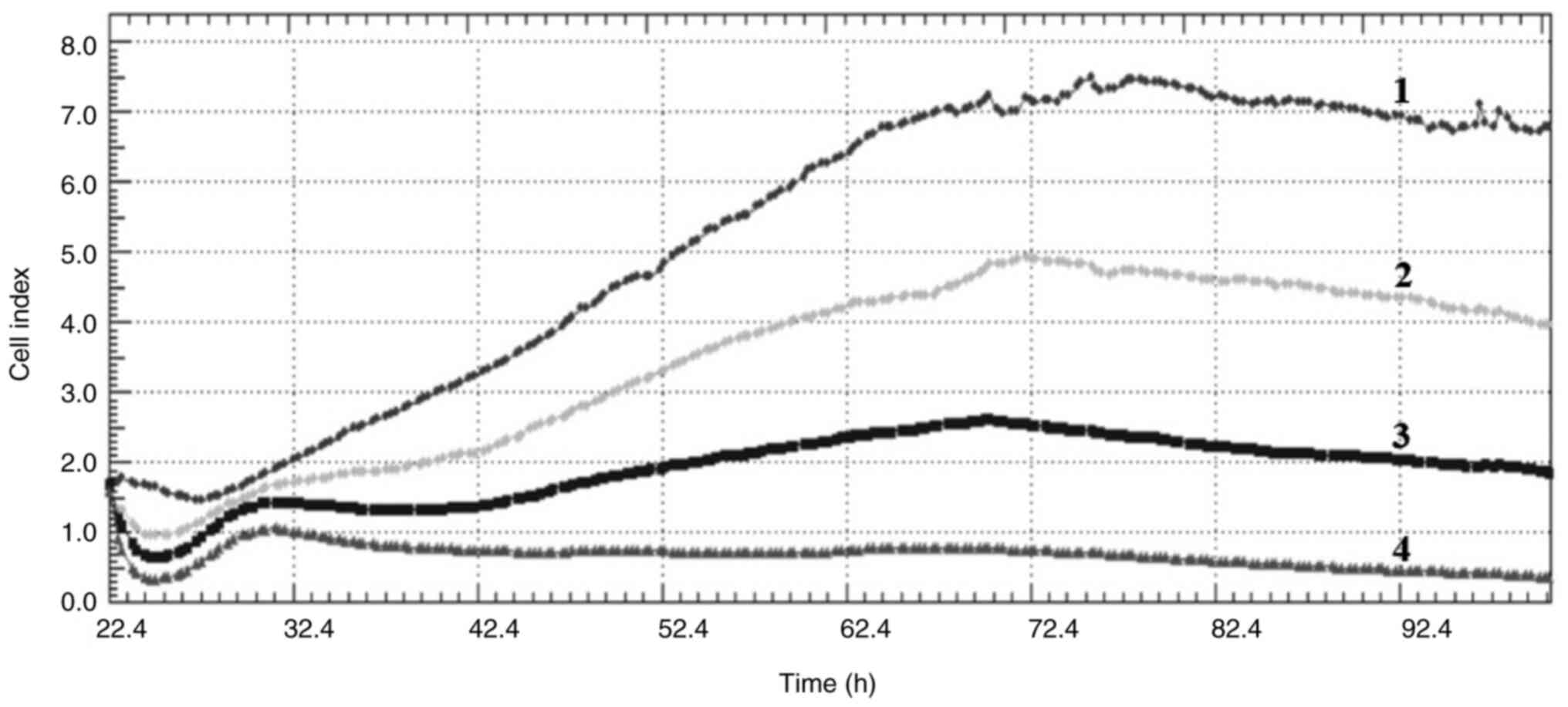
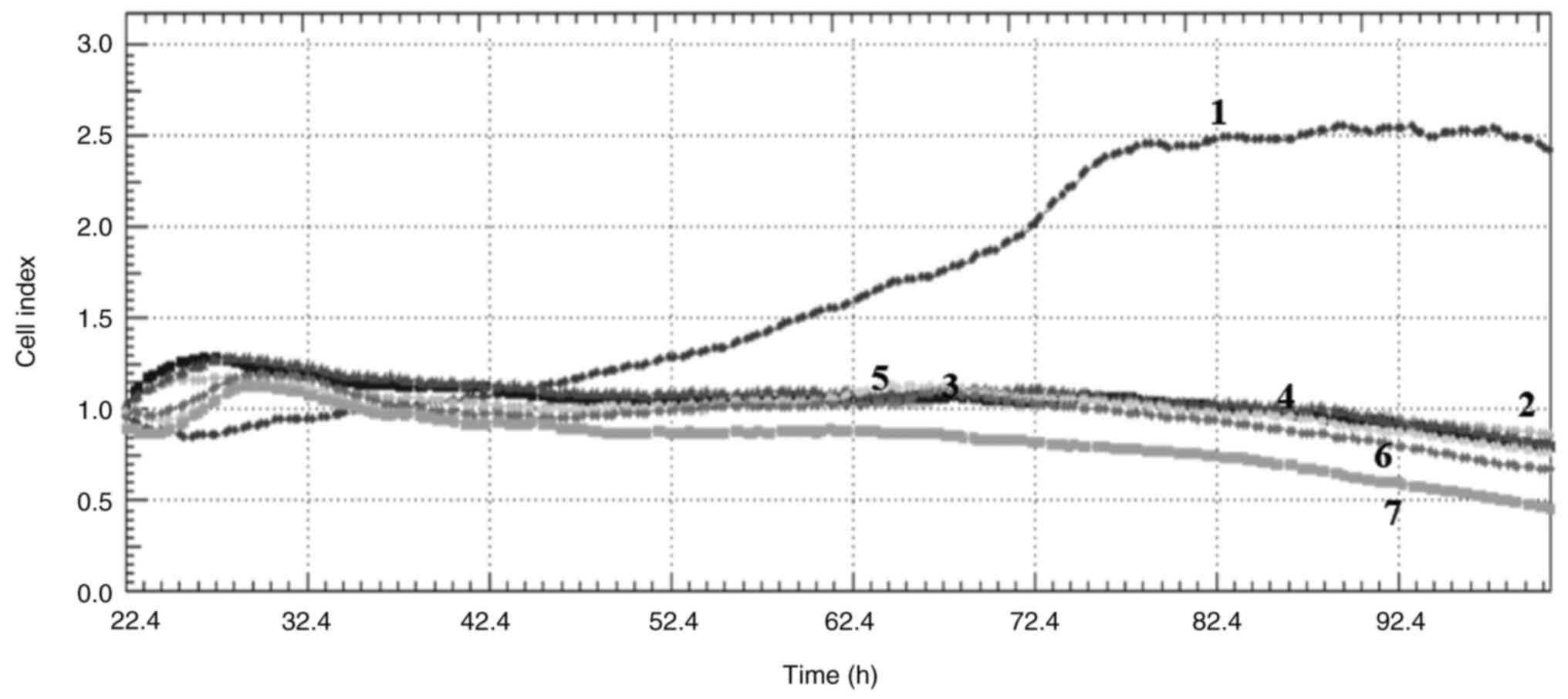
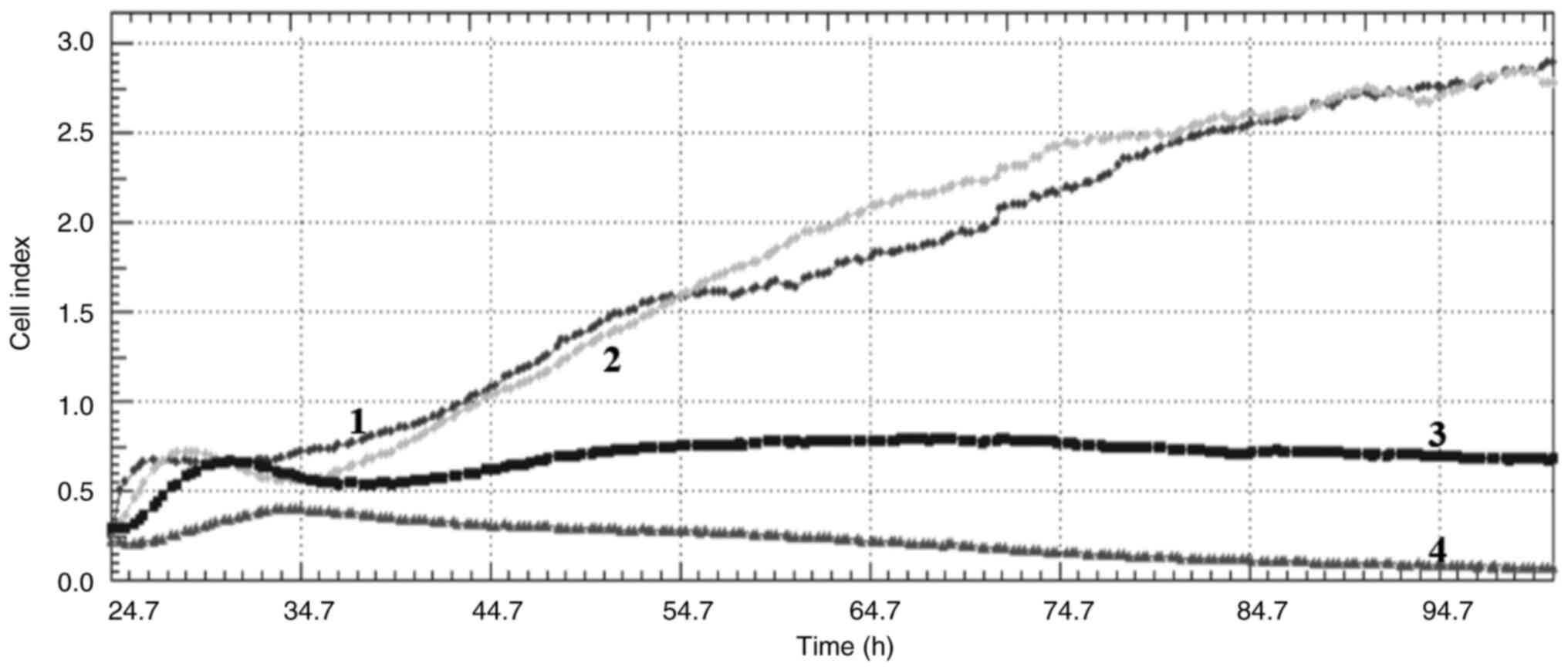
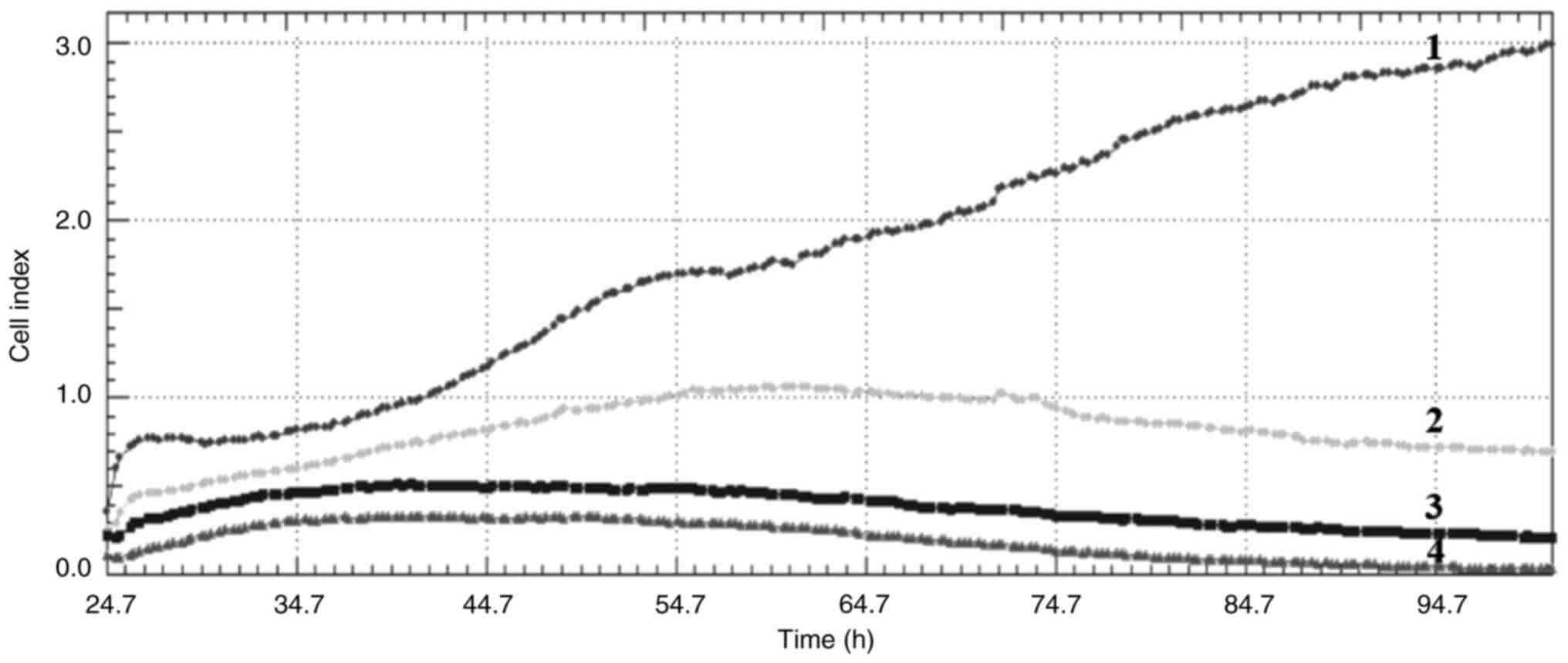
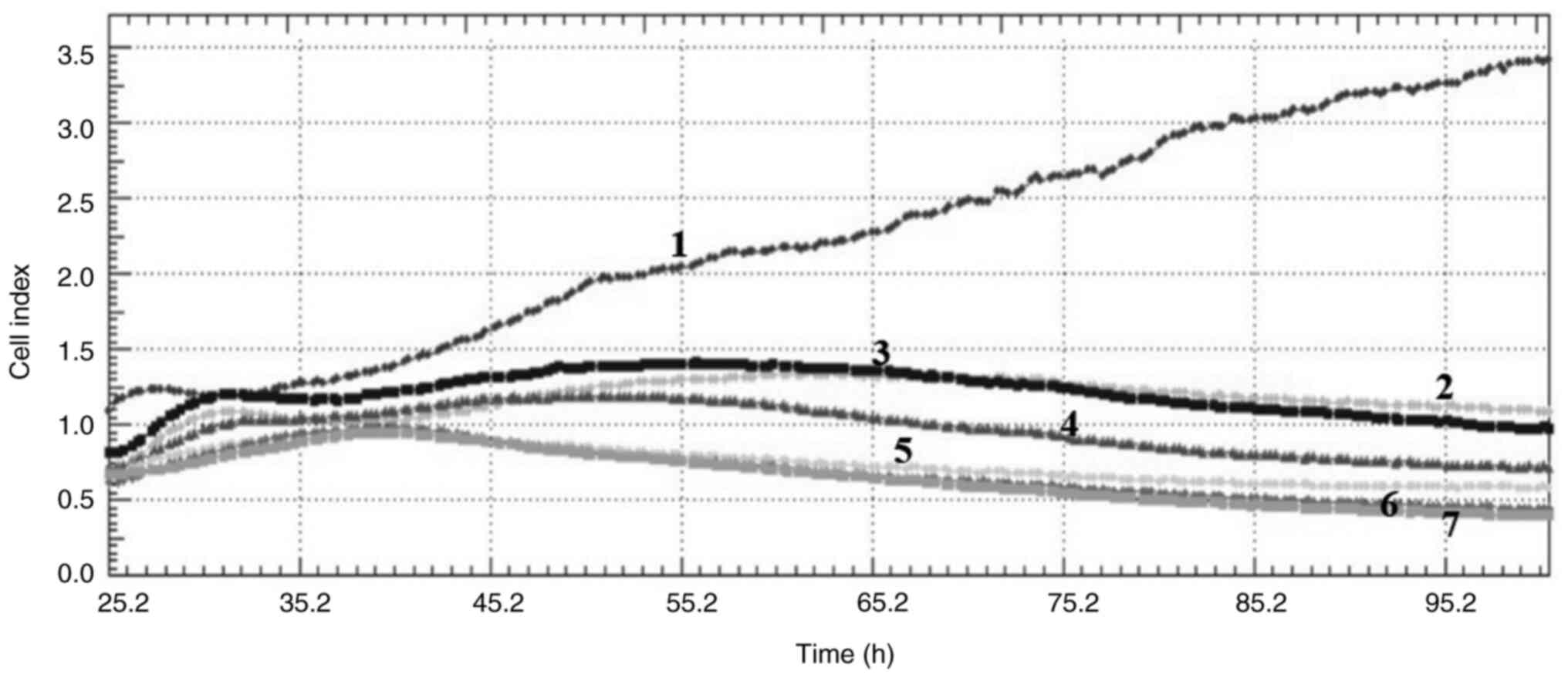
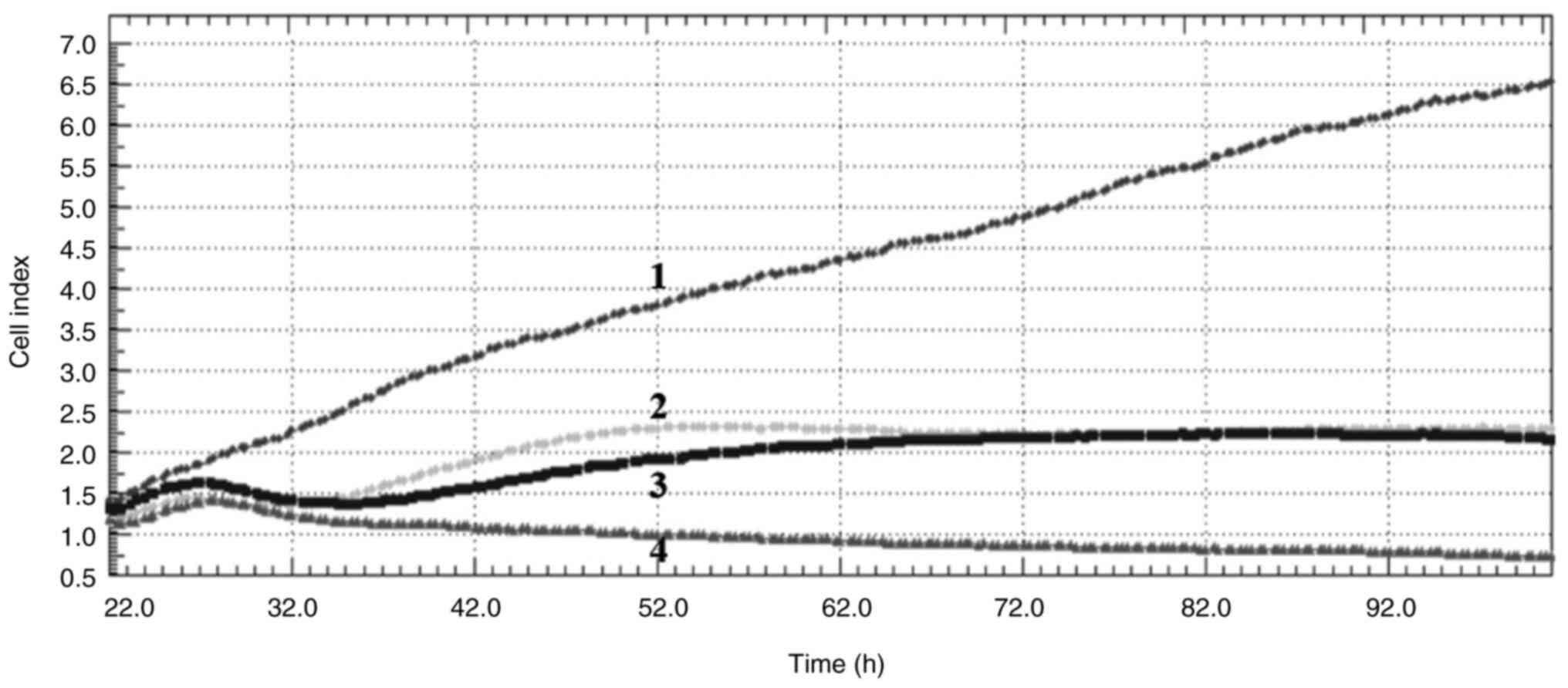
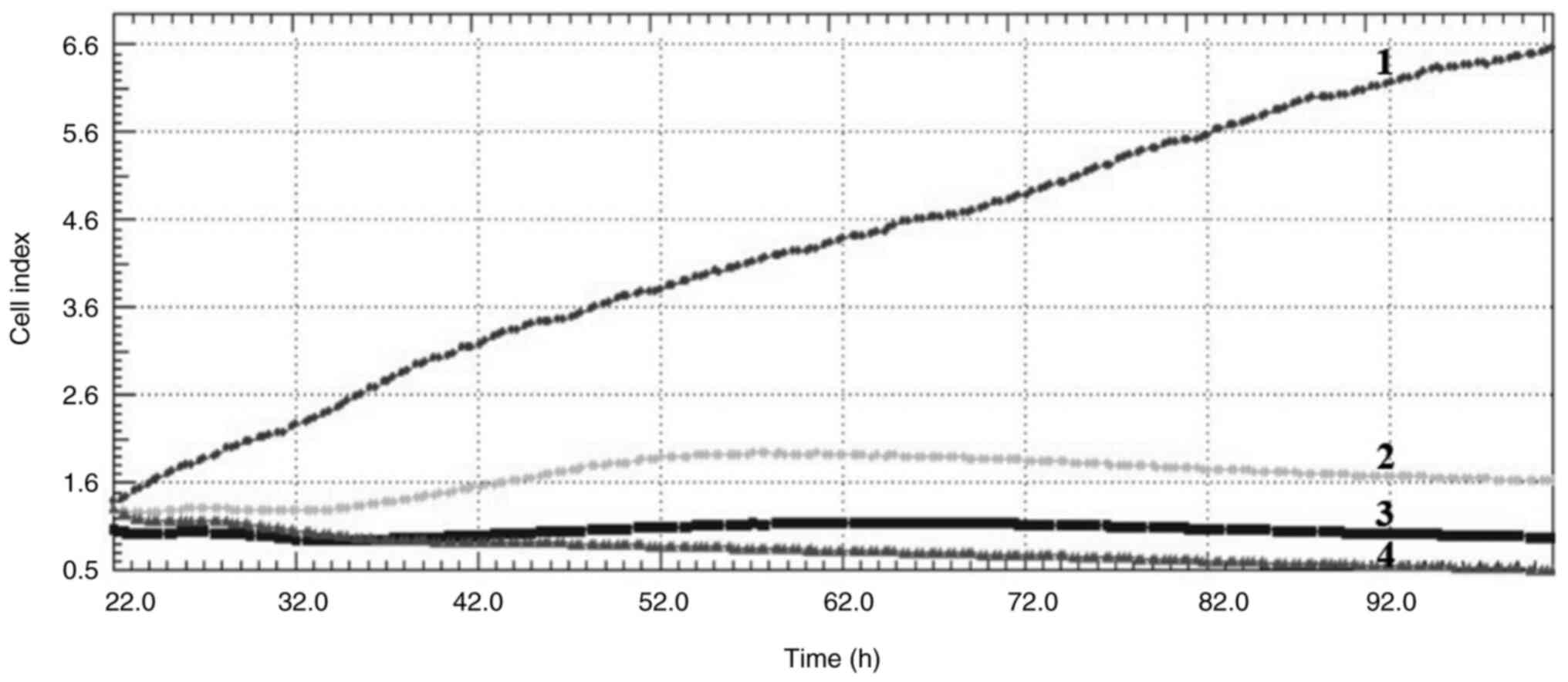
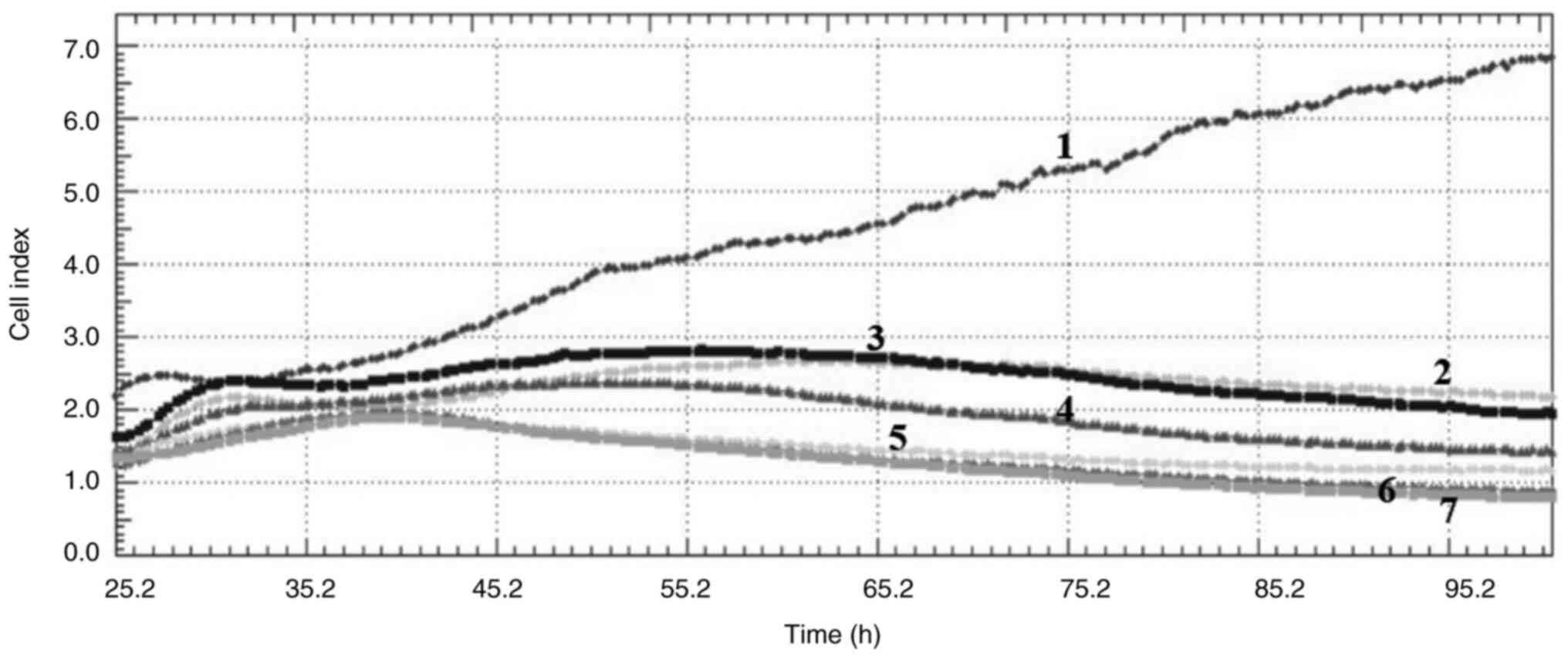
Cell index
Crizotinib and sodium butyrate were applied alone
and in combination at different concentrations on three different
cell lines: Crizotinib alone at concentrations of 5, 10 and 15 µM,
SB alone at concentrations of 300, 400 and 500 µM, and combined
concentrations of 10 µM crizotinib + 50 µM SB, 10 µM crizotinib
+100 µM SB, 10 µM crizotinib +200 µM SB, 400 µM SB + 2.5 µM
crizotinib, 400 µM SB + 5 µM crizotinib and 400 µM SB + 7.5 µM
crizotinib. The cell index was measured for each cell line. Both
single and combined applications of crizotinib and sodium butyrate
showed cytostatic effects in MCF-7 cell lines (Fig. 1, Fig.
2, Fig. 3, Fig. 4, Fig.
5, Fig. 6, Fig. 7, Fig.
8, Fig. 9).
Mitotic index
The mitotic index values obtained following the
application of crizotinib and SB alone and in combination to MCF-7,
MDA-MB-231 and SKBR-3 cells are presented in Table I. As a result of the mitotic index
application of critotinib and SB alone and in combination, mitotic
index values in all cell lines decreased significantly over time,
in comparison with the control (P<0.05). Moreover, the drug
combinations were more successful in reducing the mitotic index
values than single applications and were associated with a greater
decline in cell proliferation.
 | Table I.Mitotic index values of different
breast cancer cells. |
Table I.
Mitotic index values of different
breast cancer cells.
| A, MCF-7 cells |
|---|
|
|---|
| Time, h | Control | 10 µm Cri | 400 µm SB | 10 µm Cri + 50 µm
SB | 400 µm SB + 2.5 µm
Cri |
|---|
| 24 | 158±3 | 102±4a | 112±6a | 54±2a | 59±2a |
| 48 | 213±6 | 94±5a | 105±2a | 48±1a | 46±3a |
| 72 | 257±7 | 86±1a | 96±2a | 39±2a | 37±4a |
|
| B, MDA-MB-231
cells |
|
| Time, h | Control | 10 µm
Cri | 400 µm
SB | 10 µm Cri + 50
µm SB | 400 µm SB + 2.5
µm Cri |
|
| 24 | 121±3 | 73±2a | 68±1a | 45±2a | 41±2a |
| 48 | 134±5 | 64±4a | 54±4a | 38±3a | 33±3a |
| 72 | 149±4 | 59±6a | 47±2a | 35±4a | 28±3a |
|
| C, SKBR-3
cells |
|
| Time, h | Control | 10 µm
Cri | 400 µm
SB | 10 µm Cri + 50
µm SB | 400 µm SB + 2.5
µm Cri |
|
| 24 | 133±4 | 69±2a | 65±3a | 47±2a | 45±1a |
| 48 | 142±7 | 57±1a | 59±2a | 40±3a | 41±1a |
| 72 | 153±6 | 55±2a | 54±3a | 34±2a | 33±2a |
BrdU cell proliferation test
(labelling index)
The labeling index values obtained following the
application of crizotinib and SB alone and in combination to MCF-7,
MDA-MB-231 and SKBR-3 cells are presented in Table II. As a result of the BrdU cell
proliferation application of critotinib and SB alone and in
combination, labeling index values in all cell lines decreased
significantly over time, in comparison with the control
(P<0.05). Moreover, the drug combinations were more successful
in reducing the labeling index values compared with that of the
single applications and were associated with blockages in the
synthesis phase of the cells.
 | Table II.Bromodeoxyuridine labelling index
values of different breast cancer cells. |
Table II.
Bromodeoxyuridine labelling index
values of different breast cancer cells.
| A, MCF-7 cells |
|---|
|
|---|
| Time, h | Control | 10 µm Cri | 400 µm SB | 10 µm Cri + 50 µm
SB | 400 µm SB + 2.5 µm
Cri |
|---|
| 24 | 621±10 | 453±6a | 401±4a | 234±3a | 251±5a |
| 48 | 652±6 | 441±5a | 396±2a | 225±4a | 234±3a |
| 72 | 658±9 | 387±7a | 383±8a | 198±2a | 201±4a |
|
| B, MDA-MB-231
cells |
|
| Time, h | Control | 10 µm
Cri | 400 µm
SB | 10 µm Cri + 50
µm SB | 400 µm SB + 2.5
µm Cri |
|
| 24 | 404±7 | 296±4a | 288±3a | 199±2a | 195±4a |
| 48 | 416±8 | 287±5a | 224±4a | 145±3a | 151±3a |
| 72 | 438±6 | 265±7a | 178±5a | 121±5a | 112±6a |
|
| C, SKBR-3
cells |
|
| Time, h | Control | 10 µm
Cri | 400 µm
SB | 10 µm Cri + 50
µm SB | 400 µm SB + 2.5
µm Cri |
|
| 24 | 514±12 | 316±7a | 321±6a | 251±11a | 235±7a |
| 48 | 523±15 | 304±9a | 301±8a | 226±7a | 197±6a |
| 72 | 543±13 | 287±5a | 244±10a | 184±8a | 152±5a |
Caspase activity
The caspase activity values obtained following the
application of crizotinib and SB alone and in combination to MCF-7,
MDA-MB-231 and SKBR-3 cells are presented in Table III. As a result of the caspase
activity application of critotinib and SB alone and in combination,
caspase activity values in all cell lines increased significantly
over time, compared with the control (P<0.05). Furthermore, the
drug combinations were associated with a greater increase in
caspase activity compared with that of the single applications, and
this caused the cells to undergo apoptosis.
 | Table III.Caspase activity values of MCF-7
cells. |
Table III.
Caspase activity values of MCF-7
cells.
| A, MCF-7 cells |
|---|
|
|---|
| Time, h | Control | 10 µm Cri | 400 µm SB | 10 µm Cri + 50 µm
SB | 400 µm SB + 2.5 µm
Cri |
|---|
| 24 | 164±11 | 278±23a | 254±16a | 356±19a | 364±22a |
| 48 | 165±9 | 302±21a | 289±22a | 368±24a | 379±25a |
| 72 | 167±12 | 325±18a | 314±14a | 398±22a | 387±29a |
|
| B, MDA-MB-231
cells |
|
| Time, h | Control | 10 µm
Cri | 400 µm
SB | 10 µm Cri + 50
µm SB | 400 µm SB + 2.5
µm Cri |
|
| 24 | 216±14 | 321±13a | 343±23a | 514±38a | 523±33a |
| 48 | 214±11 | 335±16a | 544±26a | 534±41a | 544±29a |
| 72 | 215±13 | 358±12 | 365±22a | 575±37a | 565±31a |
|
| C, SKBR-3
cells |
|
| Time, h | Control | 10 µm
Cri | 400 µm
SB | 10 µm Cri + 50
µm SB | 400 µm SB + 2.5
µm Cri |
|
| 24 | 198±11 | 325±22a | 351±20a | 543±29a | 554±39a |
| 48 | 198±14 | 367±19a | 403±19a | 547±36a | 568±42a |
| 72 | 200±17 | 374±24a | 433±18a | 575±44a | 579±26a |
Discussion
In the present study, the effectiveness of
crizotinib as a monotherapy and when used with SB, which acts as a
HDACi, was evaluated in different types of breast cancer cell
lines.
Although there are different treatment approaches in
cancer treatment, a single definitive treatment method cannot be
focused on as each method has its own advantages and disadvantages,
cancer is a disease specific to each individual, and treatments may
differ from person to person (27).
Furthermore, combination treatments, which have many advantages
over monotherapy applications, are important in increasing
treatment response and reducing drug resistance (28). The combination therapy approach
allows the use of lower doses than monotherapy applications.
Therefore, it increases the effectiveness of treatment and reduces
the toxicity caused by high doses (29).
RTKs are associated with the development and
progression of most types of cancer in humans (30). Therefore, targeting RTKs has become
a therapeutic target for the treatment of cancer (30–32).
Among RTKs, MET serves an important role during tumor development
and progression (33,34). High levels of MET in breast cancer
have been associated with a high tumor grade and poor prognosis
(35–37). Results from clinical studies have
associated MET overexpression with increased recurrence rates and
decreased survival in patients with breast cancer (38,39).
Furthermore, tissue microarray analyses and expression data have
reported associations of MET overexpression with basal and
ER/HER2-negative forms of breast cancer in humans (32,35,38–40).
Animal experiments have also reported that MET activation induces
mammary tumors with different characteristics (38,39).
The therapeutic use of anti-MET drugs has attracted
interest due to the possible role of the hepatocyte growth
factor/MET axis in the development of breast carcinomas and other
malignancies (31,41). Studies have reported the development
and assessment of small molecule MET tyrosine kinase inhibitors as
a monotherapy or in combination with other targeted medicines
(42). Anaplastic lymphoma kinase,
receptor tyrosine kinase C-Ros oncogene 1 and MET are all targets
of the multitarget RTK inhibitor crizotinib (43,44).
MET activation promotes cancer cell spread,
migration and invasion (45). The
findings from the present study with MDA-MB-231 cells demonstrated
crizotinib administration dose-dependently inhibited the
proliferation of these cells. Moreover, decreased MET activation
may explain the anti-proliferative, anti-migratory and
anti-invasive effects of crizotinib in triple-negative breast
cancer (46).
Furthermore, it has been reported that MET
overexpression is associated with decreased treatment sensitivity
in different cancer types (47–50).
Therefore, it may be beneficial to use crizotinib together with
different therapeutic targets to increase treatment sensitivity
(46).
SB is a short-chain fatty acid and a byproduct of
carbohydrate metabolism in the intestine and. It is considered a
potent HDACi that has emerged as an anticancer agent for certain
cancers (51). Studies with SB have
reported that it is associated with a reduction in the rate of
breast cancer cells depending on dose and time (52,53).
The inhibitory effect of SB on hormone-dependent and independent
cell lines, and its ability to induce apoptosis through cell cycle
disruption in hormone-dependent cell lines, suggests that it may
have important implications for the treatment of human tumors,
including breast cancer (53).
Stockhammera et al (54)
reported that HDAC inhibition sensitized highly resistant PF240-PE
tumor cells to crizotinib and may overcome treatment resistance.
Moreover, Fukuda et al (55)
reported that the HDACi quinostat induced mesenchymal-epithelial
transition by reducing zinc finger E-Box binding homeobox 1
expression in clones in vitro and therefore, restoring
sensitivity to crizotinib.
The results of the present study, in which
crizotinib was used together with a HDACi, are important in terms
of creating more effective treatment strategies in different
molecular subtypes of breast cancer when crizotinib is used with
different therapeutic targets. The results demonstrated that
combined applications, in which the concentrations of both
crizotinib and SB were reduced, resulted in more effective results
in different cell kinetic parameters than monotherapy applications.
This is important in terms of reducing the side effects of the
drugs used and reducing treatment resistance. Furthermore, when
evaluated clinically, the results of the present study suggest that
the MET inhibitor crizotinib may be more effective in breast
cancers with different molecular subtypes when used together with
SB. However, the absence of healthy breast cells in the present
study makes it difficult to estimate clinical efficacy, as it could
not be observed at which dose ranges the combined applications
would cause toxic effects.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Scientific Research
Projects Coordination Unit of Istanbul University (grant no.
FBA-2022-38767).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
MT, İÇ, EP and MÇ performed the experiments. MT, İÇ,
EP and MÇ wrote and edited the manuscript. All authors confirm the
authenticity of all the raw data. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arya GC, Kaur K and Jaitak V: Isoxazole
derivatives as anticancer agent: A review on synthetic strategies,
mechanism of action and SAR studies. Eur J Med Chem.
221:1135112021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yue W, Yager JD, Wang JP, Jupe ER and
Santen RJ: Estrogen receptor-dependent and independent mechanisms
of breast cancer carcinogenesis. Steroids. 78:161–170. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
İnce E and Orhan HG: Estrogen-Induced
Breast Cancer, Therapeutical Approaches and the Role of Melatonin
in Treatment. HUJPHARM. 39:113–128. 2019.(In Türkiye).
|
|
5
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Herschkowitz JI, Simin K, Weigman VJ,
Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S,
Chandrasekharan S, et al: Identification of conserved gene
expression features between murine mammary carcinoma models and
human breast tumors. Genome Biol. 8:R762007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang E, Cheng SH, Dressman H, Pittman J,
Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM, et al: Gene
expression predictors of breast cancer outcomes. Lancet.
361:1590–1596. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sørlie T, Tibshirani R, Parker J, Hastie
T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et
al: Repeated observation of breast tumor subtypes in independent
gene expression data sets. Proc Natl Acad Sci USA. 100:8418–8423.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peng JH, Zhang X, Song JL, Ran L, Luo R,
Li HY and Wang YH: Neoadjuvant chemotherapy reduces the expression
rates of ER, PR, HER2, Ki67 and P53 of invasive ductal carcinoma.
Medicine (Baltimore). 98:e135542019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Barnes DM and Hanby AM: Oestrogen and
progesterone receptors in breast cancer: past, present and future.
Histopathology. 38:271–274. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Akoz G, Diniz G, Ekmekci S, Ekin ZY and
Uncel M: Evaluation of human epididymal secretory protein 4
expression according to the molecular subtypes (luminal A, luminal
B, human epidermal growth factor receptor 2-positive,
triple-negative) of breast cancer. Indian J Pathol Microbiol.
61:323–329. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Orrantia-Borunda E, Anchondo-Nuñez P,
Acuña-Aguilar LE, Gómez-Valles FO, Ramírez-Valdespino CA and
Mayrovitz HN: Subtypes of Breast Cancer. Exon Publications 31–42;
2022, View Article : Google Scholar
|
|
13
|
Barker S: Anti-estrogens in the treatment
of breast cancer: current status and future directions. Curr Opin
Investig Drugs. 4:652–657. 2003.PubMed/NCBI
|
|
14
|
Wong ZW and Ellis MJ: First-line endocrine
treatment of breast cancer: Aromatase inhibitor or antioestrogen?
Br J Cancer. 90:20–25. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kumar P and Aggarwal R: An overview of
triple-negative breast cancer. Arch Gynecol Obstet. 293:247–269.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hong S, Fang W, Liang W, Yan Y, Zhou T,
Qin T, Wu X, Ma Y, Zhao Y, Yang Y, et al: Risk of treatment-related
deaths with vascular endothelial growth factor receptor tyrosine
kinase inhibitors: A meta-analysis of 41 randomized controlled
trials. Onco Targets Ther. 7:1851–1867. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gurkan-Alp SA and Bozca F: Tirozin kinaz
enzim inhibitörü yeni bileşikler ve yapı aktivite ilişkilerinin
değerlendirilmesi. FABAD J Pharm Sci. 44:65–78. 2019.
|
|
18
|
De Bono JS and Yap TA: c-MET: An exciting
new target for anticancer therapy. Ther Adv Med Oncol. 3 (Suppl.
S1):S3–S5. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gastaldi S, Comoglio PM and Trusolino L:
The Met oncogene and basal-like breast cancer: Another culprit to
watch out for? Breast Cancer Res. 12:2082010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ho-Yen CM, Jones JL and Kermorgant S: The
clinical and functional significance of c-Met in breast cancer: A
review. Breast Cancer Res. 17:522015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mino-Kenudson M, Chirieac LR, Law K,
Hornick JL, Lindeman N, Mark EJ, Cohen DW, Johnson BE, Jänne PA,
Iafrate AJ and Rodig SJ: A novel highly sensitive antibody allows
for the routine detection of ALK rearranged lung adenocarcinomas by
standard immunohistochemistry. Clin Cancer Res. 16:1561–1571. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hagland HR and Søreide K: Cellular
metabolism in colorectal carcinogenesis: Influence of lifestyle,
gut microbiome and metabolic pathways. Cancer Lett. 356:273–280.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hamer HM, Jonkers D, Venema K, Vanhoutvin
S, Troost FJ and Brummer RJ: Review article: The role of butyrate
on colonic function. Aliment Pharmacol Ther. 27:104–119. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Taylor MA, Khathayer F and Ray SK:
Quercetin and sodium butyrate synergistically increase apoptosis in
rat C6 and Human T98G glioblastoma cells through inhibition of
autophagy. Neurochem Res. 44:1715–1725. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Egler V, Korur S, Failly M, Boulay JL,
Imber R, Lino MM and Merlo A: Histone deacetylase inhibition and
blockade of the glycolytic pathway synergistically induce
glioblastoma cell death. Clin Cancer Res. 14:3132–3140. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pulat E and Topçul MR: Effects of combined
use of ribociclib with PARP1 inhibitor on cell kinetics in breast
cancer. Oncol Lett. 27:2432024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Baykara O: Kanser Tedavisinde Güncel
YaklaşImlar. Balıkesir Sağlık Bilimleri Dergisi. 5:154–165.
2016.(In Türkiye).
|
|
28
|
Palmer AC and Sorger PK: Combination
cancer therapy can confer benefit via patient-to-patient
variability without drug additivity or synergy. Cell.
171:1678–1691.e13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee JH and Nan A: Combination drug
delivery approaches in metastatic breast cancer. J Drug Deliv.
2012:9153752012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Takeuchi K and Ito F: Receptor tyrosine
kinases and targeted cancer therapeutics. Biol Pharm Bull.
34:1774–1780. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu K, Kong X, Zhao D, Liang Z and Luo C:
c-MET kinase inhibitors: A patent review (2011–2013). Expert Opin
Ther Pat. 24:217–230. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Linklater ES, Tovar EA, Essenburg CJ,
Turner L, Madaj Z, Winn ME, Melnik MK, Korkaya H, Maroun CR,
Christensen JG, et al: Targeting MET and EGFR crosstalk signaling
in triple-negative breast cancers. Oncotarget. 7:69903–69915. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Blumenschein GR Jr, Mills GB and
Gonzalez-Angulo AM: Targeting the hepatocyte growth factor-cMET
axis in cancer therapy. J Clin Oncol. 30:3287–3296. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Boccaccio C and Comoglio PM: MET, a driver
of invasive growth and cancer clonal evolution under therapeutic
pressure. Curr Opin Cell Biol. 31:98–105. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gonzalez-Angulo AM, Chen H, Karuturi MS,
Chavez-MacGregor M, Tsavachidis S, Meric-Bernstam F, Do KA,
Hortobagyi GN, Thompson PA, Mills GB, et al: Frequency of
mesenchymal-epithelial transition factor gene (MET) and the
catalytic subunit of phosphoinositide-3-kinase (PIK3CA) copy number
elevation and correlation with outcome in patients with early stage
breast cancer. Cancer. 119:7–15. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Inanc M, Ozkan M, Karaca H, Berk V,
Bozkurt O, Duran AO, Ozaslan E, Akgun H, Tekelioglu F and Elmali F:
Cytokeratin 5/6, c-Met expressions, and PTEN loss prognostic
indicators in triple-negative breast cancer. Med Oncol. 31:8012014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yan S, Jiao X, Zou H and Li K: Prognostic
significance of c-Met in breast cancer: A metaanalysis of 6010
cases. Diagn Pathol. 10:622015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ponzo MG, Lesurf R, Petkiewicz S, O'Malley
FP, Pinnaduwage D, Andrulis IL, Bull SB, Chughtai N, Zuo D,
Souleimanova M, et al: Met induces mammary tumors with diverse
histologies and is associated with poor outcome and human basal
breast cancer. Proc Natl Acad Sci USA. 106:12903–12908. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Graveel CR, DeGroot JD, Su Y, Koeman J,
Dykema K, Leung S, Snider J, Davies SR, Swiatek PJ, Cottingham S,
et al: Met induces diverse mammary carcinomas in mice and is
associated with human basal breast cancer. Proc Natl Acad Sci USA.
106:12909–12914. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sierra JR and Tsao MS: c-MET as a
potential therapeutic target and biomarker in cancer. Ther Adv Med
Oncol 3 (1 suppl). S21–S35. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang X, Li E, Shen E, Wang X, Tang T,
Zhang X, Xu J, Tang Z, Guo C, Bai X and Liang T: Targeting the
HGF/MET axis in cancer therapy: Challenges in resistance and
opportunities for improvement. Front Cell Dev Biol. 8:1522020.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Goździk-Spychalska J, Szyszka-Barth K,
Spychalski Ł, Ramlau K, Wójtowicz J, Batura-Gabryel H and Ramlau R:
C-MET inhibitors in the treatment of lung cancer. Curr Treat
Options Oncol. 15:670–682. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kwak EL, Bang YJ, Camidge DR, Shaw AT,
Solomon B, Maki RG, Ou SH, Dezube BJ, Jänne PA, Costa DB, et al:
Anaplastic lymphoma kinase inhibition in non-small-cell lung
cancer. N Engl J Med. 363:1693–1703. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cui JJ, Tran-Dubé M, Shen H, Nambu M, Kung
PP, Pairish M, Jia L, Meng J, Funk L, Botrous I, et al: Structure
based drug design of crizotinib (PF02341066), a potent and
selective dual inhibitor of mesenchymal-epithelial transition
factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med
Chem. 54:6342–6363. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lawrence RE and Salgia R: MET molecular
mechanisms and therapies in lung cancer. Cell Adh Migr. 4:146–152.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ayoub NM, Al-Shami KM, Alqudah MA and
Mhaidat NM: Crizotinib, a MeT inhibitor, inhibits growth,
migration, and invasion of breast cancer cells in vitro and
synergizes with chemotherapeutic agents. Onco Targets Ther.
10:4869–4883. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Du Y, Yamaguchi H, Wei Y, Hsu JL, Wang HL,
Hsu YH, Lin WC, Yu WH, Leonard PG, Lee GR IV, et al: Blocking
c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of
PARP inhibitors. Nat Med. 22:194–201. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Raghav KP, Gonzalez-Angulo AM and
Blumenschein GR Jr: Role of HGF/MET axis in resistance of lung
cancer to contemporary management. Transl Lung Cancer Res.
1:179–193. 2012.PubMed/NCBI
|
|
49
|
Wang J and Cheng JX: c-Met inhibition
enhances chemosensitivity of human ovarian cancer cells. Clin Exp
Pharmacol Physiol. 44:79–87. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li E, Hu Z, Sun Y, Zhou Q, Yang B, Zhang Z
and Cao W: Small molecule inhibitor of c-Met (PHA665752) suppresses
the growth of ovarian cancer cells and reverses cisplatin
resistance. Tumour Biol. 37:7843–7852. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Natoni F, Diolordi L, Santoni C and
Gilardini Montani MS: Sodium butyrate sensitises human pancreatic
cancer cells to both the intrinsic and the extrinsic apoptotic
pathways. Biochim Biophys Acta. 1745:318–329. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Salimi V, Shabani M, Nourbakhsh M and
Tavakoli-Yaraki M: Involvement of 15-lipoxygenase-1 in the
regulation of breast cancer cell death induced by sodium butyrate.
Cytotechnology. 68:2519–2528. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Coradini D, Biffi A, Costa A, Pellizzaro
C, Pirronello E and Di Fronzo G: Effect of sodium butyrate on human
breast cancer cell lines. Cell Prolif. 30:149–159. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Stockhammera P, Hoc CSL, Hegedusa L, Lotzd
G, Molnárd E, Bankfalvie A, Herold T, Kalbourtzis S, Ploenes T,
Eberhardt WEE, et al: HDAC inhibition synergizes with ALK
inhibitors to overcome resistance in a novel ALK mutated lung
adenocarcinoma model. Lung Cancer. 144:20–29. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fukuda K, Takeuchi S, Katayama R, Nanjo S,
Yamada T, Suzuki T, et al: HDAC Inhibition Overcomes
Crizotinib-Resistance by Mesenchymal-Epithelial Transition (MET) in
EML4-ALK Lung Cancer Cells. J Thorac Oncol. 12:S382–S383. 2017.
View Article : Google Scholar
|