Introduction
Ovarian cancer is the deadliest gynecological
cancer, as it is often detected only at an advanced stage and shows
frequent recurrence (1). The main
histological types of epithelial ovarian cancer are serous, clear
cell, endometrioid and mucinous carcinomas. Among these,
endometrioid carcinomas account for 10–15% of all epithelial
ovarian cancers (2) and are
classified as grade 1, 2 or 3. Although >70% of endometrioid
carcinomas are diagnosed at Stage I or II, the prognosis of
patients with this type of cancer remains poor (3). In Japan and other Asian countries, the
incidence rates of endometrioid and clear cell carcinomas are
higher than in other regions (4).
The risk factors include endometriosis, Lynch syndrome and
intestinal dysbiosis (5–7). Ovarian endometrioid carcinoma (OEC) is
often associated with endometrial cancer, which is also called
‘simultaneous endometrial and ovarian cancer’ (SEOC) (8). In such cases, distinguishing whether
the cancer is an individual tumor or a metastatic case is
difficult, and controversy regarding this exists currently
(8).
In terms of epigenetics, the main factors affecting
gene expression regulation are DNA methylation and histone
modifications (9). Among histone
modifications, histone acetylation, such as H3K27Ac, promotes
transcriptional activity by transforming the chromatin state into
an open state (10). This process
is orchestrated by three factors: i) Histone acetyltransferases,
which serve the role of ‘writer’; ii) bromodomain (BRD) proteins,
which are ‘reader’; and iii) histone deacetylation, which function
as ‘eraser’ (11–13). Therefore, BRD proteins, the members
of the bromodomain and extra-terminal domain (BET) family (14), contribute to transcriptional
regulation by recognizing histone acetylation and recruiting
chromatin- and transcription-related factors (13). In particular, BRD4 promotes
transcription initiation by binding to the acetyl group of lysine
in histones H3 and H4 via its own bromodomain (15). BET proteins, particularly BRD4, have
been implicated in human diseases, particularly cancer (16). c-Myc, which is often
upregulated in cancer, is the main downstream gene regulated by
BRD4 (17). BRD4 inhibition has
been reported to downregulate c-Myc expression in several
tumor types. Accordingly, BET inhibitors, including JQ1, have been
reported as novel therapeutic agents in the treatment of several
cancers (18). However, no studies
have reported on the effects of JQ1 in OEC, to the best of our
knowledge. Therefore, the present study aimed to assess the
antitumor effect of JQ1 in OEC and endometrial endometrioid
carcinoma (EEC) to develop a novel treatment for SEOC.
Materials and methods
Cell lines
A total of three OEC cell lines (A2780, TOV112D and
OVK18) and three EEC cell lines (HEC265, human endometrioid
adenocarcinoma G1; HEC151, human endometrioid adenocarcinoma G2;
and HEC50B, human endometrioid adenocarcinoma G3) we used in the
present study.
A2780 cells (European Collection of Authenticated
Cell Cultures) were cultured in RPMI-1640 medium (FUJIFILM Wako
Pure Chemical Corporation) supplemented with 10% heat-inactivated
fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.). TOV112D
cells (American Type Culture Collection) were cultured in MCDB 105
medium (Sigma-Aldrich; Merck KGaA) supplemented with 15%
heat-inactivated FBS. OVK18 cells (RIKEN BioResource Center) were
cultured in minimum essential medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% heat-inactivated FBS.
HEC265, HEC151 and HEC50B cell lines (JCRB Cell
Bank) were cultured in Eagle's minimum essential medium (FUJIFILM
Wako Pure Chemical Corporation) containing 10% heat-inactivated
FBS. All cell lines were maintained at 37°C in a humidified
atmosphere with 5% CO2. The mutation status of OEC and
EEC cell lines was searched using the Cancer Cell Line Encyclopedia
data (https://sites.broadinstitute.org/ccle/).
Small interfering (si)RNA
transfection
A2780 and HEC50B cells were transfected with 10 nM
siRNAs at 37°C for 3.5 h using Lipofectamine™ RNAiMAX Transfection
Reagent (cat. no. 13778150; Invitrogen™; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. A total of 72 h
after siRNA transfection, RNA extraction, protein extraction and
cell viability assay were performed. siBRD4 #1 (sense:
5′-GUGCUGAUGUCCGAUUGAU-3′ and antisense:
5′-AUCAAUCGGACAUCAGCAC-3′), siBRD4 #2 (cat. no. NM_058243,
SASI_Hs01_00126965) and a negative control siRNA (siNC;
MISSION® siRNA Universal Negative Control; cat. no.
SIC001) were purchased from Sigma-Aldrich (Merck KGaA).
RNA extraction and reverse
transcription(RT)-quantitative (q)PCR
After siRNA transfection, total RNA from A2780 and
HEC50B cells was extracted using RNeasy® Mini Kit (cat.
no. 74104; Qiagen, Inc.). cDNA synthesis from mRNA was performed
using ReverTra Ace™ qPCR Master Mix with gDNA Remover (cat. no.
FSQ-301; Toyobo Co., Ltd.) with the following steps: 37°C for 15
min, 50°C for 5 min and 98°C for 5 s. The mRNA expression levels
were measured using qPCR using the One-Step SYBR Prime Script
RT-PCR Kit (cat. no. RR064A; Takara Bio, Inc.) and the QuantStudio™
1 Real-Time PCR System (Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: Initial denaturation step
at 98°C for 2 min, followed by 40 cycles at 98°C for 10 s, 60°C for
10 s and 72°C for 30 s. The mRNA expression levels were normalized
to the mRNA levels of β-actin. The relative mRNA expression level
was calculated using the 2−ΔΔCq method (19). The sequences of primers were as
follows: β-actin, (forward) 5′-CACACTGTGCCCATCTACGA-3′ and
(reverse) 5′-CTCCTTAATGTCACGCACGA-3′; and BRD4, (forward)
5′-GTGGTGCACATCATCCAGTC-3′ and (reverse)
5′-CCGACTCTGAGGACGAGAAG-3′.
Cell viability assay
The OEC cell lines (A2780, TOV112D and OVK18;
1×104 cells/well) and EEC cell lines (HEC265, HEC151 and
HEC50B; 4×103 cells/well) were seeded on 48-well plate
and treated with JQ1 (cat. no. HY-13030; MedChemExpress) for 72 h.
After treatment, cells were incubated in a 10% Cell Count Kit-8
solution (Dojindo Laboratories, Inc.) for 2 h. The optical density
at 450 nm was measured using a microplate reader (BioTek; Agilent
Technologies, Inc.). Cell viability was normalized using 0.1%
dimethyl sulfoxide (DMSO; Sigma-Aldrich) as the control.
Colony formation assay
The OEC cell lines (A2780, TOV112D and OVK18) and
EEC cell lines (HEC265, HEC151 and HEC50B) were seeded at a density
of 2×103 cells/well on 6-well plates. After overnight
incubation at 37°C, the cells were treated with JQ1 (0.1, 0.2 and
0.5 µM) or 0.1% DMSO for 10 days to assess colony formation. The
medium was replaced every 3–4 days. The plates were then washed
with phosphate-buffered saline (PBS). Colonies were fixed with 100%
methanol at room temperature (RT) for 2 h and stained with Giemsa
stain (FUJIFILM Wako Pure Chemical Corporation) at RT for 1 h. The
colonies (>50 cells) were counted manually under a microscope
and normalized to the number of colonies treated with 0.1%
DMSO.
Protein extraction and western
blotting
The OEC cell lines (A2780, TOV112D and OVK18;
1×105 cells/dish) and EEC cell lines (HEC265, HEC151 and
HEC50B; 4×104 cells/dish) were plated onto a 6-cm dish
and treated with JQ1 (1 µM) or 0.1% DMSO at 37°C for 72 h. Protein
was extracted using a lysis buffer [0.1 M Tris-HCl; pH 7.5; 10%
glycerol and 1% sodium dodecyl sulfate (SDS)]. The extracted
proteins were boiled for 5 min and centrifuged at 4°C for 10 min at
20,000 × g. The protein concentration was measured using a BCA
protein assay kit (cat. no. 06385-00; Nacalai Tesque, Inc.). Each
sample (15 µg/lane) was separated using SDS-PAGE
(Mini-PROTEAN® TGX™ Precast Protein Gels (Any kD™);
Bio-Rad Laboratories, Inc.) and transferred to polyvinylidene
difluoride (PVDF) membranes using Trans-Blot Turbo Mini PVDF
transfer packs (Bio-Rad Laboratories, Inc.). Blocking was performed
with 5% skim milk at RT for 1 h. The membrane was incubated with
the primary antibodies at 4°C overnight and incubated with the
secondary antibodies at RT for 1 h. Protein expression was measured
using the Amersham™ ECL™ Select (Cytiva), and the emitted signals
were imaged using the ImageQuant™ LAS 4000 system (Cytiva). The
following primary antibodies were used for immunoblotting: Rabbit
anti-BRD4 (1:1,000; cat. no. 13440; Cell Signaling Technology,
Inc.), rabbit anti-cleaved poly ADP ribose polymerase (PARP;
1:1,000; cat. no. 5625; Cell Signaling Technology, Inc.), rabbit
anti-c-Myc (D84C12; 1:1,000; cat. no. 5605; Cell Signaling
Technology, Inc.), PARP (1:1,000; cat/ no. 9542; Cell Signaling
Technology, Inc.) and mouse anti-β-actin (1:7,000; cat. no. A2228;
Sigma-Aldrich; Merck KGaA). For all primary antibodies, the
incubation condition was at 4°C overnight. The secondary antibodies
used were as follows: Anti-mouse IgG HRP-linked (1:5,000; cat. no.
7076; Cell Signaling Technology, Inc.) and anti-rabbit IgG
HRP-linked (1:3,000; cat. no. 7074; Cell Signaling Technology,
Inc.). The semi-quantified values of the target protein were
normalized by dividing them by the semi-quantified values of each
β-actin in the same sample. Subsequently, using the value of DMSO
as 1, the values of JQ1 were normalized. The values were
semi-quantified using ImageJ 1.53 (National Institutes of Health).
Graphs were subsequently constructed using the normalized values
obtained from three independent experiments.
Cell cycle assay
The OEC cell lines (A2780, TOV112D and OVK18;
2×106 cells/dish) and EEC cell lines (HEC265, HEC151 and
HEC50B; 1×106 cells/dish) were plated on a 10-cm dish
with JQ1 (5 µM) or 0.1% DMSO and incubated at 37°C for 96 h.
Subsequently, the cells were harvested with trypsin, washed with
PBS and fixed in 70% ethanol at −20°C overnight. After washing
twice with PBS, the samples were stained with propidium iodide (PI;
Sigma-Aldrich; Merck KGaA) at 4°C for 15 min. Cell cycle analysis
was performed using flow cytometry with a BD FACSCalibur HG Flow
Cytometer (BD Biosciences) and Cell Quest Pro software v. 6.1 (BD
Biosciences). Data were assessed using FlowJo software, version 10
(BD Biosciences).
Kaplan-Meier survival analysis
The overall survival (OS) was analyzed using the
Kaplan-Meier method with TCGA datasets in cBioPortal for Cancer
Genomics (www.cbioportal.org). The dataset
TCGA-OV (20) for ovarian cancer
and PanCanAtlas (21) for
endometrioid cancer were analyzed. The cases were categorized into
high and low groups, based on BRD4 expression levels. Statistical
significance was determined using the log-rank test.
Statistical analysis
Data are presented as the mean ± standard deviation
of >3 independent experiments. Data were analyzed using
Microsoft Excel 2016 (Microsoft Corporation) and GraphPad Prism 10
(Dotmatics). For comparisons between two groups, the unpaired
Student's t-test was used. For comparisons among ≥3 groups, one-way
analysis of variance followed by Dunnett's post hoc test was used.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of BRD4 is associated with
prognosis in patients with ovarian and endometrial carcinoma
Taking into account previous reports suggesting an
association between the expression levels of BRD4 and prognosis in
other cancer types (22–24), the present study assessed the
association between BRD4 expression and prognosis in patients with
epithelial ovarian carcinoma and endometrioid endometrial carcinoma
using RNA-seq data from TCGA. OS was significantly worse in the
BRD4-High than in the BRD4-Low group for both ovarian carcinoma
(P=0.016; Fig. S1A) and
endometrial carcinoma (P=0.033; Fig.
S1B).
JQ1 suppresses cell proliferation in
OEC and EEC cell lines
To evaluate the antitumor effect of JQ1 on OEC and
EEC, the present study performed cell viability assays on three OEC
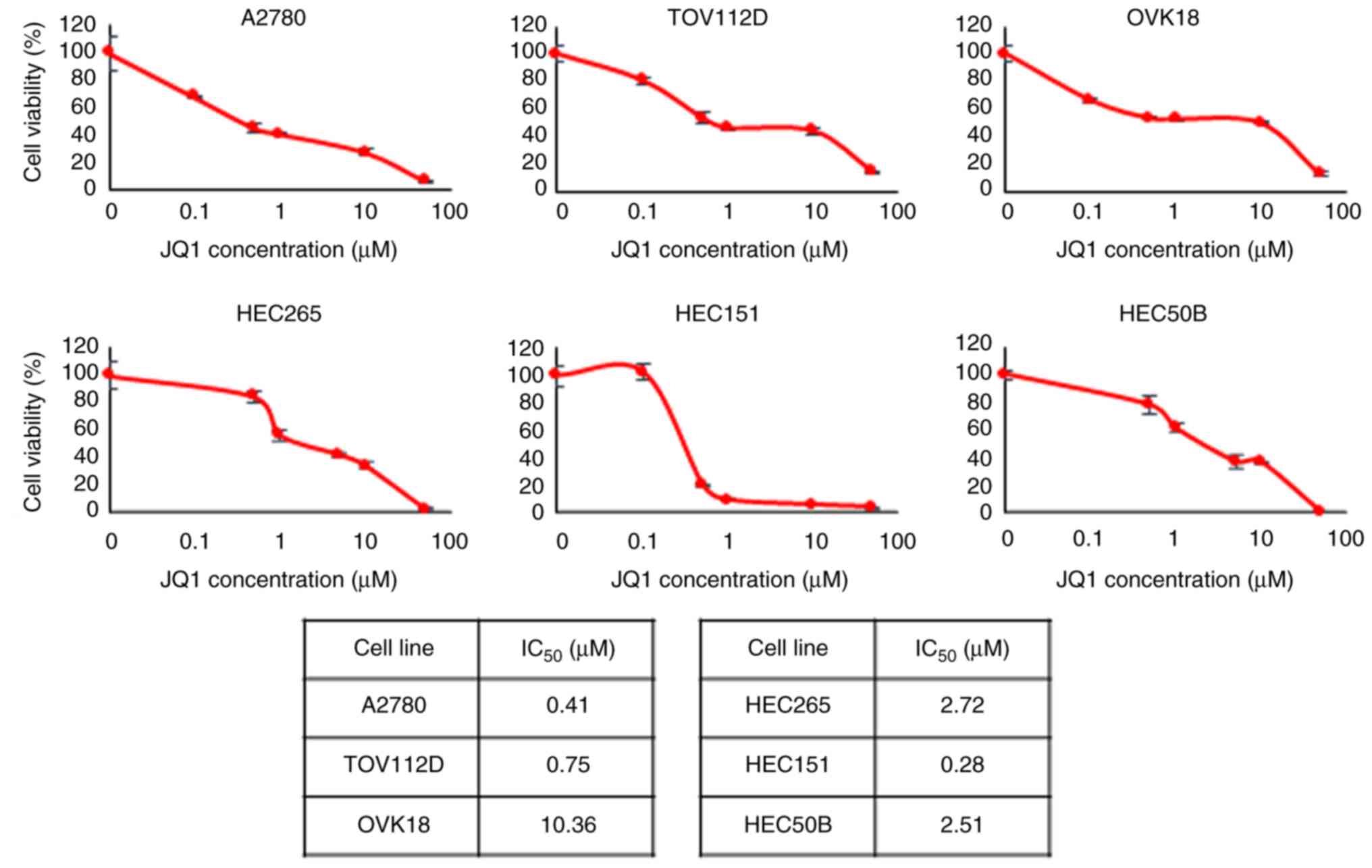
and three EEC cell lines treated with 0.01–50 µM JQ1 for 72 h. Cell
viability decreased in a dose-dependent manner in response to JQ1
treatment (Fig. 1). The
IC50, half maximal inhibitory concentration
(IC50) of JQ1 for A2780, TOV112D, OVK18, HEC265, HEC151
and HEC50B cells was 0.41, 0.75, 10.36, 2.72, 0.28 and 2.51 µM,
respectively.
Furthermore, the common mutation status for each
cell line is presented in Table
SI. No mutation status indicative of JQ1 sensitivity was
observed.
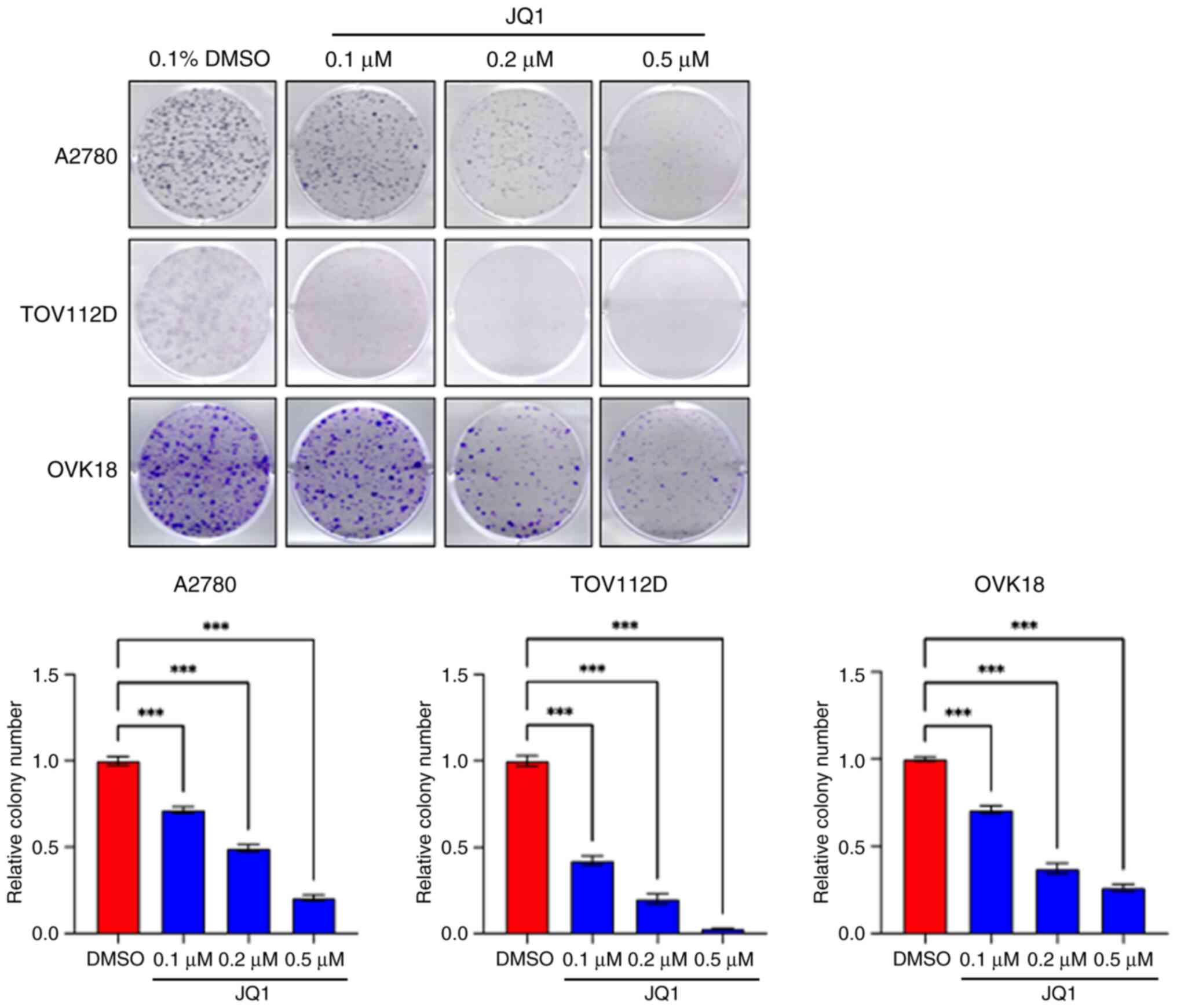
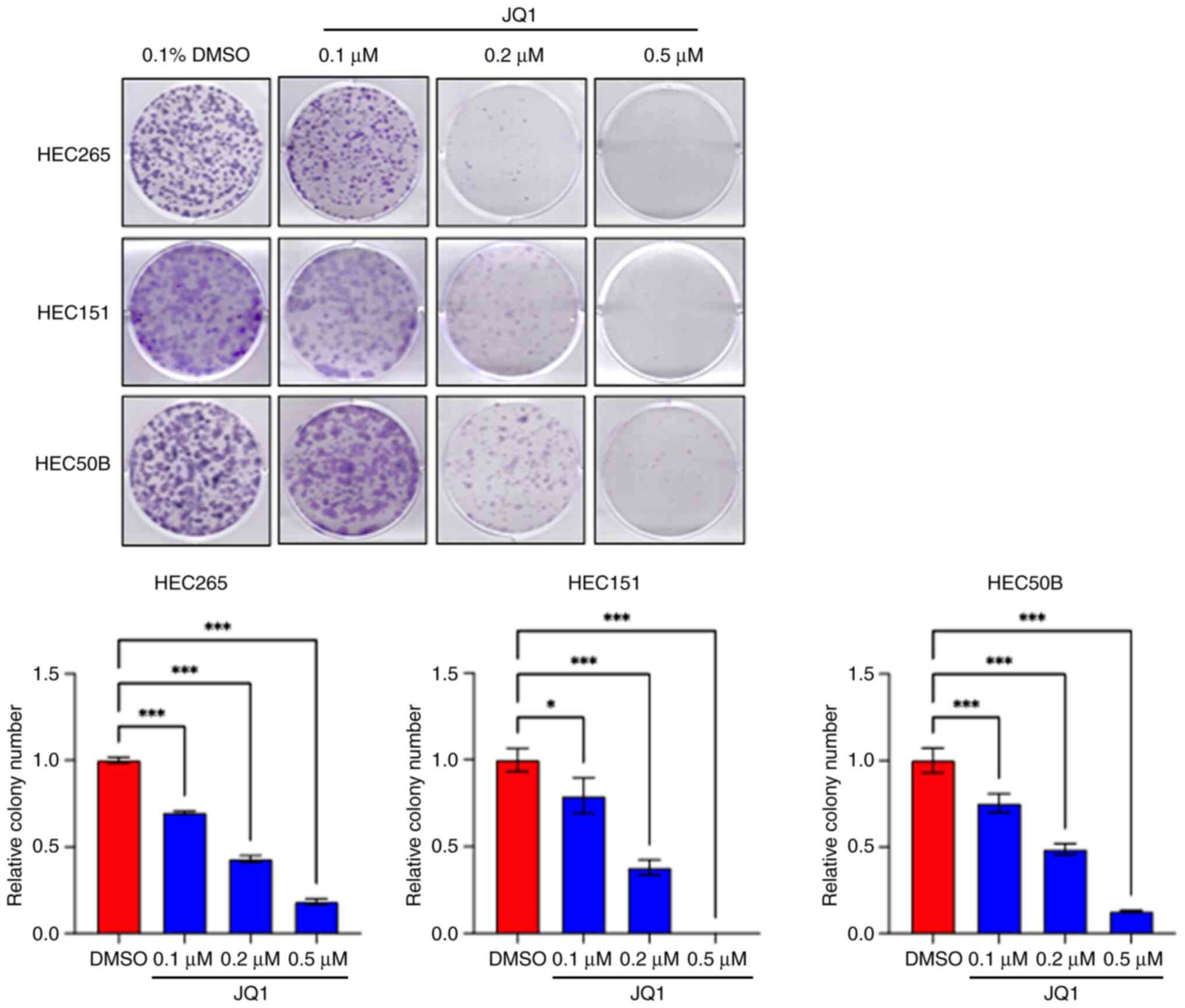
Colony formation assays were also performed to
evaluate the long-term cytotoxicity of JQ1. Colony formation was
significantly suppressed in a dose-dependent manner in all cell
lines, in comparison with the control (Figs. 2 and 3). A low dose of JQ1 (0.1 µM) was
sufficient to affect the long-term clonogenicity of OEC and EEC
cell lines. The number of colonies in each treatment group is
presented in Table SII.
JQ1 suppresses c-Myc expression
Western blot analyses were performed to assess the
effect of JQ1 treatment on c-Myc expression in OEC and EEC
cells. The c-Myc expression significantly decreased in all
six cell lines after treatment with 1 µM JQ1 for 72 h, in
comparison with the control (Fig.
4).
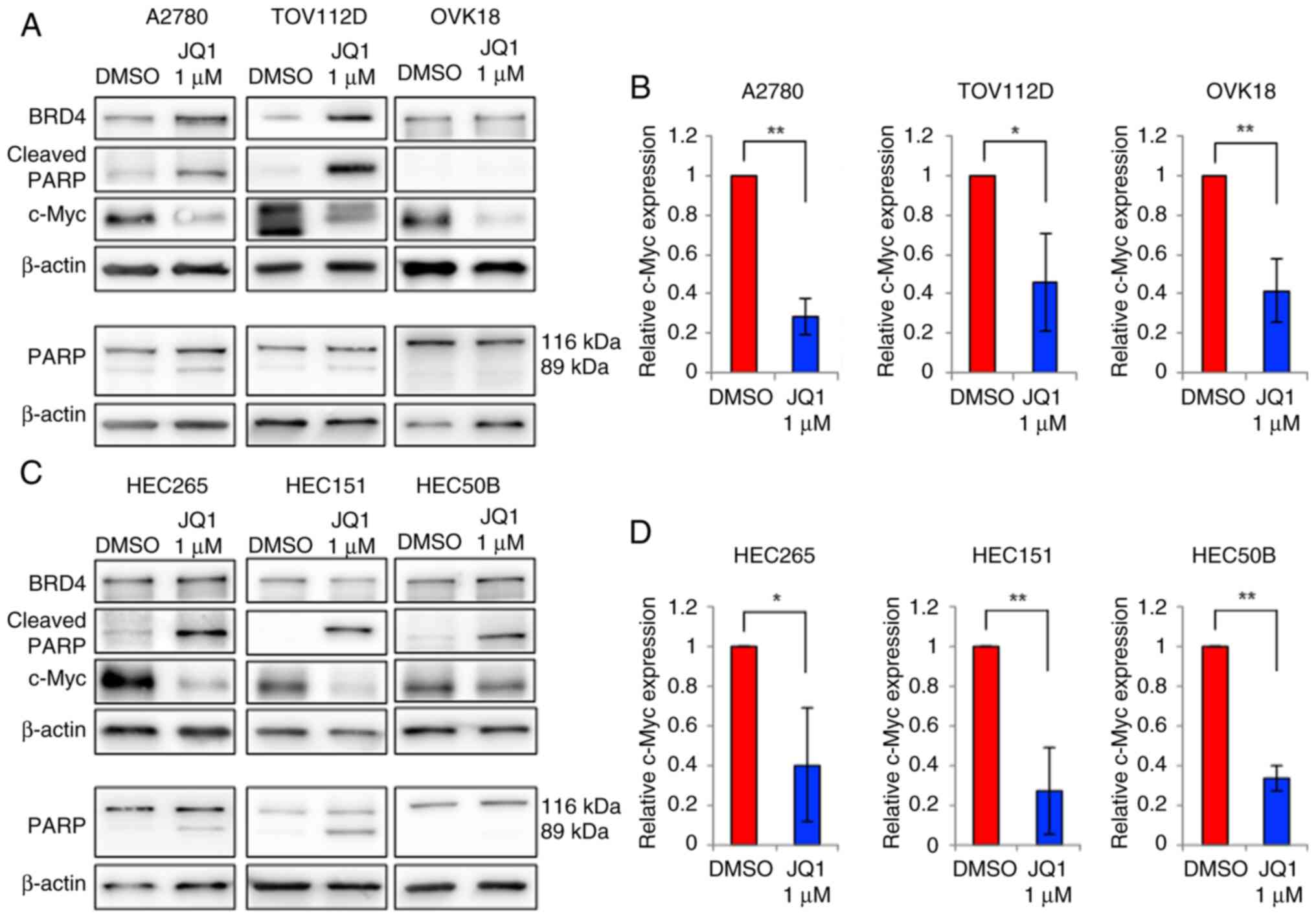 | Figure 4.Western blotting analysis of the
protein expression levels after JQ1 treatment. (A) BRD4, c-Myc,
cleaved PARP, PARP and β-actin protein expression levels, and (B)
relative c-Myc protein expression level in A2780, TOV112D and OVK18
cells treated with 0.1% DMSO or 1 µM JQ1 for 72 h. (C) BRD4, c-Myc,
cleaved PARP, PARP and β-actin protein expression levels, and (D)
relative c-Myc protein expression level in HEC265, HEC151 and
HEC50B cells treated with 0.1% DMSO or 1 µM JQ1 for 72 h. The
expression level of c-Myc decreased and that of cleaved PARP
increased with the JQ1 treatment in all cell lines. *P<0.05;
**P<0.01. BRD4, bromodomain 4; PARP, poly ADP ribose
polymerase. |
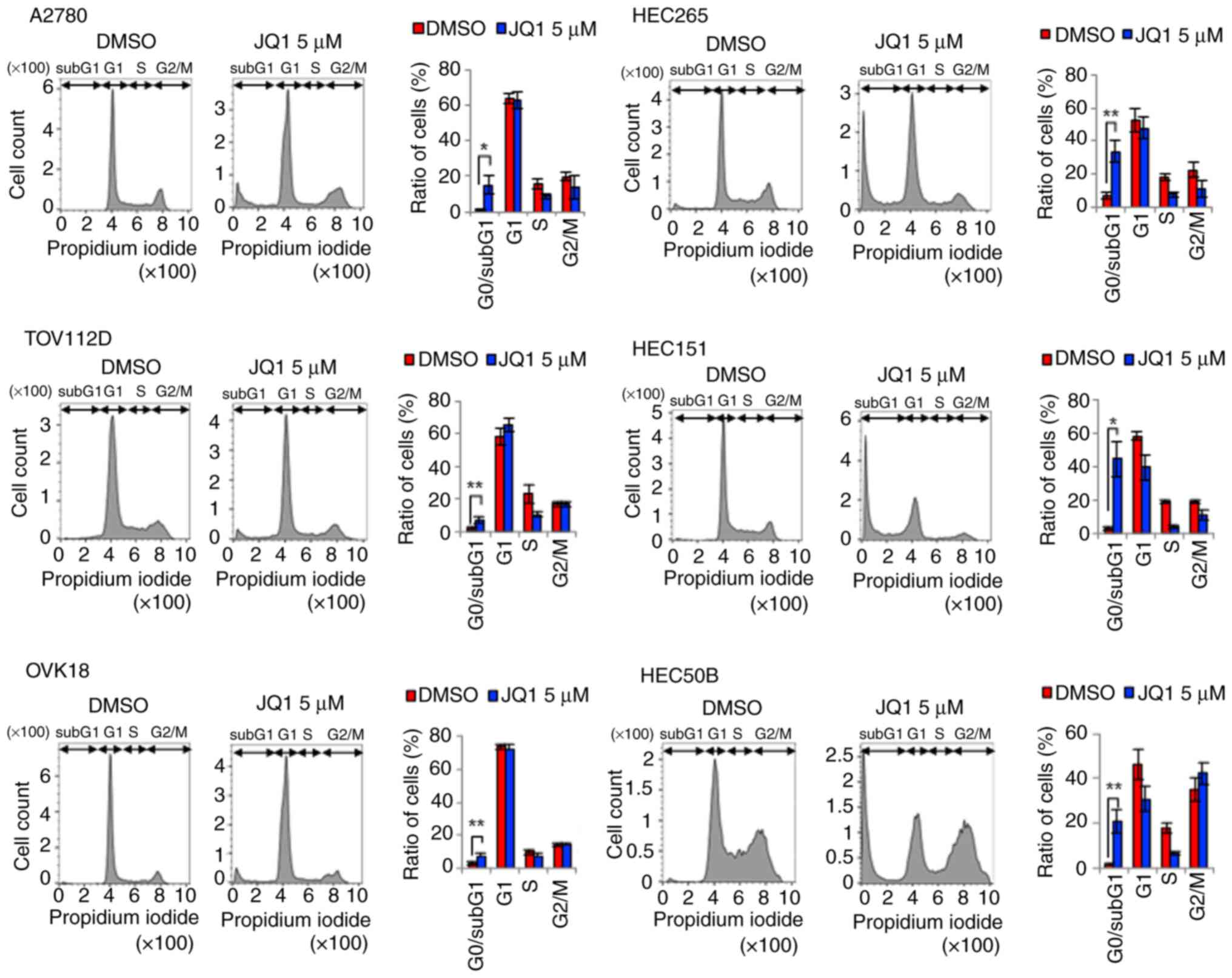
JQ1 induces apoptosis
Immunoblotting and cell cycle assays were performed
to determine whether the decrease in cell viability was associated
with apoptosis. As indicated by the western blotting results
(Fig. 4), the expression levels of
cleaved PARP (an apoptosis marker) were increased in A2780,
TOV112D, HEC265, HEC151 and HEC50B cells treated with 1 µM JQ1 for
72 h. However, in OVK18 cells, for which a relatively high JQ1 IC50
value was obtained, the cleaved PARP level was not notably elevated
with 1 µM, but was markedly elevated with 5 and 10-µM JQ1 treatment
(Figs. 4A and S2). Furthermore, to demonstrate the
increase in cleaved PARP, the present study confirmed that the
expression level of the PARP protein itself did not increase
(Figs. 4A, C and S2). The same processed protein samples
were used for this experiment.
Moreover, the cell cycle analysis results revealed
that the 5-µM JQ1 treatment significantly increased the population
of cells in the sub-G1 phase in all six cell lines (Fig. 5). Consistent with the expression of
cleaved PARP (Fig. 4), the cell
population was lowest in OVK18 cells among all the cell lines
(Fig. 5). Collectively, these
results indicate that JQ1 induced apoptosis and suppressed the
proliferation of OEC and EEC cells.
BRD4 knockdown suppresses cell
proliferation and induces apoptosis
The effect of BRD4 knockdown on cell proliferation
was also evaluated. BRD4 knockdown tended to suppress cell
proliferation in both OEC and EEC cell lines, compared with the
negative control (Fig. S3A and B).
Similar to the inhibitor experiment findings, BRD4 knockdown was
also associated with a marked decrease in c-Myc and an increase in
cleaved PARP expression levels (Fig.
S3C).
Discussion
c-Myc is involved in several functions such
as cell proliferation, cell immortalization and promotion of
metastasis. Most of these functions reflect the function of c-Myc
as a transcription factor. Furthermore, previous studies using
chromatin immunoprecipitation experiments with BRD4 protein
reported that c-Myc gene transcription is regulated by BET.
Transcription of the c-Myc gene was also repressed following
treatment with the BET inhibitor JQ1 (23,25).
Histone acetylation is a crucial epigenetic
regulatory mechanism. The BET family and histone deacetylase (HDAC)
family both regulate the expression of important cancer genes and
tumor suppressor genes (26,27).
HDAC inhibitors are clinically used as anticancer agents, and
synergistic effects of combining BET inhibitors with HDAC
inhibitors have been reported (28). BET inhibitors are currently
undergoing clinical trials and are anticipated as promising new
cancer therapeutics.
Although many studies on BET inhibitors for ovarian
cancer have been published (29–32),
the present study is the first in which JQ1 has been assessed in an
OEC context, to the best of our knowledge. OEC is one of the four
main histological types of epithelial ovarian cancer, with a high
incidence in Japan and other Asian countries (4). Although OEC is often diagnosed at an
early stage (stage I or II), the prognosis of certain patients
remains poor (33). Endometrioid
and clear cell carcinoma of the ovary have often been associated
with endometriosis (5), and
patients with endometriosis are at a higher risk of developing
ovarian endometrioid and clear cell carcinoma than those without
endometriosis (34). A popular
hypothesis for endometriosis development is the retrograde
menstruation theory, which implies that the endometrium expelled as
menstrual blood retrogradely enters the abdominal cavity and
implants itself in pelvic organs, such as the peritoneum and
ovaries, thus causing endometriosis (35,36).
Notably, OEC is often associated with EEC of the endometrium
(namely, SEOC) (37). Our previous
study demonstrated a higher SEOC rate in patients with
endometriosis than in those without (34). Moreover, the molecular and
pathological features of low-grade OEC and EEC are similar
(8,37). For example, mutations in
cancer-related genes, such as AT-rich interaction domain 1A
(ARID1A), tumor protein P53,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α,
PTEN, KRAS and DNA polymerase ε catalytic subunit A, are
similar in OEC and EEC (38). These
results support the hypothesis that endometria with carcinogenic
changes may backflow from the uterus to the abdominal cavity,
causing endometriosis in the ovaries. Consequently, the ovaries may
become a reservoir for the further development of endometrial
carcinoma (34). Therefore, the
present study assessed JQ1 for the treatment of OEC and EEC.
The results of the present study indicate that JQ1
has antitumor effects in almost all the endometrial carcinoma cell
lines evaluated. Moreover, exposing OEC and EEC cell lines to JQ1
inhibited cell proliferation. Notably, a high IC50 value
was obtained in OVK18 cells treated with JQ1. In the colony
formation assay, JQ1 showed efficacy in all cell lines, suggesting
that long-term administration may be effective in all OECs and
EECs. Furthermore, JQ1 induced apoptosis and suppressed
c-Myc expression in ovarian and endometrial cancers.
Different concentrations of JQ1 were used in each experiment as the
optimal concentration of the target to be confirmed varied in each
experiment. Moreover, the present study also performed experiments
using the lowest feasible concentrations of JQ1 to demonstrate its
effects. There are previous reports using JQ1 in A2780 cells
(39,40); however, there are no previous
reports in other cell lines, to the best of our knowledge.
A total of 1 µM was used for the western blot
analysis (Fig. 4) as 1 µM JQ1
markedly reduced cell viability across all cell lines in the
inhibitor experiment (Fig. 1).
Previous studies using A2780 cells reported decreased c-Myc
expression with 0.3 µM (39) and
2.5 µM (40) JQ1. In the colony
formation assay (Figs. 2 and
3), lower concentrations of JQ1
exhibited effects, likely due to prolonged JQ1 administration.
Considering previous research indicating colony formation
suppression even at 0.1 µM (41,42),
the present study assessed concentrations of 0.1, 0.2 and 0.5 µM to
evaluate outcomes at lower concentrations. Furthermore, regarding
the cell cycle assay (Fig. 5), a
previous study (42) was referred
to and pilot experiments at 2.5 and 5 µM concentrations were
performed. As a significant increase in the sub-G1 phase was
demonstrated at 5 µM across all cell lines, this concentration was
selected for subsequent experiments.
Although JQ1 was initially designed as a selective
inhibitor of BRDs, aiming to displace BRDs from chromatin and
disrupt their role in regulating gene transcription (43), it is difficult to demonstrate that
JQ1 inhibits BET function as there are no definite biomarkers that
indicate that JQ1 inhibits BET protein. However, as JQ1 selectively
binds to BET proteins to regulate transcription (43), and it is known to suppress
c-Myc transcription (23,25),
it can indirectly be inferred that when treatment with JQ1 leads to
reduced c-Myc levels, it acts through BET proteins. Many studies
have reported that JQ1 inhibits c-Myc and induces apoptosis
(17,23,41,44).
The findings of the present study were similar, suggesting that JQ1
inhibits BET protein and leads apoptosis via the inhibition of
c-Myc.
Previous studies have reported the antitumor effects
of BET inhibitors in ovarian and endometrial cancers. For example,
Karakashev et al (30) reported
that BET inhibitors enhance the sensitivity to PARP inhibitors in
homologous recombination-proficient ovarian cancer. Liu et al
(31) reported that i-BET151 (a BET
inhibitor), when administered to ovarian cancer cell lines,
promoted CD8-positive T cell infiltration and exhibited antitumor
effects. Furthermore, i-BET151 was reported to demonstrate a
synergistic effect with cisplatin by reducing survivin and B-cell
lymphoma 2 levels in ovarian cancer cell line (32). Notably, the BET inhibitors GS-5829
and GS626510 have demonstrated antitumor effects against serous
carcinoma cell lines, which belong to a special histological type
of endometrial cancer (45). BET
proteins regulate c-Myc levels, which are dysregulated in several
cancers (46). c-Myc is known to be
a critical transcription factor that regulates cell proliferation,
differentiation and apoptosis (47,48).
Previous studies have reported that suppression of c-Myc induces
apoptosis in cancer (17,23,47,48).
BET inhibitors, such as JQ1, have also been reported to suppress
c-Myc expression in several cancers (including ovarian cancer) and
exhibit antitumor effects (44,49).
Qiu et al (41) reported that
treating PTEN-positive uterine carcinoma with JQ1 inhibited cell
growth and decreased c-Myc expression, which is consistent with the
results of the present study.
Furthermore, a total of >50% of OECs have
ARID1A mutations (3). Berns
et al (40) reported that BET
inhibitors were associated with synthetic lethality in
ARID1A mutant ovarian cancer cells by reducing the available
levels of SWI/SNF members, such as ARID1B. However, the
present study revealed no differences in the antitumor effects of
BET inhibitors on the ARID1A status of EEC and OEC cell
lines.
The present study has certain limitations: i) As
experiments on c-Myc overexpression were not performed, the
mechanism of action of JQ1 was not elucidated. Future studies
should perform additional experiments using BRD4 siRNA and
c-Myc overexpression; ii) as the inhibition of c-Myc is
known to induce apoptosis, the findings of the present study
suggest that JQ1 may induce apoptosis via inhibition of c-Myc, but
further studies are needed to prove this; and iii) analyses using
TCGA database were performed, but not experiments using clinical
samples. Therefore, future studies should assess the expression of
BRD4 and its association with prognosis using clinical samples.
In conclusion, several reports on the antitumor
effects of BET inhibitors in endometrial carcinoma have been
published. Considering the findings of the present study, the BET
inhibitor JQ1 may be effective in SEOC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present work was supported by a Grant-in-Aid for Scientific
Research (B) from the Ministry of Education, Culture, Sports,
Science and Technology of Japan (grant no. 20H03820). The present
study was also partially supported by BRIDGE programs for Bridging
the gap between R&d and the IDeal society (society 5.0) and
Generating Economic and social value (grant no.
J0125252401h0001).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
ST and KS conceived and designed the study. ST, KS,
RHac, ES, NT, YT and FI designed the experiments. Experiments were
performed by ST and YJ. YJ cooperated with the additional
experiment. ST and KS confirm the authenticity of all the raw data.
ST, KS, HH, TF, AT, YM, TI, MM, KA, MK, SK, RHam, OWH, KO, YH and
YO contributed to the analysis and interpretation of the results.
KS, KA, MK, SK, RHam, OWH, KO, YH and YO reviewed and revised the
manuscript for important intellectual content. Technical and
material support was provided by RH, ES, NT, YT, FI, TF and AT. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
KO was supported by grants from Daiichi Sankyo Co.,
Ltd. and AstraZeneca plc, and lecture fees from Chugai
Pharmaceutical Co., Ltd. and AstraZeneca plc. All other authors
declare that they have no competing interests.
Glossary
Abbreviations
Abbreviations:
|
BET
|
bromodomain and extra-terminal
domain
|
|
BRD
|
bromodomain
|
|
DMSO
|
dimethyl sulfoxide
|
|
EEC
|
endometrial endometrioid carcinoma
|
|
FBS
|
fetal bovine serum
|
|
OEC
|
ovarian endometrioid carcinoma
|
|
PARP
|
poly ADP ribose polymerase
|
|
PBS
|
phosphate-buffered saline
|
|
PVDF
|
polyvinylidene difluoride
|
|
SDS
|
sodium dodecyl sulfate
|
|
SEOC
|
simultaneous endometrial and ovarian
cancer
|
References
|
1
|
Karst AM and Drapkin R: Ovarian cancer
pathogenesis: A model in evolution. J Oncol.
2021:9323712010.PubMed/NCBI
|
|
2
|
Seidman JD, Horkayne-Szakaly I, Haiba M,
Boice CR, Kurman RJ and Ronnett BM: The histologic type and stage
distribution of ovarian carcinomas of surface epithelial origin.
Int J Gynecol Pathol. 23:41–44. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Winterhoff B, Hamidi H, Wang C, Kalli KR,
Fridley BL, Dering J, Chen HW, Cliby WA, Wang HJ, Dowdy S, et al:
Molecular classification of high grade endometrioid and clear cell
ovarian cancer using TCGA gene expression signatures. Gynecol
Oncol. 141:95–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Coburn SB, Bray F, Sherman ME and Trabert
B: International patterns and trends in ovarian cancer incidence,
overall and by histologic subtype. Int J Cancer. 140:2451–2460.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Murakami K, Kotani Y, Nakai H and
Matsumura N: Endometriosis-associated ovarian cancer: The origin
and targeted therapy. Cancers (Basel). 12:16762020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakamura K, Banno K, Yanokura M, Iida M,
Adachi M, Masuda K, Ueki A, Kobayashi Y, Nomura H, Hirasawa A, et
al: Features of ovarian cancer in Lynch syndrome (Review). Mol Clin
Oncol. 2:909–916. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Łaniewski P, Ilhan ZE and Herbst-Kralovetz
MM: The microbiome and gynaecological cancer development,
prevention and therapy. Nat Rev Urol. 17:232–250. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rodolakis A, Thomakos N, Akrivos N,
Sotiropoulou M, Ioannidis I, Haidopoulos D, Vlachos G and Antsaklis
A: Clinicopathologic insight of simultaneously detected primary
endometrial and ovarian carcinomas. Arch Gynecol Obstet.
285:817–821. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Lu Q and Chang C: Epigenetics in
health and disease. Adv Exp Med Biol. 1253:3–55. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Margueron R, Trojer P and Reinberg D: The
key to development: Interpreting the histone code? Curr Opin Genet
Dev. 15:163–176. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brownell JE, Zhou J, Ranalli T, Kobayashi
R, Edmondson DG, Roth SY and Allis CD: Tetrahymena histone
acetyltransferase A: a homolog to yeast Gcn5p linking histone
acetylation to gene activation. Cell. 84:843–851. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zaware N and Zhou MM: Bromodomain biology
and drug discovery. Nat Struct Mol Biol. 26:870–879. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Taunton J, Hassig CA and Schreiber SL:
Mammalian histone deacetylase related to the yeast transcriptional
regulator Rpd3p. Science. 272:408–411. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu SY and Chiang CM: The double
bromodomain-containing chromatin adaptor brd4 and transcriptional
regulation. J Biol Chem. 282:13141–13145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Belkina AC and Denis GV: BET domain
co-regulators in obesity, inflammation and cancer. Nat Rev Cancer.
12:465–477. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang L, Zhang Y, Shan W, Hu Z, Yuan J, Pi
J, Wang Y, Fan L, Tang Z, Li C, et al: Repression of BET activity
sensitizes homologous recombination-proficient cancers to PARP
inhibition. Sci Transl Med. 9:eaal16452017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mertz JA, Conery AR, Bryant BM, Sandy P,
Balasubramanian S, Mele DA, Bergeron L and Sims RJ III: Targeting
MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl
Acad Sci USA. 108:16669–16674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Doroshow DB, Eder JP and LoRusso PM: BET
inhibitors: A novel epigenetic approach. Ann Oncol. 28:1776–1787.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expres- sion data using real-time quantitative PCR
and the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cancer Genome Atlas Research Network, .
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
The Cancer Genome Atlas Research Network,
. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou S, Zhang S, Wang L, Huang S, Yuan Y,
Yang J, Wang H, Li X, Wang P, Zhou L, et al: BET protein inhibitor
JQ1 downregulates chromatin accessibility and suppresses metastasis
of gastric cancer via inactivating RUNX2/NID1 signaling.
Oncogenesis. 9:332020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Delmore JE, Issa GC, Lemieux ME, Rahl PB,
Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et
al: BET bromodomain inhibition as a therapeutic strategy to target
c-Myc. Cell. 146:904–917. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen YR, Ouyang SS, Chen YL, Li P, Xu HW
and Zhu SL: BRD4/8/9 are prognostic biomarkers and associated with
immune infiltrates in hepatocellular carcinoma. Aging (Albany NY).
12:17541–17567. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dhanasekaran R, Deutzmann A,
Mahauad-Fernandez WD, Hansen AS, Gouw AM and Felsher DW: The MYC
oncogene-the grand orchestrator of cancer growth and immune
evasion. Nat Rev Clin Oncol. 19:23–36. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Donati B, Lorenzini E and Ciarrocchi A:
BRD4 and Cancer: Going beyond transcriptional regulation. Mol
Cancer. 17:1642018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hai R, He L, Shu G and Yin G:
Characterization of histone deacetylase mechanisms in cancer
development. Front Oncol. 11:7009472021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ren Q and Gao W: Current status in the
discovery of dual BET/HDAC inhibitors. Bioorg Med Chem Lett.
31:1276712021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Andrikopoulou A, Liontos M, Koutsoukos K,
Dimopoulos MA and Zagouri F: Clinical perspectives of BET
inhibition in ovarian cancer. Cell Oncol (Dordr). 44:237–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Karakashev S, Zhu H, Yokoyama Y, Zhao B,
Fatkhutdinov N, Kossenkov AV, Wilson AJ, Simpkins F, Speicher D,
Khabele D, et al: BET bromodomain inhibition synergizes with PARP
inhibitor in epithelial ovarian cancer. Cell Rep. 21:3398–3405.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu A, Fan D and Wang Y: The BET
bromodomain inhibitor i-BET151 impairs ovarian cancer metastasis
and improves antitumor immunity. Cell Tissue Res. 374:577–585.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Momeny M, Eyvani H, Barghi F, Ghaffari HS,
Javadikooshesh S, Hassanvand Jamadi R, Esmaeili F, Alishahi Z,
Zaghal A, Bashash D, et al: Inhibition of the bromodomain and
extra-terminal domains reduces the growth and invasive
characteristics of chemoresistant ovarian carcinoma cells.
Anticancer Drugs. 29:1011–1020. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen S, Li Y, Qian L, Deng S, Liu L, Xiao
W and Zhou Y: A review of the clinical characteristics and novel
molecular subtypes of endometrioid ovarian cancer. Front Oncol.
11:6681512021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ishizaka A, Taguchi A, Tsuruga T, Maruyama
M, Kawata A, Miyamoto Y, Tanikawa M, Ikemura M, Sone K, Mori M, et
al: Endometrial cancer with concomitant endometriosis is highly
associated with ovarian endometrioid carcinoma: A retrospective
cohort study. BMC Women's Health. 22:3322022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Symons LK, Miller JE, Kay VR, Marks RM,
Liblik K, Koti M and Tayade C: The immunopathophysiology of
endometriosis. Trends Mol Med. 24:748–762. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sampson JA: Metastatic or embolic
endometriosis, due to the menstrual dissemination of endometrial
tissue into the venous circulation. Am J Pathol. 3:93–110.43.
1927.PubMed/NCBI
|
|
37
|
Wendel JRH, Wang X and Hawkins SM: The
endometriotic tumor microenvironment in ovarian cancer. Cancers
(Basel). 10:2612018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
McConechy MK, Ding J, Senz J, Yang W,
Melnyk N, Tone AA, Prentice LM, Wiegand KC, McAlpine JN, Shah SP,
et al: Ovarian and endometrial endometrioid carcinomas have
distinct CTNNB1 and PTEN mutation profiles. Mod Pathol. 27:128–134.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bauer K, Berger D, Zielinski CC, Valent P
and Grunt TW: Hitting two oncogenic machineries in cancer cells:
Cooperative effects of the multi-kinase inhibitor ponatinib and the
BET bromodomain blockers JQ1 or dBET1 on human carcinoma cells.
Oncotarget. 9:26491–26506. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Berns K, Caumanns JJ, Hijmans EM,
Gennissen AMC, Severson TM, Evers B, Wisman GBA, Jan Meersma G,
Lieftink C, Beijersbergen RL, et al: ARID1A mutation sensitizes
most ovarian clear cell carcinomas to BET inhibitors. Oncogene.
37:4611–4625. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qiu H, Li J, Clark LH, Jackson AL, Zhang
L, Guo H, Kilgore JE, Gehrig PA, Zhou C and Bae-Jump VL: JQ1
suppresses tumor growth via PTEN/PI3K/AKT pathway in endometrial
cancer. Oncotarget. 7:66809–66821. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pang Y, Bai G, Zhao J, Wei X, Li R, Li J,
Hu S, Peng L, Liu P and Mao H: The BRD4 inhibitor JQ1 suppresses
tumor growth by reducing c-Myc expression in endometrial cancer. J
Transl Med. 20:3362022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Filippakopoulos P, Qi J, Picaud S, Shen Y,
Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et
al: Selective inhibition of BET bromodomains. Nature.
468:1067–1073. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qiu H, Jackson AL, Kilgore JE, Zhong Y,
Chan LL, Gehrig PA, Zhou C and Bae-Jump VL: JQ1 suppresses tumor
growth through downregulating LDHA in ovarian cancer. Oncotarget.
6:6915–6930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bonazzoli E, Predolini F, Cocco E, Bellone
S, Altwerger G, Menderes G, Zammataro L, Bianchi A, Pettinella F,
Riccio F, et al: Inhibition of BET bromodomain proteins with
GS-5829 and GS-626510 in uterine serous carcinoma, a biologically
aggressive variant of endometrial cancer. Clin Cancer Res.
24:4845–4853. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sarnik J, Popławski T and Tokarz P: BET
proteins as attractive targets for cancer therapeutics. Int J Mol
Sci. 22:111022021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dang CV, Le A and Gao P: MYC-induced
cancer cell energy metabolism and therapeutic opportunities. Clin
Cancer Res. 15:6479–6483. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Vita M and Henriksson M: The Myc
oncoprotein as a therapeutic target for human cancer. Semin Cancer
Biol. 16:318–330. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Luan W, Pang Y, Li R, Wei X, Jiao X, Shi
J, Yu J, Mao H and Liu P: Akt/mTOR-mediated autophagy confers
resistance to BET inhibitor JQ1 in ovarian cancer. Onco Targets
Ther. 12:8063–8074. 2019. View Article : Google Scholar : PubMed/NCBI
|