Introduction
Oral mucosal cancer is a common subtype of head and
neck cancer, and >90% of oral cancer cases present
pathologically as oral squamous cell carcinoma (OSCC) (1). Recently, patients with OSCC,
particularly adolescents and young adults, have been increasing as
newly diagnosed with this cancer, at approximately 20,000 annually
in Japan, for instance (2). OSCC
generally exhibits as a red and/or white patched lesion sometimes
accompanied by ulceration and bleeding when irritated during
progression. These symptoms resemble oral stomatitis; thus,
detecting OSCC at an early stage is sometimes difficult. As a
result, those with OSCC tend to be diagnosed with locoregionally
advanced stages, which carry higher risks of local relapse and
metastasis, resulting in poor prognosis with a 5-year survival rate
of approximately 50% (3–5). Conversely, patients with OSCC at early
stage show favorable outcomes by conventional combined treatment
including surgery, chemotherapy, and radiotherapy with their 5-year
survival rate of >80% (6).
Therefore, detecting this notorious cancer at an early stage is
crucial.
MicroRNAs (miRNAs) are endogenous noncoding RNAs
with approximately 19–25 nucleotides in length (7). MiRNAs regulate the expression of
target mRNAs at the transcriptional and post-transcriptional levels
by binding to their 3′-untranslated regions (UTRs) of the mRNAs
(7). These processes result in
targeted mRNA degradation, and miRNAs function eventually as
regulators of biological processes including cell proliferation,
differentiation, and apoptosis induction (8). Intriguingly, the miRNAs are reported
to have oncogenic and tumor-suppressive functions (9,10).
Moreover, each cancer possesses a unique miRNA expression profile,
and miRNAs take on an oncogenic or tumor-suppressive role depending
on the cancer type (11). This
evidence has accelerated the use of miRNAs not only as a diagnostic
biomarker but also as a therapeutic and prognostic marker and even
as a therapeutic target (8,12). In collecting miRNAs, a previous
study reported that miRNAs are protected from degradation by being
capsulized with extracellular vesicles such as exosomes, and oral
swirl samples enabled isolation of adequate miRNAs (13). Moreover, a recent study demonstrated
that miRNAs isolated from oral swirl samples are potential
biomarkers for classifying the risk of OSCC development among
patients with oral potentially malignant disorders (OPMDs)
(14,15).
Herein, this case report aimed to demonstrate
further potential availability of salivary miRNAs as therapeutic
and prognostic markers of OSCC. To end this, this report presented
the case of a patient with advanced OSCC of the tongue and assessed
the patient's salivary miRNAs during each clinical course.
Case report
A 33-year-old Japanese woman was referred to our
department by her family dentist for an examination of her
right-sided tongue lesion. Her medical history included congenital
anomaly syndrome, presenting with cranial deformity, brachydactyly,
esotropia, and micrognathia. She did not smoke nor drink alcohol
regularly. She had noticed the lesion a few months before the first
visit to our department and recently noticed it had gotten larger,
resulting in difficulty speaking and eating. On extraoral
examination, no lymph nodes were swollen in her head and neck
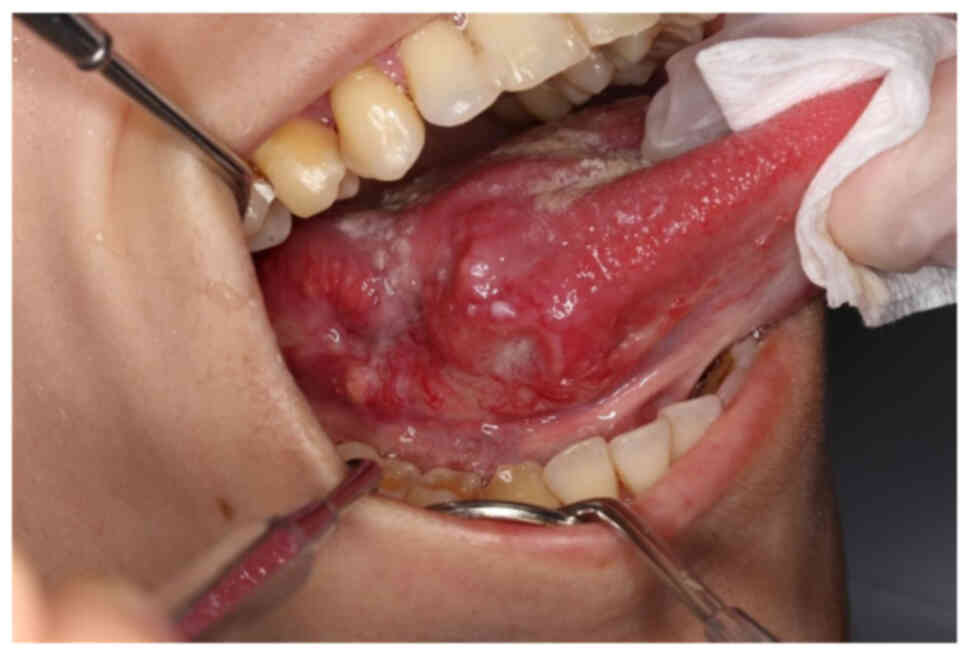
region. On intraoral examination, an indurative and hemorrhagic
mass measuring 20×30 mm accompanied by ulceration on the right side
of the tongue was identified (Fig.
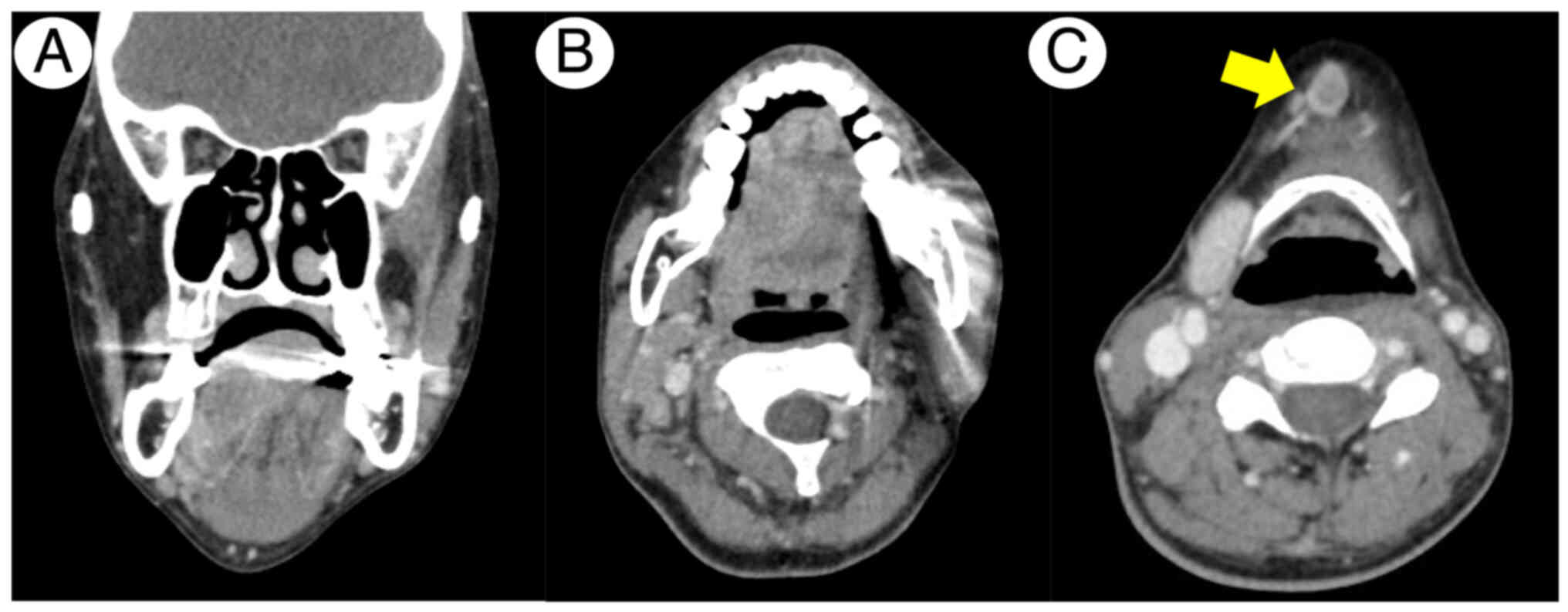
1). Contrast-enhanced computed tomography (CECT) identified the
lesion extending beyond the lingual septum presenting 24 mm depth
of invasion without any signs of bone invasion (Fig. 2A and B). CECT also detected signs of
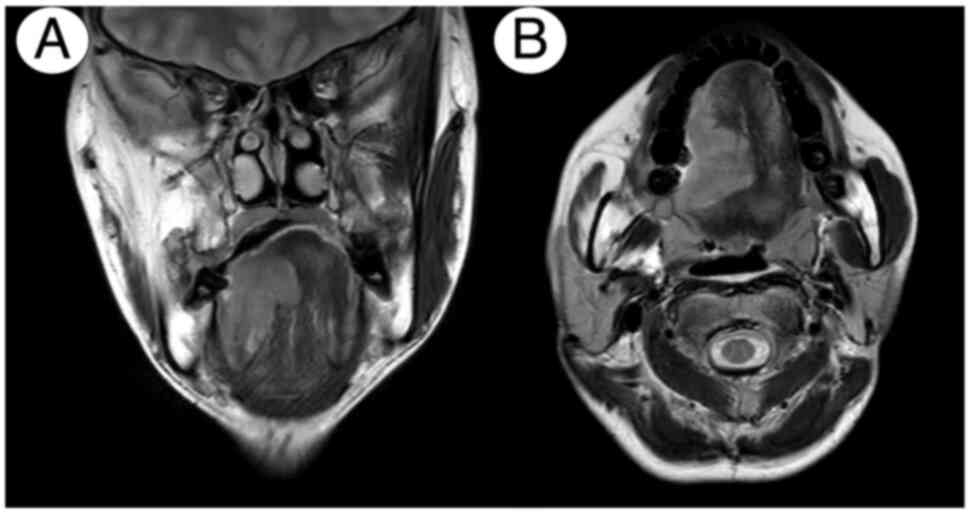
metastases in a submandibular lymph node (Fig. 2C). Furthermore, CE magnetic
resonance imaging identified the gadolinium-enhanced lesion on the
right side of the tongue (Fig. 3A and
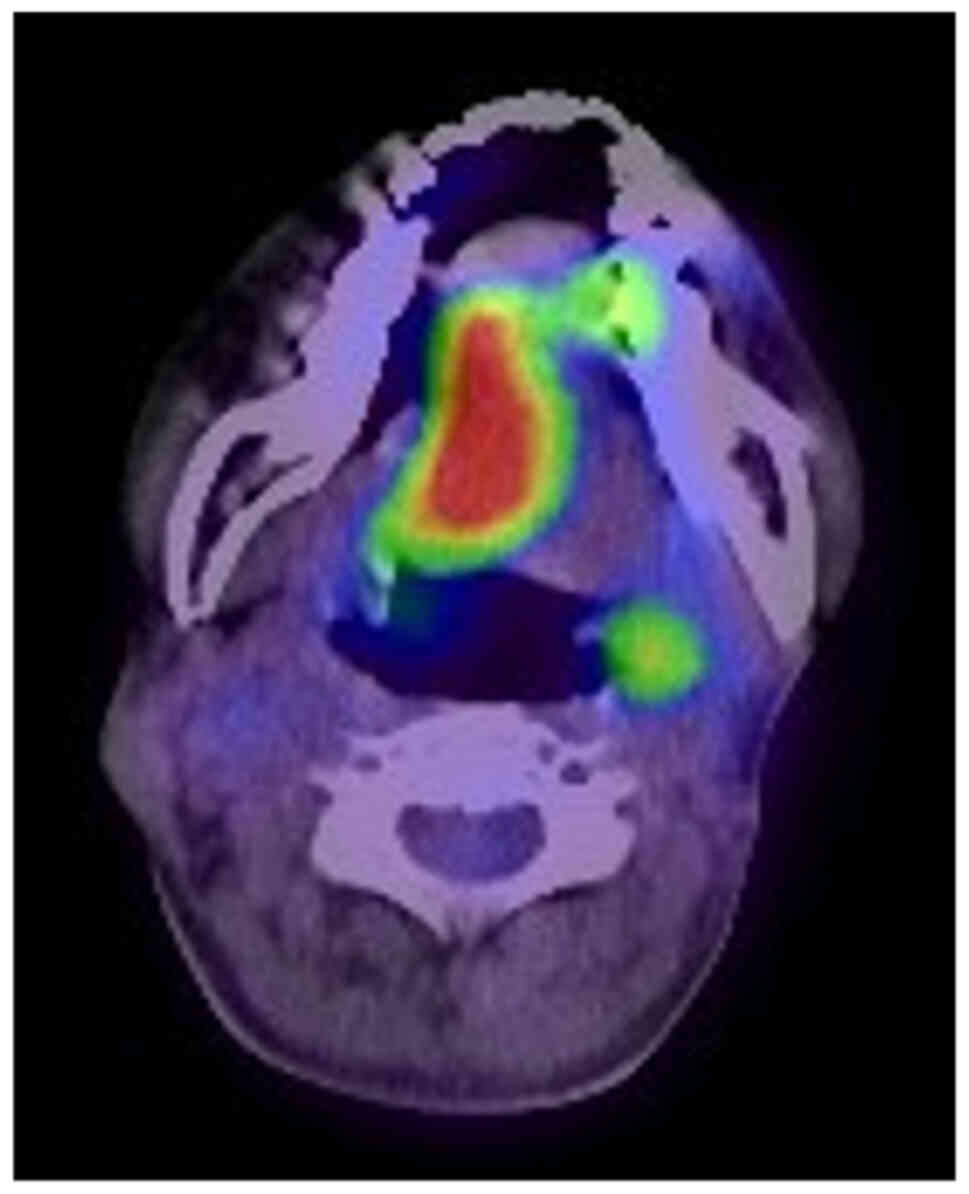
B). Positron emission tomography (PET)/CT found increased
uptake of 18fluoro-deoxyglucose in the right-sided
tongue, with standard uptake valuemax of 12.893 without
apparent increased uptake in bilateral cervical lymph nodes
(Fig. 4). An incision biopsy from
the right-sided tongue revealed neoplastic proliferation with
nuclear atypia and atypical mitoses, representing the findings of
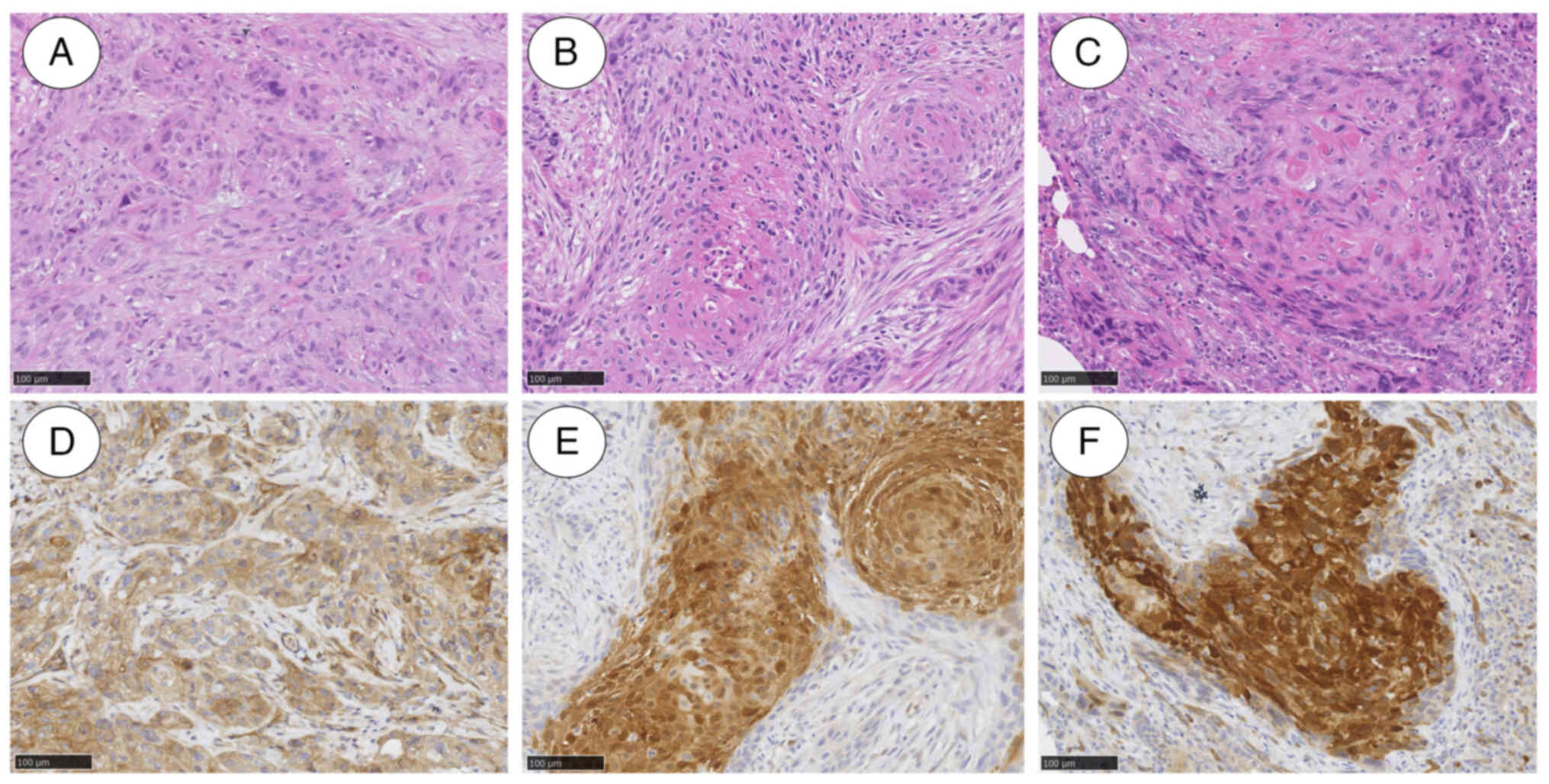
SCC (Fig. 5A). Finally, she was
diagnosed with right-sided tongue SCC with suspicion of
submandibular lymph node metastasis, cT3N1M0.
Subsequently, she received neoadjuvant chemotherapy
with tegafur-gimeracil-oteracil potassium 40 mg/day for 2 weeks,
showing a stable disease. Thereafter, she underwent subtotal
glossectomy, right-sided modified radical neck dissection type III
with levels I–V, and left-sided supraomohyoid neck dissection. She
further underwent reconstructive surgery with rectus abdominis
musculocutaneous flap. Pathological examination revealed SCC with
negative surgical margin and four positive nodes (total 4/51
including right-sided level 1B; 1/7 with extranodal extension
(ENE), left-sided level 1A; 1/6 with ENE and left-sided lateral
retropharyngeal lymph node 2/9) (Fig.
5B). This led to her diagnosis of right-sided tongue SCC with
lymph node metastasis, pT4aN3bM0. Subsequently, she received
concurrent chemoradiotherapy (CCRT) with 6-week cisplatin (50
mg/m2) and intensity-modulated radiation therapy (total
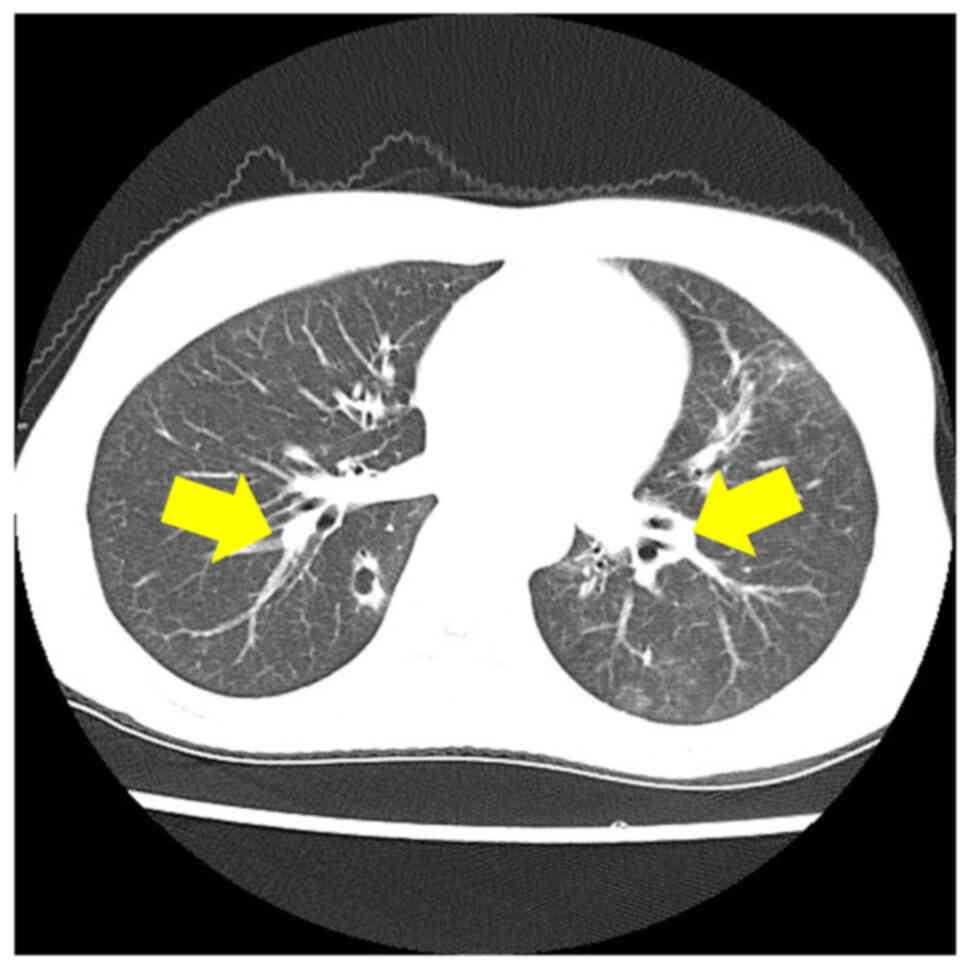
dose: 60 Gy in 30 fractions). Two months after her regimen, regular
CT identified aberrant cavitary nodules in both the right and left
lungs, suspicious of multiple lung metastases (Fig. 6). During further examinations for
the lung lesions (Fig. 5C), she
died because of multiple-organ failure 11 months after the initial
diagnosis.
For miRNA profiling, resting saliva samples were
obtained from the patient at her first visit (day 0), after
neoadjuvant chemotherapy (day 12), after surgery (day 57), after
postoperative CCRT completion (day 119), at the time of her
discharge from our hospital (day 148), and when lung metastasis was
detected (day 205) (Fig. 7A). Each
sample was collected in the morning before brushing her teeth or
rinsing her mouth. Thereafter, total RNA was extracted from the
exosome in each salivary sample using 3D-Gene® RNA
extraction reagent supplied with a liquid sample kit (Toray
Industries, Inc., Tokyo, Japan) (Fig.
7B). To perform miRNA microarray, the total RNA was first
labeled with a 3D-Gene® miRNA labeling kit (Toray
Industries, Inc., Tokyo, Japan) and hybridized with
3D-Gene® Human miRNA Oligo chips (Toray Industries
Inc.). This kit enables the evaluation of 2,632 miRNAs with the
microarray chip device. After washing the device, fluorescent
signals were scanned with 3D-Gene® Scanner (Toray
Industries, Inc.), and their intensity was analyzed using
3D-Gene® Extraction software (Toray Industries, Inc.).
Raw data were adjusted by the mean signal intensity of the
background, which was calculated based on the mean signal
intensities among whole blank spots with 95% confidence intervals.
The signal intensity of each spot was verified referring to the
signal intensity greater than two standard deviations of the
background signal intensity. In addition, global normalization was
applied to each verified signal intensity to adjust the median to
25. Furthermore, data were converted into log2 data by the 75th
percentile normalization so that the top 25% of its values (75th
percentile) become 1. Then, ‘log2 ratio’ was calculated by
subtracting each of the 75th percentile of the log-converted data
among each sample point. Moreover, the ‘ratio’ was calculated by
the antilog transformation referring to the log2 ratio. Finally,
this study compared the ‘ratio’ and defined the upregulation of
miRNA of interest as the ratio >2 and the downregulation as the
ratio <0.5. In this study, the miRNAs that meet following both
criteria were determined as candidate biomarkers for OSCC: miRNA
displaying 1) downregulation in the postoperative sample compared
with the sample obtained on patient's first visit, 2) upregulation
in the sample obtained when the lung metastasis was detected
compared with the postoperative sample. This study was approved by
Hokkaido University Hospital's independent clinical research review
committee (Approval No. 020-0085).
As a result, hsa-miR-6798-5p, miR-6803-5p,
miR-6805-5p, and miR-6845-5p were significantly upregulated at the
first visit to our department (Table
I; Fig. 8). Then, these miRNA
levels decreased after the neoadjuvant chemotherapy and surgery
(day 57) and increased gradually until lung metastasis was detected
(day 205). In addition, these four miRNAs were found to contain
specific sequences each of which are complementary to those in
3′UTR of kinase insert domain receptor (KDR) gene
encoding the vascular endothelial growth factor receptor-2,VEGFR2
(Data S1) (16). Hence, these four miRNAs have a
potential to directly recognize the 3′UTR, resulting in controlling
the expression level of VEGFR2. Furthermore, immunohistochemical
examination showed that VEGFR-2 stained positive in both tongue and
lung metastatic cells (Fig.
5D-F).
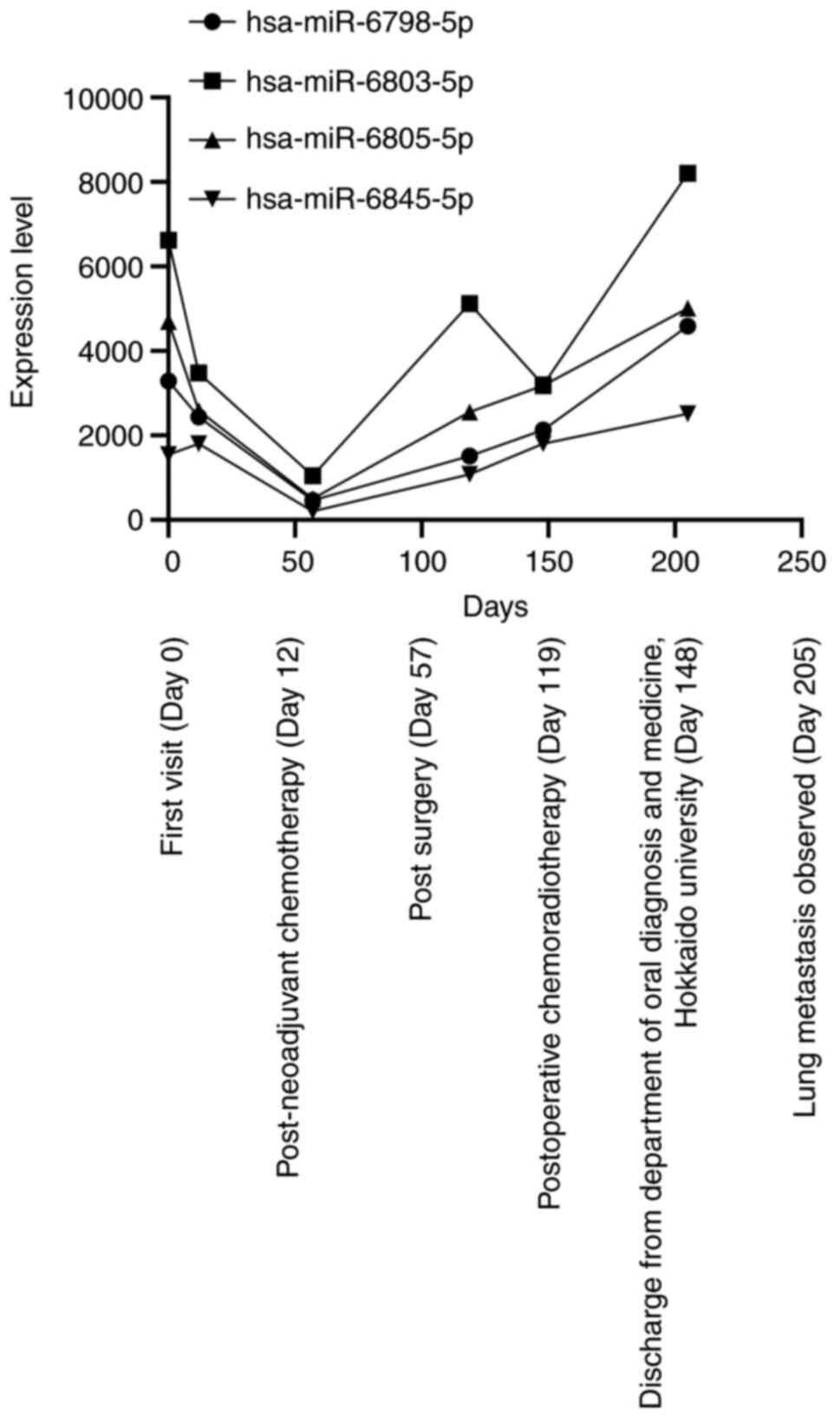 | Figure 8.Microarray results. Microarray
analysis of the saliva identified high levels of hsa-miR-6798-5p,
miR-6803-5p, miR-6805-5p and miR-6845-5p at the first visit to the
Department of Oral Diagnosis and Medicine, Hokkaido University
(Sapporo, Japan; day 0), and the neoadjuvant chemotherapy and
surgical procedure reduced the level of the four miRNAs (days 12
and 57). Thereafter, each level was increased when the patient
completed the postoperative concurrent chemoradiotherapy (day 119),
and the levels gradually increased until the lung metastasis was
detected (day 205). Among the four miRNAs, the level of
hsa-miR-6803-5p was decreased after the postoperative chemotherapy;
however, the level was markedly increased when the lung metastasis
was detected. miRNA/miR, microRNA. |
 | Table I.miRNA sequences. |
Table I.
miRNA sequences.
| miRNA | Sequence
(5′-3′) |
|---|
|
hsa-miR-6798-5p |
CCAGGGGGAUGGGCGAGCUUGGG |
|
has-miR-6803-5p |
CUGGGGGUGGGGGGCUGGGCGU |
|
has-miR-6805-5p |
UAGGGGGCGGCUUGUGGAGUGU |
|
has-miR-6845-5p |
CGGGGCCAGAGCAGAGAGC |
Discussion
To our knowledge, this is one of the few reports
describing the availability of salivary miRNAs as biomarkers for
monitoring the therapeutic effect and predicting the prognosis of a
patient with advanced OSCC. Herein, the comprehensive microarray
targeting miRNA revealed that the patient exhibited remarkable
changes in the level of four miRNAs, namely, hsa-miR-6798-5p,
miR-6803-5p, miR-6805-5p, and miR-6845-5p, isolated from the
salivary samples. In addition, the expression profile of VEGFR-2
regulated by the four miRNAs was observed in both tongue and lung
lesions. Moreover, this study revealed that the levels of the four
miRNAs dramatically changed in accordance with the clinical course
of the patients.
Currently, cancer biomarkers for diagnosis,
evaluating the therapeutic effect and predicting patients'
prognosis, generally include DNAs, RNAs, and proteins such as
metabolites (17). As for OSCC,
serum SCC antibody has been employed as a diagnostic and
therapeutic biomarker; however, it requires invasive procedure and
still has low specificity (18). In
addition, collecting sufficient circulating tumor cells that are
rarely detected at the early stage and mainly detected at the
advanced stages is still challenging (19,20).
Recently, miRNAs have been gaining attention as biomarkers given
their reliable stability. miRNAs are released by normal and tumor
cells as extracellular vesicles such as exosomes, which protect the
miRNAs from degradation by RNase (19). So far, several miRNAs have been
identified as potential candidates for OSCC biomarkers including
miR-137 and miR-29a/b/c in the tumor for diagnostic marker
(21), miR-1275 upregulation and
miR-222-3p and miR-423-5p downregulation in the plasma for regional
lymph node invasion (22), miR-196a
and miR-21 upregulation in the tumor for the radiotherapy-resistant
marker (23), all of which require
further assessment to determine their sensitivity and specificity
in a large cohort. However, these markers require invasive
procedure, hampering further investigations in a large cohort.
Conversely, salivary miRNA offers a noninvasive and straightforward
procedure. Indeed, previous papers demonstrated the availability of
salivary miRNA to employ as a biomarker for classifying the risk of
OSCC development among the patients with OPMDs (14,15).
Among the miRNAs mentioned above, our patient also displayed the
significant downregulation of miR-423-5p and miR-24-3p after the
surgery compared with those obtained from the sample at the
patient's first visit. Although these miRNAs did not meet our
criteria, they also have a possibility to contribute to OSCC
progression. Other than the two miRNAs, our report did not find
remarkable changes of miRNA mentioned above during the clinical
course. The discrepancy between previous reports and our results
should be due to differences in race, ethnicity, sample collection
method and sample quality. Previously, it has been demonstrated
that the miRNA profile obtained from serum sample varies depending
on the host factors (24). Hence,
it should be necessary to evaluate the miRNA candidate considering
the patients' background.
In our report, miR-6798-5p, miR-6803-5p,
miR-6805-5p, and miR-6845-5p levels were remarkably changed; that
is, their levels decreased after the neoadjuvant chemotherapy and
surgery compared with those at the first visit, and the levels
increased when lung metastasis was detected compared with observed
after the surgery (Fig. 8). These
results support the hypothesis that the four miRNAs were derived
from the tumor cells themselves. Hence, these miRNAs have a
potential as therapeutic targets of OSCC. Owing to the ease with
miRNA inhibitor development and its usability that miRNA inhibitor
can affect simultaneously on the multiple genes that are controlled
by the targeted miRNA, miRNA-based therapy has been highlighted in
treating various diseases (25). To
date, several miRNA-based therapies are now under clinical trials
for the treatment of various types of solid tumors however, there
are no trials among the patients with OSCC (25,26).
Of note, a recent paper proposed miR-31-5p as a therapeutic target
for OSCC treatment. In the paper, the authors identified that the
serum miR-31-5p, which was not determined as therapeutic target
candidate in our report, was significantly upregulated among the
patients with OSCC (27). The
authors also demonstrated that introducing miR-31-5p into normal
epithelial cells, HaCaT cells, resulted in accelerating
proliferation and a miR-31-5p inhibitor showed remarkable antitumor
effect on both OSCC cells in vitro and OSCC patient-derived
xenograft mouse models (27).
Accordingly, the four miRNAs presented in this report are also
worth consideration as the therapeutic target. To prove this,
further experiments including investigation of the miRNA function
in OSCC cell lines by introducing the overexpression of the miRNA
candidate and by administrating the anti-miRNA oligonucleotide to
inhibit the miRNA function are required. Further, validating the
antitumor effect in mammalian OSCC models by administrating miRNA
inhibitors targeting the miRNA candidates should be necessary to
support the in vitro experiment. Lastly, it is crucial to
perform OSCC tissue-based analysis to confirm the expression level
of the miRNA candidate in the specimen, proposing the candidates as
the potent therapeutic target.
Moreover, this report proposed the possibility to
employ the four miRNAs as a predicting biomarker for distant
metastasis of OSCC. This is because the expression level of the
four miRNAs were remarkably upregulated when the lung metastasis
was observed (Fig. 8). However,
further study should be required to conclude the usability of the
biomarker by confirming how the miRNAs contribute to OSCC
tumorigenesis and by evaluating the availability among a large
number of patients with OSCC. Thus far, the four miRNAs have
already been reported to have a potential as a biomarker in various
types of diseases. As for miR-6805-5p, a previous study reported
that miR-6805-5p extracted from urine sample is a potential
biomarker for predicting cisplatin-induced nephrotoxicity among
patients with head and neck cancer (28). In this study, the authors found that
the miR-6805-5p contributes to renal repair and regeneration
through the appropriate regulation of the Wnt pathway. In addition,
they proposed that miR-6805-5p downregulation may result in the
aberrant activation of this pathway, causing kidney fibrosis and
damage (28). Moreover, miR-6798
and miR-6845-5p are reported to play roles in hepatocellular
tumorigenesis by regulating macrophage polarization in the tumor
microenvironment and activating autophagy, leading to
chemoresistance to 5-fluorouracil (FU), oxaliplatin, and
pirarubicin (29,30). Besides that, miR-6803-5p level was
upregulated in the serum sample extracted from patients with
colorectal cancer, and the higher level of miR-6803-5p correlated
with poor prognosis, proposing miR-6803-5p as a potential
diagnostic and prognostic marker (31). In addition, miR-6803-5p is reported
to enhance tumor proliferation and provokes inflammation by
regulating the protein tyrosine phosphatase receptor type O/nuclear
factor-κB axis among the patients with colorectal cancer (31,32).
As for our patient, she displayed miR-6845-5p upregulation prior to
the chemotherapy and after that, the patient received chemotherapy
with tegafur-gimeracil-oteracil potassium, which included 5-FU
prodrug. Contrary to the fact that miR-6845-5p upregulation
attributes to the resistance to 5-FU (29,30),
the patient showed a stable disease with her miR-6845-5p
downregulated after the treatment. This outcome may be due to the
difference in cancer context, hence miR-6845-5p may not be employed
as chemotherapeutic sensitivity in OSCC patients.
Focusing on the function of the miRNAs, our report
revealed that each miRNA is associated with the regulation of
VEGFR-2 expression. VEGFRs are tyrosine kinase receptors and
interact with their ligands, VEGFs, and placental growth factor,
promoting angiogenesis. VEGFR includes VEGFR-1 and VEGFR-2
expressed in vascular endothelial cells (ECs), and VEGFR-3 is
expressed in the lymphatic ECs (33). Among them, VEGFR-2 largely
contributes to mediating VEGF effects such as EC proliferation,
migration, survival, and angiogenesis by activating its downstream
signaling pathways including phosphoinositide 3
kinase/AKT/mechanistic target of rapamycin and
Ras/Raf/mitogen-activated protein kinase/extracellular
signal-regulated kinase (34). Thus
far, previous studies have demonstrated that tumor cells secrete
proangiogenic growth factors to induce tumor-associated
angiogenesis (34,35). In this process, VEGF/VEGFR-2 is the
most crucial signaling pathway. Hereby, targeting the pathway is
being highlighted as a therapeutic target. Currently, several drugs
including bevacizumab, an anti-VEGF antibody, and ramucirumab, an
anti-VEGFR-2 monoclonal antibody have been approved to tackle
exacerbating cancers such as non-small cell lung cancer,
adenocarcinoma, and metastatic colorectal cancer (34). As for OSCC, VEGF and VEGFR-2 are
generally expressed in OSCC cells, and previous studies have
demonstrated that VEGF/VEGFR-2 should be implicated in OSCC
development (36–38). Moreover, the OSCC group exhibited
higher VEGF levels in their serum than the healthy group, and in
addition, high VEGF levels correlated with the development of lymph
node metastasis and showed higher clinical stages (37). Consequently, VEGF in the serum
should aid in diagnosing OSCC and predicting the risk of lymph node
metastasis. As regards the relationship between the miRNA profile
and VEGFR-2 expression status in the tumor specimen, in
silico analysis identified multiple putative locations in
3′-UTR on the VEGFR-2 mRNA complementary to the four miRNAs
(Data S1). Thus, the four miRNAs
are proposed to inhibit the VEGFR-2 translation. Intriguingly, this
report identified high VEFGR-2 expression levels in the tumor cells
in the tongue and lung metastatic lesions (Fig. 5D-F). This result may be due to a
certain mutation or deletion in the 3′-UTR that hinder the miRNAs
to bind to VEFGR-2 mRNA, resulting in VEGFR-2 overexpression in the
tumor specimens. Recently, with the as the advancement of sequence
technologies, patients with certain types of cancer have reported
to harbor mutations in the 5′ and 3′ UTR regions, which assumes to
contribute to cancer pathogenesis (39). Although our report could not confirm
the mutations in the patient's KDR gene this time, further
whole genome sequencing would reveal the associations between the
four miRNAs and VEGFR-2. Therefore, not only the exact interaction
between the four miRNAs and VEGFR-2 mRNA but also the mutation
profile of VEGFR-2 must be confirmed to employ these factors as
OSCC biomarkers. This report presents remarkable changes in the
salivary miRNA levels that are associated with controlling VEGFR-2
in accordance with the patient's clinical course, which indicates
that VEGFR-2 expression is also a candidate OSCC biomarker. Indeed,
the expression of VEGFR-2 appears upregulated in the lung
metastatic lesion compared with that observed in the tongue lesion
(Fig. 5E and F). Therefore, our
results proposed that not only the four miRNA profiles but also
VEGFR-2 expression status may be employed as a prognostic biomarker
for OSCC.
However, some limitations must be addressed to
support the availability of salivary miRNAs. First, this report
examined the miRNA profile of a single patient with OSCC and
identified the four miRNA candidates; thus, it is not guaranteed
that the miRNAs are also applicable to other patients as
biomarkers. Considering the genomic heterogeneity of OSCC, more
studies are needed to validate the four miRNAs and investigate
their availability for other patients with OSCC (40). Focusing on the gene mutation for
instance, the expression of miRNAs is reported to be regulated by
p53 during miRNA maturation, which implies that TP53 somatic
mutation should influence on the miRNA profile particularly among
patients with OSCC whose TP53 generally functions aberrantly
due to its gene mutations (1,41). In
our report, in silico analysis revealed the potential
interaction with the four miRNAs and 3′-UTR of p53 mRNA. This
result proposed that the four miRNAs contribute to the inhibition
of p53 function, promoting tumor cells to cause aberrant
proliferation and finally metastasis. Therefore, combining the
genetic alteration profile and salivary miRNA profile should lead
to the establishment of sophisticated biomarkers. Second, this
study could not conclude that the four miRNAs are responsible for
the OSCC tumorigenesis, and the four miRNAs would be potent
therapeutic and prognostic biomarkers of OSCC. This is mainly
because the patient presented in this study had a history of
congenital anomaly syndrome. Some miRNAs were reported to play
roles in stabilizing the phenotype of organisms, meaning that they
are responsible for controlling individual normal development
(42,43). Thus, miRNA dysregulation may
influence the normal development and may cause malformations
(44). Therefore, further
examinations are needed to conclude that the four miRNAs are
bona fide biomarkers by investigating the association
between the miRNA profile and background of congenital anomalies
syndrome. Third, this report did not elucidate the underlying
mechanisms of action of the four miRNAs in OSCC development and
metastasis. Of note, this report could not identify the miRNAs in
the tumor specimens, which make it difficult to conclude that the
miRNAs are indeed derived from the tumor cells. In addition, since
a single miRNA targets multiple mRNAs, we could not conclude that
the four miRNAs and VEGFR-2 solely are the key factors for OSCC
development. Accordingly, further analysis for establishing
salivary miRNAs as a biomarker is required by confirming the
localization of the four miRNAs in the specimen and examining the
relationship between the four miRNAs and OSCC tumorigenesis in a
large cohort.
In conclusion, this case report proposed that the
salivary miRNA should provide biological signatures of OSCC cells
and may be used as a novel biomarker for early diagnosis,
evaluating the therapeutic effect and predicting the prognosis of
patients with OSCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The microarray data generated in the present study
are not publicly available due to containing information that could
compromise patient privacy but may be requested from the
corresponding author. The other data generated in the present study
may be requested from the corresponding author.
Authors' contributions
TK collected the clinical data and wrote the
manuscript. TK, KIS, NO, JS, TI, TMu, TMa, MH and HI acquired and
interpreted clinical data. AYM, KCH and YH performed the
pathological examination. KCH, YH and HI performed and analyzed the
experimental examinations. KIS, JS and HI revised the manuscript.
KCH, YH and HI confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present report was approved by Hokkaido
University Hospital Independent Clinical Research Review Committee
(approval no. 020-0085; Sapporo, Japan). All study procedures were
conducted according to the principles of the Declaration of
Helsinki.
Patient consent for publication
Written informed consent for the publication of the
clinical data, including photos and images, was obtained from the
patient.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCRT
|
concurrent chemoradiotherapy
|
|
CECT
|
contrast-enhanced computed
tomography
|
|
EC
|
endothelial cell
|
|
ENE
|
extra-nodal extension
|
|
FU
|
fluorouracil
|
|
OPMD
|
oral potentially malignant
disorders
|
|
OSCC
|
oral squamous cell carcinoma
|
|
UTR
|
untranslated region
|
|
VEGFR-2
|
vascular endothelial growth factor
receptor-2
|
References
|
1
|
Mody MD, Rocco JW, Yom SS, Haddad RI and
Saba NF: Head and neck cancer. Lancet. 398:2289–2299. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yoshioka Y, Sakaue T, Matsui K, Tsushima
K, Obayashi F, Hamada A, Yamasaki S, Hamana T, Sumi K, Kanda T, et
al: Clinical investigation of oral cancer in adolescent and young
adult generation. Oral Sci Int. 18:126–134. 2021. View Article : Google Scholar
|
|
3
|
Braakhuis BJM, Brakenhoff RH and Leemans
CR: Treatment choice for locally advanced head and neck cancers on
the basis of risk factors: Biological risk factors. Ann Oncol. 23
(Suppl 10):x173–x177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carreras-Torras C and Gay-Escoda C:
Techniques for early diagnosis of oral squamous cell carcinoma:
Systematic review. Med Oral Patol Oral Cir Bucal. 20:e305–e315.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chow LQM: Head and neck cancer. N Engl J
Med. 382:60–72. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ho PS, Wang WC, Huang YT and Yang YH:
Finding an oral potentially malignant disorder in screening program
is related to early diagnosis of oral cavity cancer-experience from
real world e. Oral Oncol. 89:107–114. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim T and Croce CM: MicroRNA: Trends in
clinical trials of cancer diagnosis and therapy strategies. Exp Mol
Med. 55:1314–1321. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calin G, Dumitru C, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
O'Donnell KA, Wentzel EA, Zeller KI, Dang
CV and Mendell JT: c-Myc-regulated microRNAs modulate E2F1
expression. Nature. 435:839–843. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yap T, Vella L, Seers C, Nastri A,
Reynolds E, Cirillo N and McCullough M: Oral swirl samples-a robust
source of microRNA protected by extracellular vesicles. Oral Dis.
23:312–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yap T, Seers C, Koo K, Cheng L, Vella LJ,
Hill AF, Reynolds E, Nastri A, Cirillo N and McCullough M:
Non-invasive screening of a microRNA-based dysregulation signature
in oral cancer and oral potentially malignant disorders. Oral
Oncol. 96:113–120. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Balakittnen J, Ekanayake Weeramange C,
Wallace DF, Duijf PHG, Cristino AS, Hartel G, Barrero RA, Taheri T,
Kenny L, Vasani S, et al: A novel saliva-based miRNA profile to
diagnose and predict oral cancer. Int J Oral Sci. 16:142024.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Agrawal A, Balcı H, Hanspers K, Coort SL,
Martens M, Slenter DN, Ehrhart F, Digles D, Waagmeester A, Wassink
I, et al: WikiPathways 2024: Next generation pathway database.
Nucleic Acids Res. 52(D1): D679–D689. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Passaro A, Al Bakir M, Hamilton EG, Diehn
M, André F, Roy-Chowdhuri S, Mountzios G, Wistuba II, Swanton C and
Peters S: Cancer biomarkers: Emerging trends and clinical
implications for personalized treatment. Cell. 187:1617–1635. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
van Schaik JE, Muller Kobold AC, van der
Laan BFAM, van der Vegt B, van Hemel BM and Plaat BEC: Squamous
cell carcinoma antigen concentration in fine needle aspiration
samples: A new method to detect cervical lymph node metastases of
head and neck squamous cell carcinoma. Head Neck. 41:2561–2565.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dar GM, Agarwal S, Kumar A, Nimisha
Apurva, Sharma AK, Verma R, Sattar RSA, Ahmad E, Ali A, et al: A
non-invasive miRNA-based approach in early diagnosis and
therapeutics of oral cancer. Crit Rev Oncol Hematol.
180:1038502022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Prakash N and Pradeep G: Circulating
biomarkers in oral cancer: Unravelling the mystery. J Oral
Maxillofac Pathol. 26:300–306. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kinoshita T, Nohata N, Hanazawa T, Kikkawa
N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto
Y and Seki N: Tumour-suppressive microRNA-29s inhibit cancer cell
migration and invasion by targeting laminin-integrin signalling in
head and neck squamous cell carcinoma. Br J Cancer. 109:2636–2645.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Manikandan M, Deva Magendhra Rao AK,
Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R and
Munirajan AK: Oral squamous cell carcinoma: microRNA expression
profiling and integrative analyses for elucidation of
tumourigenesis mechanism. Mol Cancer. 15:282016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suh YE, Raulf N, Gäken J, Lawler K, Urbano
TG, Bullenkamp J, Gobeil S, Huot J, Odell E and Tavassoli M:
MicroRNA-196a promotes an oncogenic effect in head and neck cancer
cells by suppressing annexin A1 and enhancing radioresistance. Int
J Cancer. 137:1021–1034. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Alimena S, Stephenson BJK, Webber JW,
Wollborn L, Sussman CB, Packard DG, Williams M, Comrie CE, Wang JY,
Markert T, et al: Differences in serum miRNA profiles by race,
ethnicity, and socioeconomic status: implications for developing an
equitable ovarian cancer screening test. Cancer Prev Res (Phila).
17:177–185. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Diener C, Keller A and Meese E: Emerging
concepts of miRNA therapeutics: From cells to clinic. Trends Genet.
38:613–626. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z and Rana TM: Therapeutic targeting of
microRNAs: Current status and future challenges. Nat Rev Drug
Discov. 13:622–638. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu Z, He Q, Liang J, Li W, Su Q, Chen Z,
Wan Q, Zhou X, Cao L, Sun J, et al: miR-31-5p is a potential
circulating biomarker and therapeutic target for oral cancer. Mol
Ther Nucleic Acids. 16:471–480. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Torso NDG, Quintanilha JCF, Cursino MA,
Pincinato EC, Loren P, Salazar LA, Lima CSP and Moriel P:
miR-6805-5p as a biomarker of cisplatin-induced nephrotoxicity in
patients with head and neck cancer. Front Pharmacol.
14:12752382023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiong H, Ni Z, He J, Jiang S, Li X, He J,
Gong W, Zheng L, Chen S, Li B, et al: LncRNA HULC triggers
autophagy via stabilizing Sirt1 and attenuates the chemosensitivity
of HCC cells. Oncogene. 36:3528–3540. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li LB, Yang L, Xie GQ, Zhou XC, Shen XB,
Xu QL, Ma ZY and Guo XD: The modulation relationship of genomic
pattern of intratumor heterogeneity and immunity microenvironment
heterogeneity in hepatocellular carcinoma. Oncol Lett. 20:2332020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan S, Jiang Y, Liang C, Cheng M, Jin C,
Duan Q, Xu D, Yang L, Zhang X, Ren B and Jin P: Exosomal
miR-6803-5p as potential diagnostic and prognostic marker in
colorectal cancer. J Cell Biochem. 119:4113–4119. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yan S, Cheng M, Duan Q, Wang Z, Gao W, Ren
B and Xu D: MiR-6803-5p promotes cancer cell proliferation and
invasion via PTPRO/NF-κB axis in colorectal cancer. Mediators
Inflamm. 2019:81285012019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ferrara N, Gerber HP and LeCouter J: The
biology of VEGF and its receptors. Nat Med. 9:669–676. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu ZL, Chen HH, Zheng LL, Sun LP and Shi
L: Angiogenic signaling pathways and anti-angiogenic therapy for
cancer. Signal Transduct Target Ther. 8:1982023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Carmeliet P and Jain RK: Molecular
mechanisms and clinical applications of angiogenesis. Nature.
473:298–307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Araki-Maeda H, Kawabe M, Omori Y, Yamanegi
K, Yoshida K, Yoshikawa K, Takaoka K, Noguchi K, Nakano Y and
Kishimoto H: Establishment of an oral squamous cell carcinoma cell
line expressing vascular endothelial growth factor a and its two
receptors. J Dent Sci. 17:1471–1479. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Edirisinghe ST, Weerasekera M, De Silva
DK, Devmini MT, Pathmaperuma S, Wijesinghe GK, Nisansala T,
Maddumage A, Huzaini H, Rich AM, et al: Vascular endothelial growth
factor A (VEGF-A) and vascular endothelial growth factor receptor 2
(VEGFR-2) as potential biomarkers for oral squamous cell carcinoma:
A Sri Lankan study. Asian Pac J Cancer Prev. 24:267–274. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pekarek L, Garrido-Gil MJ, Sánchez-Cendra
A, Cassinello J, Pekarek T, Fraile-Martinez O, García-Montero C,
Lopez-Gonzalez L, Rios-Parra A, Álvarez-Mon M, et al: Emerging
histological and serological biomarkers in oral squamous cell
carcinoma: Applications in diagnosis, prognosis evaluation and
personalized therapeutics (review). Oncol Rep. 50:2132023.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schuster SL and Hsieh AC: The untranslated
regions of mRNAs in cancer. Trends Cancer. 5:245–262. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Q, Tie Y, Alu A, Ma X and Shi H:
Targeted therapy for head and neck cancer: Signaling pathways and
clinical studies. Signal Transduct Target Ther. 8:312023.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hermeking H: MicroRNAs in the p53 network:
Micromanagement of tumour suppression. Nat Rev Cancer. 12:613–626.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gibson G and Wagner G: Canalization in
evolutionary genetics: A stabilizing theory? Bioessays. 22:372–380.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hornstein E and Shomron N: Canalization of
development by microRNAs. Nat Genet. 38 (Suppl):S20–S24. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Amiel J, de Pontual L and Henrion-Caude A:
miRNA, development and disease. Adv Genet. 80:1–36. 2012.
View Article : Google Scholar : PubMed/NCBI
|