Beyond its role in regulating normal cellular
activity, FAT1 is one of the most commonly mutated genes in types
of human cancer (14–17). Over the past 20 years, studies have
shown that FAT1 regulates various signaling pathways (18–20),
including the Wnt/β-catenin, Hippo and MAPK/ERK pathways, thereby
affecting tumor-cell proliferation, migration, invasion (21–24),
stemness and epithelial-mesenchymal transition (EMT) (25,26).
Given the large size of FAT1 mRNA and the 49.2 kDa protein it
encodes, understanding the function of FAT1 protein is challenging
(27). Currently, the understanding
of FAT1′s biological functions and the precise downstream signaling
pathways that it mediates is limited, but increasing interest in
its role in cancer suggests that FAT1 is an emerging cancer
biomarker and a potential target for new therapies or monitoring
(28).
A recent study conducted a comprehensive pan-cancer
analysis of FAT1, utilizing data from The Cancer Genome Atlas and
Gene Expression Omnibus, to explore its potential oncogenic
mechanisms across 33 types of cancer (29). It was found that FAT1 is highly
expressed in a large proportion of tumors, significantly associated
with prognosis and has a mutation rate of >10% in >10 types
of cancers (29), such as lymphoid
neoplasm diffuse large B-cell lymphoma, lung adenocarcinoma, lung
squamous cell, uterine corpus endometrial, bladder urothelial, and
head and neck squamous cell carcinoma (HNSCC). In numerous types of
cancer, FAT1 mRNA expression levels are significantly associated
with EMT phenotype-related marker genes, as well as with tumor
exosomes (29), immune cells
(30), methylation (31), hypoxia-related mutations (32) and autophagy marker genes (29). Considering the critical role of FAT1
in tumorigenesis and progression, this review discusses current
research on FAT1 in both solid tumors and hematological
malignancies. It focuses particularly on tumor types most closely
associated with FAT1 in solid tumors, aiming to deepen the
understanding of its role in cancer and provide insights for future
research directions.
In 2020, it was estimated that there would be over
600,000 new cases of ESCC and 544,000 deaths worldwide, with nearly
half of these cases occurring in China (33,34).
Whole-genome sequencing studies have identified FAT1 as one of the
frequently mutated genes in ESCC (35,36). A
Chinese study involving 225 patients with ESCC reported a FAT1
mutation frequency of 16% (37),
which is consistent with results from another study (36), indicating that FAT1 is one of the
most commonly mutated genes in ESCC and may be a key driver of
tumorigenesis and progression. Studies have shown that most FAT1
mutations occur in the cadherin domain and FAT1 expression is
significantly reduced in ESCC tissues (38,39).
In vitro studies have demonstrated that knockdown of FAT1
reduces cell adhesion, increases cell elasticity and accelerates
cell migration and invasion (39),
which suggests that FAT1 may serve a key role in inhibiting cell
proliferation, migration and invasion in ESCC, potentially acting
as a tumor suppressor gene (40).
Through chromatin immunoprecipitation and luciferase
reporter gene assays, it has been demonstrated that FAT1
transcription is regulated by E2F transcription factor 1 (E2F1),
which binds to the FAT1 promoter region. Depletion of E2F1 reduces
FAT1 transcription activity and mRNA expression levels, indicating
that FAT1 is a direct transcriptional target of E2F1 (41). A further study has shown that FAT1
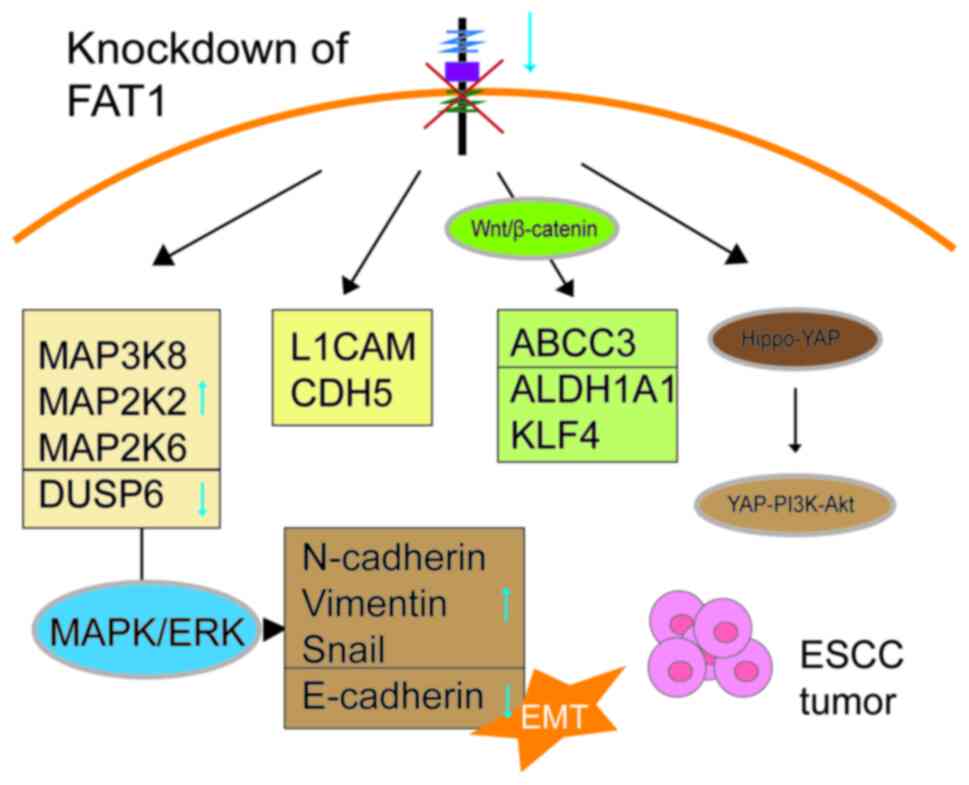
regulates multiple pathways in ESCC, including the MAPK signaling
pathway (42). Knockdown of FAT1 in
ESCC cells increases mRNA expression levels of MAPK kinase kinase 8
(MAP3K8), MAP2K2, MAP2K6 and L1 cell adhesion molecule and cadherin
5 involved in cell adhesion processes, and decreases mRNA
expression levels of the MAPK signaling pathway inactivator dual
specificity phosphatase 6, demonstrating the regulatory role of
FAT1 in the MAPK signaling pathway and cell adhesion (41). In addition, FAT1 influences EMT in
ESCC cells through the MAPK pathway. FAT1 knockdown reduces
E-cadherin expression levels, while increasing N-cadherin, vimentin
and Snail expression levels, suggesting that FAT1 regulates EMT in
ESCC cells via the MAPK/ERK pathway (42–44). A
study has also found that FAT1 downregulation enhances stemness and
cisplatin resistance in ESCC cells through the Wnt/β-catenin
signaling pathway. Therefore, FAT1 and its downstream gene ATP
binding cassette subfamily C member 3 may be potential targets to
overcome cisplatin resistance in ESCC (45).
In addition to the MAPK and Wnt/β-catenin signaling
pathways, studies have found that FAT1 mutations influence ESCC
drug resistance and prognosis through the Hippo-Yes1-associated
transcriptional regulator (YAP) signaling pathway. A targeted
sequencing study of 201 patients with ESCC identified a specific
molecular subtype called FAT/FRY, characterized by mutations in
FAT1, FAT3 and FRY microtubule binding protein (FRY). The FAT/FRY
subtype showed poor prognosis in multiple ESCC cohorts,
characterized by Hippo pathway inactivation, hypoxia, chemotherapy
resistance and high infiltration of CD8+ T cells and activated
dendritic cells (46). Furthermore,
a drug response analysis from the Genomics of Drug Sensitivity in
Cancer database conveyed that ESCC cell lines with FAT/FRY
mutations were more sensitive to the PIK3Ca inhibitor alpelisib.
Alpelisib mitigates tumor growth by inhibiting the phosphorylation
of PI3K downstream targets such as AKT and the interaction between
the PI3K/AKT pathway and other pathways, such as the Hippo pathway,
may affect drug efficacy, warranting further research to determine
whether FAT/FRY-type ESCC is more sensitive to alpelisib (46). Another study found that
downregulation of FAT1 and protein tyrosine phosphatase
non-receptor type 14 (PTPN14) was associated with upregulation of
YAP1 in ESCC tissues, indicating that FAT1 may suppress ESCC
progression and chemotherapy resistance through upregulation of
PTPN14 and inhibition of YAP1 and MYC, thus involving the Hippo-YAP
signaling pathway in the malignant progression and chemotherapy
resistance of ESCC (47) (Fig. 1).
HNSCC is a severe and often fatal disease that
affects the upper respiratory and digestive functions of patients,
accounting for ~4.6% of cancer-associated deaths worldwide
(48). As the sixth most common
cancer globally, HNSCC has the highest FAT1 gene mutation rate
among various solid tumors. However, the role of FAT1 gene
mutations in the pathogenesis and progression of HNSCC and the
mechanisms of associated signaling pathway activation remain
limited (49–54). The mutation rate of the FAT1 gene in
patients with oral squamous cell carcinoma (OSCC), a subtype of
HNSCC, is ~17% (55). A study
conducted in Korea detected genetic alterations in 44 cases of
advanced oral tongue squamous cell carcinoma, with a FAT1 mutation
rate of 9.1% (56). In addition, a
study conducted in Taiwan performed whole-exome sequencing on 120
samples of OSCC tumors and corresponding normal tissues and
identified inactivating FAT1 mutations in 35% of tumors (57). These findings suggest that FAT1 gene
mutations may serve a carcinogenic or driver role in OSCC and other
HNSCCs (58–61). The differences in reported FAT1 gene
mutation rates among different studies may be due to tumor
heterogeneity or variations in patient cohorts. Furthermore,
studies indicate differences in tumor biology and genomics between
different ethnic populations. For instance, Chaudhary et al
(62) identified increased mutation
frequencies in key driver genes such as FAT1 and TP53 in African
American patients with HNSCC compared with human papillomavirus
(HPV)-positive or negative white patients. The higher FAT1 mutation
frequency in African American patients was significantly associated
with decreased survival rates, partially explaining the worse
prognosis of HNSCC in this population compared with white
patients.
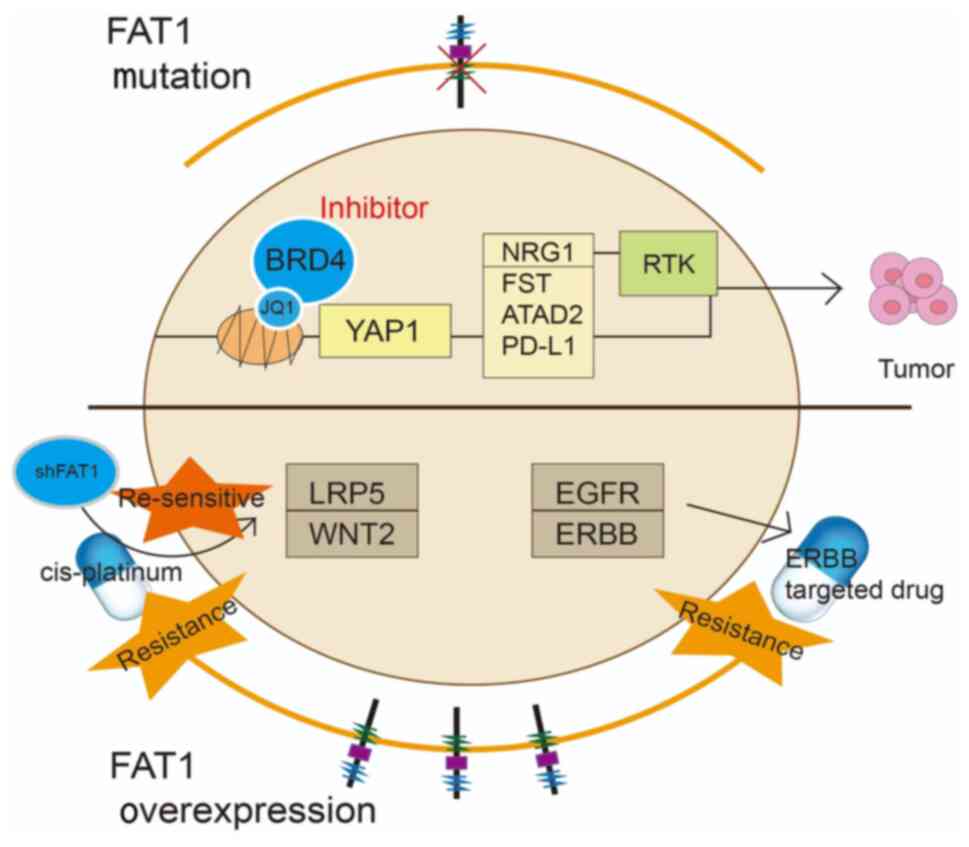
Studies have found that ~50% of patients with HNSCC
have somatic alterations in the Hippo-YAP pathway (63,64).
In particular, FAT1 gene mutations contribute to the activation of
YAP1 transcription, with the FAT1/YAP1 signaling axis directly
involved in the development of HNSCC. Proteogenomic and drug
screening studies across various types of cancer models have shown
that FAT1 mutations sensitize HNSCC cells to JQ1, a bromodomain and
extra-terminal domain (BET) family (BRD2, BRD3 and BRD4) inhibitor.
In contrast to other types of cancer with Hippo pathway variations,
such as ESCC and lung squamous cell carcinoma, FAT1 mutations in
HNSCC confer high specificity and sensitivity to BET inhibitors.
Further studies have demonstrated that FAT1 knockdown increases
cell sensitivity to JQ1 and lowers the IC50. Epigenomic
analyses demonstrated that FAT1 mutations in HNSCC lead to
increased YAP1 nuclear translocation and activation of multiple
cancer-related genes such as neuregulin 1 (NRG1), follistatin,
ATPase family AAA domain containing 2 and programmed cell death
ligand 1 (PD-L1). Persistent activation of NRG1 mediates receptor
tyrosine kinase pathway activation, promoting tumor development and
drug resistance. Therefore, combining BET inhibitors,
erythroblastic leukemia viral oncogene (ErbB) inhibitors or immune
checkpoint inhibitors (ICIs) may offer potential therapeutic
opportunities for patients with HNSCC with FAT1 mutations (65).
FAT1 mutations in head and neck cancer are closely
associated with tumor progression and survival. Knockout of
endogenous FAT1 expression and exogenous expression of key domains
of FAT1 demonstrate that FAT1 can inhibit the migration and
invasion abilities of HNSCC cells (66). Further functional analysis suggests
that nonsense mutations in FAT1 result in the loss of its tumor
suppressive function, while FAT1 mutations and low expression
levels are significantly associated with lymph node involvement,
lymphovascular invasion and tumor recurrence (67). Treatment of the HNSCC cell line
HO-1-u-1 with PTC124 (also known as Ataluren), a drug used for
treating genetic diseases mediated by nonsense mutations,
demonstrated that PTC124 could re-express functional FAT1 and
thereby rescue FAT1 function in HNSCC cells with nonsense mutations
and inhibit cell proliferation (68). Another study used two small
interfering RNAs (siRNAs) to reduce FAT1 expression levels in OSCC
cell lines in vitro to demonstrate that FAT1 silencing
inhibited OSCC cell proliferation, stemness, cell cycle and
migration, while promoting early and late apoptosis (69). The discrepancy between these
findings and aforementioned reports may be due to the different
biological functions of FAT1 mutations in contrast to FAT1
expression. Bioinformatics and clinical analyses indicate that
although the four most common FAT1 mutation sites were detected in
various types of cancer, these variants were not significantly
associated with FAT1 expression levels. Thus, the correlation
between FAT1 mutations and lower FAT1 expression in tumors remains
controversial.
Lung cancer is the leading cause of cancer-related
death in both men and women worldwide. It is primarily classified
into two types: SCLC, which accounts for ~15% of lung cancer cases
and NSCLC, which accounts for ~85% of cases. NSCLC is further
divided into lung adenocarcinoma (LUAD) and lung squamous cell
carcinoma (LUSC) as the main subtypes. Despite significant
therapeutic advances over the past few decades, the recurrence and
metastasis rates of NSCLC remain high at 30–40%, with a 5-year
overall survival rate of <15%. Therefore, there is a pressing
need to explore the genetic mechanisms underlying NSCLC, identify
prognostic biomarkers and discover new therapeutic targets.
Research on FAT1 in lung cancer has primarily focused on NSCLC. A
recent study utilized next-generation sequencing (NGS) technology
to identify high-frequency mutant genes in 110 Chinese patients
with NSCLC. The results showed a FAT1 mutation rate of 12.90%, one
of the frequently mutated genes of those analyzed (73). Another study used paired tumor and
adjacent lung tissue samples from 112 surgically resected patients
with initial treatment for comprehensive proteogenomic
characterization of SCLC, further demonstrating the role of FAT1
mutations in carcinogenesis with same findings as above (74). Recent findings suggest that FAT1
deletion in LUSC may lead to an enhanced EMT state, tumor stemness
and metastatic ability (25),
providing further insight into the potential role and therapeutic
targets of FAT1 in lung cancer.
Over the past decade, the identification of key
mutations and the introduction of immune checkpoint blockade drugs
have revolutionized the therapeutic landscape of NSCLC. Biomarkers
such as tumor mutation burden (TMB), T-cell infiltration and PD-L1
protein expression levels in tumor tissues have been proposed as
indicators of immune therapy response (75). Studies have indicated that
co-mutations of low-density lipoprotein receptor-related protein 1B
and FAT1 may serve as a set of potential predictive factors to
guide immunotherapy in NSCLC (76).
It was reported that patients with FAT1-mutated NSCLC may have
higher sustained clinical benefits and objective response rates
than FAT1-nonmutated (77). These
results were validated in other independent datasets, suggesting
that FAT1 mutations could be a robust biomarker for predicting
immunotherapy efficacy (77). A
Chinese study also reported that patients with NSCLC with FAT1
mutations might be associated with improved ICI treatment outcomes.
Genomic and immunological analyses showed that patients with NSCLC
with FAT1 mutations often had a high TMB, increased
immune-responsive cell infiltration, decreased immune-suppressive
cell infiltration and enrichment of IFN and cell cycle-associated
pathways. FAT1 mutations are associated with improved
immunogenicity and ICIs efficacy, making it a potential biomarker
for the selection of patients for immunotherapy (78). A study proposed a model using lung
cancer patient genetic mutation profiles, including FAT1 mutations,
to predict the survival of patients with various types of cancer
using immunotherapy. This predictive model effectively identifies
patients with various types of cancer who can benefit from ICIs
treatment, potentially providing notable assistance in clinical
oncology treatment (79).
HCC is a common type of cancer and the third leading
cause of cancer-related death worldwide. HCC poses significant
treatment challenges with a 10–20% 5-year overall survival rate,
necessitating further research to elucidate the molecular
mechanisms of HCC progression and identify new therapeutic targets
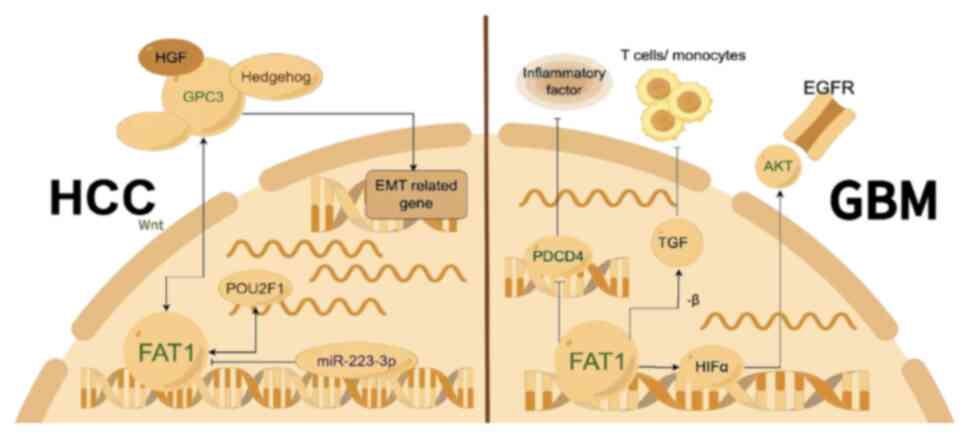
(32,80). Zhu et al (81) found that the POU class 2 homeobox 1
(POU2F1) transcription factor is significantly upregulated in HCC
tumor tissues and cell lines compared with healthy tissues,
promoting HCC cell growth and metastasis, with FAT1 acting
downstream of POU2F1. It was demonstrated that FAT1 is strongly
positively expressed in HCC and weakly expressed in the normal
liver, with FAT1 upregulation positively associated with lower
overall survival rates. In vitro experiments demonstrated
that transfection of targeted FAT1 shRNA into HepG2 and SNU-423
cells significantly reduced their migration and invasion. In
addition, reducing FAT1 levels could reverse POU2F1
overexpression-mediated HCC cell proliferation, colony formation,
migration and invasion, suggesting that FAT1 independently
regulates HCC metastasis and is a potential new therapeutic target
for HCC. Further research indicated that FAT1 is highly expressed
in liver cancer tissues and human liver cancer cell lines, whereas
miR-223-3p is lowly expressed. Dual-luciferase assay results showed
that miR-223-3p inhibits HCC proliferation, migration, invasion and
EMT by targeting and downregulating FAT1 expression (82,83).
Glypican-3 (GPC3) is a cell surface heparan sulfate
proteoglycan that interacts with several extracellular signaling
molecules, including Wnt, hepatocyte growth factor (HGF) and
Hedgehog, making it an emerging therapeutic target for HCC
(84). A study indicated that FAT1,
as a novel GPC3-interacting protein, binds to the C-terminal region
of GPC3 (Q14517, residues 3,662-4,181), which contains a putative
receptor tyrosine phosphatase-like domain, a laminin G-like domain
and five EGF-like domains. GPC3 and FAT1 were found to have similar
expression patterns in HCC cells, including enhanced expression and
upregulation under hypoxic conditions, and can regulate EMT-related
genes such as Snail, vimentin and E-cadherin, promoting HCC cell
migration. This research provides preliminary evidence for a novel
mechanism by which GPC3 and FAT1 can promote HCC cell migration
(85). Overall, FAT1 expression
levels are closely associated with HCC occurrence and development.
Further exploration of FAT1 mechanisms and its associations with
factors such as hypoxia, HGF and methyl donor
S-adenosyl-L-methionine is crucial for HCC diagnosis, treatment and
prognosis.
Glioblastoma (GBM) invasiveness is influenced by a
hypoxic microenvironment through hypoxia-inducible factor (HIF)1α,
while the tumor microenvironment is significantly affected by FAT1
(86,87). A study under severe hypoxic
conditions explored the interaction between FAT1 and HIF1α in
primary tumors. Findings in GBM tumor specimens indicated a
positive association between FAT1 and HIF1α and its target genes,
highlighting the importance of the FAT1-HIF1α signaling axis in
glioma cells (88). Specific FAT1
siRNA-transfected GBM cell lines were cultured under hypoxia and it
was found that reducing endogenous FAT1 expression significantly
decreased HIF1α and its downstream target gene expression levels,
which also notably reduced the invasiveness of GBM cells. This
reduction is attributed to impaired EGFR-AKT signaling and
increased von Hippel-Lindau-dependent proteasomal degradation of
HIF1α, further suggesting that FAT1 could be a novel potential
target for GBM treatment (89). A
study also found that FAT1, along with EMT markers (such as Snail,
lysyl oxidase, vimentin and N-cadherin), stemness markers (such as
sex-determining region y-box 2, POU class 5 homeobox 1, nestin and
RE1-silencing transcription factor) and hypoxia markers (such as
HIF1α, VEGF, phosphoglycerate kinase 1 and carbonic anhydrase IX)
are upregulated in at least 39% of GBM cases. The glioma cell lines
U87MG and A172 that were exposed to severe hypoxia (0.2%
O2) showed increased mRNA expression levels of FAT1,
EMT, stemness and hypoxia markers compared with cells cultured
under normoxia (20% O2). Furthermore, FAT1 knockdown in
U87MG and A172 cells cultured under both severe hypoxia and
normoxia conditions significantly reduced the expression of EMT and
stemness markers, suggesting that FAT1 may regulate these markers
through independent action from HIF1α, thus suggesting a novel
mechanism by which FAT1 regulates EMT/stemness in hypoxic GBM
(90).
In GBM, high expression of FAT1 affects the
expression of inflammatory factors. Research using high
FAT1-expressing grade IV glioma cell lines, such as U87MG and A172,
showed that reducing FAT1 expression levels using an siRNA
decreased cell migration and invasion capabilities, and also
increased the expression levels of the tumor suppressor gene
programmed cell death 4 (PDCD4). Increased PDCD4 expression levels
suppress the phosphorylation of c-Jun, thereby weakening activator
protein (AP)-1 transcriptional activity, which leads to decreased
expression levels of AP-1 target genes such as MMP3, VEGF-C and
plasminogen activator, urokinase, inflammatory factor
cyclooxygenase-2 and cytokines IL-1β and IL-6. This demonstrated a
novel FAT1-mediated signaling mechanism that acts as an upstream
regulator of oncogenic and inflammatory pathways in GBM by
modulating PDCD4 activity (91). A
recent study has found that FAT1 is involved in regulating the
expression of anti-inflammatory mediators TGF-β1/2 in resected
human gliomas, primary glioma cultures and other cancer cell lines,
with FAT1 expression correlating positively with TGF-β1/2 level in
various tumors. FAT1 knockdown using an siRNA led to reduced
expression and secretion of TGF-β1/2, increasing the chemotacticity
of THP-1 monocytes to the supernatants of tumor cells transfected
with siFAT1, which resulted in immune suppression. Additionally,
FAT1 expression was positively correlated with the expression of
myeloid-derived suppressor cell (MDSC) markers in gliomas,
suggesting that FAT1 may serve a role in MDSC-mediated
immunosuppression. Therefore, FAT1 expression levels in various
types of cancer are inversely associated with the infiltration of
tumor-suppressing immune cells (such as monocytes and T cells) and
positively correlated with tumor-promoting immune cells (such as
MDSCs). FAT1 serves a significant role in cancer immune evasion,
particularly through promoting an immunosuppressive
microenvironment in GBM and other types of cancer via TGF-β1/2
(Fig. 3) (92).
Significant progress has been made in understanding
the role of FAT1 in breast cancer resistance (93,94).
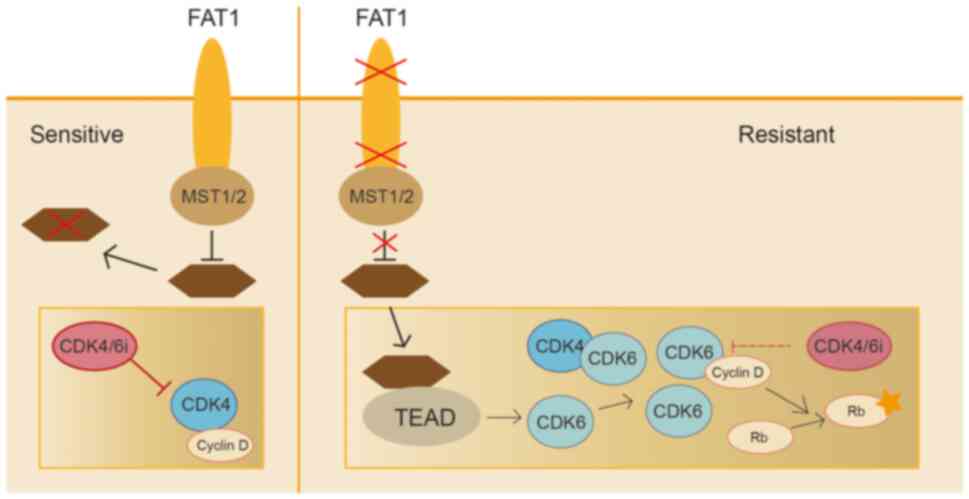
Studies indicated that cyclin-dependent kinase (CDK4/6) inhibitors
are somewhat effective against breast cancer, but resistance is
notably high. Genomic analysis of 348 patients with estrogen
receptor-positive (ER+)/HER2- breast cancer showed that the absence
of FAT1 leads to notable resistance to CDK4/6 inhibitors. It was
found that loss of FAT1 significantly increases CDK6 expression
levels, while inhibition of CDK6 could restore sensitivity to
CDK4/6 inhibitors. Further research indicated that the induction of
CDK6 is mediated by the Hippo pathway, with the accumulation of YAP
and TAZ transcription factors on the CDK6 promoter enhancing
resistance to CDK4/6 inhibitors. These findings highlight the
anticancer role of the Hippo signaling pathway in ER+ breast cancer
and identify the absence of FAT1 as a mechanism leading to
resistance to CDK4/6 inhibitors (95,96)
(Fig. 4).
Aldehyde dehydrogenase 1 (ALDH1) is considered a
marker of breast cancer stem cells and its enzymatic activity is
crucial for the regulation of cancer stem cells. A recent study
found that KK-LC-1 (also known as CT83 or Cxorf61), a type of
testicular cancer antigen, can interact directly with FAT1, leading
to its ubiquitin-mediated proteasomal degradation. This process
regulates the expression levels of FAT1, which in turn influences
the stemness of ALDH+ cells in triple-negative breast cancer
(TNBC). Degradation of FAT1 affected the Hippo pathway and led to
YAP1 nuclear translocation and ALDH1A1 transcription. These
findings identified the KK-LC-1-FAT1-Hippo-ALDH1A1 pathway as a
potential therapeutic target in TNBC, providing a novel research
direction for the treatment of breast cancer (97).
A study indicated that, in addition to the role in
breast cancer, FAT1 is involved in the development and progression
of various other malignancies, including bladder, prostate,
uterine, colorectal and gastric cancer (GC) (98,99).
Early genome-wide sequencing identified recurrent
protein-inactivating mutations in FAT1 among 14 different grades
and stages of bladder cancer (100). An in vitro study reported
that S100 calcium binding protein A14 (S100A14) promotes the
expression of FAT1 and activates the Hippo pathway, thereby
inhibiting the growth and EMT of prostate cancer. In vivo
results confirmed that S100A14, mediated through the FAT1-driven
Hippo pathway, inhibits tumor growth in mouse prostate cancer cells
(101). Evidence also closely
associates FAT1 with the progression of GC (102). A study showed that FAT1 is
upregulated in GC tissues and silencing FAT1 inhibits the oncogenic
phenotype of GC cells. A further mechanistic study indicated that
LINC00857 serves an oncogenic role in GC by regulating the
transcription factor AP-2 gamma/FAT1/AP-1 pathway (103).
Colorectal cancer (CRC) consists of tumors of the
colon, rectum and anus and represents the third most common cancer
type, accounting for 10% of new cancer cases globally with 935,173
deaths in 2020. Studies suggest that FAT1 is a key gene promoting
cancer cell migration and growth, and, compared with normal colon
tissues, is highly expressed on the plasma membrane of colon cancer
cells (104–106). The discovery of novel molecules
that can inhibit the expression of FAT1 and its downstream
signaling pathways is crucial for the development of new anti-CRC
drugs. Dehydroabietic acid (DIAP) is a specific natural product
mainly found in the Hypericum perforatum Linn. HPLC-UV
screening identified 46 DIAPs in H. perforatum Linn roots,
with compounds 2 and 6 showing potent and selective cytotoxicity
against colon cancer cells, significantly inhibiting NF-κB and FAT1
expression in HCT116 cells and promoting the novel tumor suppressor
gene PDCD4. These effects are mediated through the FAT1 signaling
pathway. Therefore, DIAPs may be further studied as a new type of
anti-CRC lead drug targeting FAT1 (107).
NGS analysis of 111 patients diagnosed with CRC
highlighted the complex heterogeneity of genetic alterations within
CRC (108). Currently,
immunotherapy is approved for CRC tumors with high microsatellite
instability. Targeted sequencing using the tumor tissues of 161
patients with CRC demonstrated that, compared with the wild-type
FAT1 gene, FAT1 gene mutation of CRC with microsatellite
instability events often occur simultaneously and showed a higher
TMB. Kyoto Encyclopedia of Genes and Genomes pathway analysis
showed that the PI3K-AKT pathway and immune pathways were altered
in CRC tissue samples with mutant FAT1. Tumor samples with FAT1
mutations from patients with CRC showed improved characteristics
for immunotherapy. Although the studies were conducted
retrospectively and further in vitro experiments are
necessary to verify the association between FAT1 mutations and the
immune environment of CRC tumors, this suggested that tumors with
FAT1 mutations may define a new subtype of CRC immunocompetence
(30,109). Therefore, in future immunotherapy
trials, FAT1 gene mutations in patients with CRC may be considered
a specific subgroup for further study.
FAT1 was initially cloned from a human T-cell ALL
cell line, indicating its expression in ALL (110,111). Feng et al (112) used targeted NGS to analyze 112
genes from 121 adult patients with ALL. In the group studied, 110
patients (90.9%) carried at least one mutation, including the five
most common mutated genes, with FAT1 at the top. In B-cell ALL
(B-ALL), FAT1 mutations are among the most common (10.75%),
suggesting that gene mutations are prevalent in adult patients with
ALL, with FAT1 mutations potentially being a pathogenic factor.
Another study involving 147 adolescent and adult patients with ALL
analyzed by NGS showed that 91.2% of the patients carried at least
one mutation, with 67.35% carrying multiple (≥2) mutations. FAT1
mutations are more common in B-ALL compared with T-ALL (113). In addition, a study on genetic
variations in pediatric T-ALL identified 302 mutations across 60
genes, with FAT1 (32.81%) showing a higher mutation frequency,
suggesting that FAT1 mutations are more common in pediatric
patients with ALL (114). These
results indicate that FAT1 mutations are common genetic alterations
in both children and adults with ALL, potentially driving the
disease's progression and possibly affecting prognosis.
In addition to the presence of FAT1 mutations, the
expression of FAT1 also serves a significant role in ALL. de Bock
et al (115) found that
FAT1 protein is expressed in various leukemia cell lines, but not
in healthy peripheral blood and bone marrow cells. Further clinical
leukemia data analysis showed that in 11% of AML, 29% of B-cell ALL
(B-ALL) and 63% of T-cell ALL (T-ALL), FAT1 transcription levels
rise significantly, and normal peripheral blood or bone marrow
cells show little or no FAT1 transcription. Furthermore, in two
independent studies using matching diagnosis-relapse samples from
children with precursor B-cell (pre-B)-ALL, high FAT1-mRNA
expression at diagnosis predicted shorter relapse-free and overall
survival. Data suggest that the expression of FAT1 in pre-B-ALL is
associated with the occurrence of relapse and can provide a
suitable therapeutic target for high-risk pre-B-ALL. Another study
on adult acute leukemia analyzed the expression levels of FAT1 in
samples from healthy donors, patients with AML, adult T-ALL and
pre-B-ALL, and various leukemia cell lines (116). In bone marrow from healthy donors,
CD34+ progenitor cells, peripheral blood and CD3+ T cells were
found not to express FAT1, whereas FAT1 was highly expressed in
bone marrow mesenchymal stromal cells from healthy donors. By
contrast, adult leukemic samples showed abnormal FAT1 expression
and FAT1 expression was associated with a more mature leukemia
immune phenotype. Further investigation demonstrated that FAT1
mutations were present in early T-ALL (25%) and thymic T-ALL (12%),
but not in T-ALL with a mature immunophenotype. No differences in
overall survival rates or duration of response were observed
between patients with mutant and normal FAT1. Although FAT1 was not
of significant prognostic value, FAT1 may be considered a potential
candidate for disease monitoring, targeted therapy and insight into
the pathogenesis of leukemia in different ALL subgroups. In
addition to the potential role of leukemia, FAT1 is also involved
in cell migration, polarity and intercellular adhesion and interact
with β-serial protein directly. High FAT1 expression levels were
identified in bone marrow mesenchymal stromal cells, suggesting
that FAT1 may serve a role in stabilizing the interaction of
leukemic cells with bone marrow niches and thymic homing.
A recent study reported that FAT1 suppresses
autophagy and proliferation levels in AML by downregulating
autophagy-related 4B (ATG4B) expression. The mutation rate of all
mutated genes in 22 patients with AML were analyzed and a high FAT1
mutation rate of 40.90% was found, which is notably higher
(124). Further analysis using the
Gene Expression Profiling Interactive Analysis database indicated
that FAT1 mRNA expression levels in AML were significantly lower
compared with the control group. These results suggest that FAT1
may serve an anti-tumor role in AML. A study using the AML cell
lines KG-1a and THP-1 demonstrated that FAT1 suppresses autophagy
in AML by inhibiting TGF-β-SMAD2/3 signal activity, thereby
reducing the expression of ATG4B and consequently inhibiting AML
proliferation. These findings suggest that the
FAT1-TGF-β-SMAD2/3-ATG4B-autophagy pathway may represent a novel
target for the development of therapeutic drugs for AML (125).
Although the FAT1 gene is described as a tumor
suppressor in various types of cancer, FAT1 mutations are
infrequently found in lymphoma entities. Peripheral T-cell lymphoma
(PTCL), not otherwise specified, is the most common subtype among
nodal peripheral T-cell lymphomas and is a tumor with strong
clinical, histological and molecular heterogeneity. However, its
genetic landscape has remained to be fully clarified. A study has
shown that a subset of patients with PTCL-NOS exhibit recurrent
mutations in the FAT1 gene, which is significant for understanding
the pathogenesis of this type of lymphoma. A large proportion of
mutations in the FAT1 gene are missense mutations rather than
frameshift insertions/deletions or nonsense mutations. Further
analysis indicated that tumors in patients with FAT1 mutations are
associated with characteristics related to growth, apoptosis, cell
migration and invasion. Patients with FAT1 mutations have a shorter
overall survival compared with those with wild-type FAT1 (126,127). Furthermore, FAT1 mutations have
also been found to be associated with poor prognosis in
angioimmunoblastic T-cell lymphoma (AITL). A study involving
detailed genetic analysis of blood samples from 64 patients with
AITL found that combinations of mutations in FAT1 with RHOA and
KDM5A are associated with poor prognosis. This emphasizes the
importance of cell-free DNA as a liquid biopsy in AITL and
demonstrates new molecular markers that may help guide molecular
diagnosis and treatment plans for patients with AITL (128).
Besides T-cell lymphoma, FAT1 mutations have also
been found in B-cell lymphomas (129). Zhao et al (130) conducted whole-exome sequencing
studies on cases of ocular adnexal mucosa-associated lymphoid
tissue lymphoma (OAML) and found that ~10% of the patients had FAT1
gene mutations, indicating that FAT1 may be involved in additional
or alternative lymphomagenesis pathways in OAML. In a retrospective
study on blastoid or pleomorphic mantle cell lymphoma (B/P-MCL),
NGS performed on samples from patients with blastoid and
pleomorphic variants was conducted and it was found that FAT1
mutations are one of the most common genetic changes in B/P-MCL. It
could be considered that FAT1 mutations may be a pathogenic factor
contributing to the aggressive manifestation in patients with
mantle cell lymphoma (131).
FAT1 is a transmembrane protein considered to serve
a significant role in the occurrence and development of tumors. The
inclusion of a large proportion of studies conducted in China in
the present review is primarily due to the increased attention
given to the FAT1 gene and the related publications in recent
years. However, the pan-cancer analysis data on FAT1 (29,31),
including a recent study identifying FAT1 as a target antigen in a
subset of de novo allograft membranous nephropathy
associated with antibody-mediated rejection (133), originate from research conducted
worldwide. These studies demonstrate that FAT1 is of interest to
the global research community as a therapeutic target and
immunotherapy biomarker for various types of cancer. Studies
indicate that FAT1 acts as a relay for signals from the
extracellular environment to the inside of the cell, regulating
various signaling pathways such as Wnt/β-catenin, Hippo and
MAPK/ERK, which affect tumor cell proliferation, migration,
invasion, stemness and EMT. In addition, FAT1 also serves key roles
in precancerous lesions, driving factors, immune escape, tumor
microenvironment, drug sensitivity, prognosis, disease monitoring,
biomarkers and target development. However, there is still
uncertainty regarding the function and clinical significance of
FAT1 in tumors. Research on the functional impact of FAT1 mutations
and expression levels has shown that FAT1 may exhibit carcinogenic
or tumor-suppressive properties in various types of tumors, with
specific effects depending on the tumor type (Table I). Although in a large proportion of
cases, mutations and expression levels of FAT1 are inversely
related, biological functions between mutations and expression
levels of FAT1 differ in certain types of tumors, such as HNSCC and
ALL. Furthermore, since FAT1 is a large cadherin, there are
operational limitations in therapeutic targeting at the protein
level and in molecular therapeutic perspectives. Also, as a gene
without clearly defined hotspots for mutations, the specific
functional changes caused by mutations in FAT1 require further
exploration.
The current understanding of FAT1 remains
incomplete, particularly concerning the functions of its large
extracellular domain. It is still unclear which upstream signals
trigger the Wnt, Hippo and MAPK/ERK pathways in relation to FAT1,
which receptors are involved in detecting these signals and how the
34 cadherin repeat sequences regulate cell-cell contact. In
addition, whether FAT1 primarily acts as an adhesion molecule or a
signaling protein and how these functions are coordinated remain to
be fully elucidated. Furthermore, the mechanisms that lead to the
release and transport of the FAT1 intracellular region to the
nucleus, whether FAT1 is localized to mitochondria in cell types
other than vascular smooth muscle cells and the impact of FAT1 on
cellular metabolism also require further research. The
identification of transcription factors and target genes that
mediate FAT1 functions and the molecular mechanisms underlying
dysregulated FAT1 expression are also key for future investigation.
Further research on the aforementioned issues and increasing the
understanding of the role of FAT1 in tumor genesis and development
may help highlight the importance of FAT1 as a diagnostic,
therapeutic and prognostic biomarker and target in clinical
applications. These studies will aid in identifying more functions
and mechanisms of FAT1, providing more theoretical support for
future development of FAT1-based cancer treatment strategies.
Although there are no studies focusing on FAT1 small molecule
targeted drugs, to the best of our knowledge, the prospects for
such drugs targeting FAT1 in the future are promising. The drug
binding, metabolic specificity and adverse events of FAT1 small
molecule targeted drugs across various organs warrant future
investigation. A recent study demonstrated that the tumor
suppressor FAT1 is dispensable for normal murine hematopoiesis
(134), suggesting that it may be
a safe and viable target for therapeutic interventions,
particularly in disease contexts where FAT1 is dysregulated or
plays a pathogenic role. It is important to address the potential
risk of adverse events in various organs when applying
anti-molecular targeted drugs systemically, highlighting the need
for additional information on the organ-specific reachability of
these treatments. Different organs may exhibit varying
pharmacokinetics and pharmacodynamics for similar drugs; therefore,
the distribution, metabolism and clearance rates of medications in
specific organs need to be given special attention.
In conclusion, the current literature demonstrates
the potential of FAT1 as a promising therapeutic target. The
potential use of FAT1 as a therapeutic target requires further
elucidation through research including not only mechanistic in
vitro investigation, but also through pre-clinical and clinical
studies in the future.
Not applicable.
Funding: No funding was received.
Not applicable.
GL and WZ contributed to the conception and design
of the study, with GL overseeing the overall direction of the
project. GL and WZ also played a key role in formulating the
research questions and establishing the scope of the review. GL and
WZ supervised the manuscript writing and provided critical guidance
throughout the research process. TW and JL contributed to the data
analysis, including the synthesis of data from existing studies and
the development of the figures. TW was responsible for performing
the quantitative and qualitative analyses, while JL provided
statistical support and assisted in interpreting the findings. Both
TW and JL were instrumental in writing the initial draft of the
manuscript and revising the text for clarity and precision. JD
played a role in the interpretation of the data, ensuring the
results were correctly contextualized within the existing
literature. JD also refined the analysis approach critically,
offering suggestions for improving the methodological framework.
Additionally, JD contributed to the revision of the manuscript,
particularly in areas related to the discussion and conclusions. JD
provided feedback on the design and methodology of the review,
helping to strengthen the overall structure of the manuscript. Data
authentication is not applicable. All authors have read and
approved the final version of the study.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Tanoue T and Takeichi M: New insights into
Fat cadherins. J Cell Sci. 118:2347–2353. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Irshad K, Malik N, Arora M, Gupta Y, Sinha
S and Chosdol K: The quest for ligands and binding partners of
atypical cadherin FAT1. Transl Oncol. 14:1010972021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dunne J, Hanby AM, Poulsom R, Jones TA,
Sheer D, Chin WG, Da SM, Zhao Q, Beverley PC and Owen MJ: Molecular
cloning and tissue expression of FAT, the human homologue of the
Drosophila fat gene that is located on chromosome 4q34-q35 and
encodes a putative adhesion molecule. Genomics. 30:207–223. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sadeqzadeh E, de Bock CE, Zhang XD,
Shipman KL, Scott NM, Song C, Yeadon T, Oliveira CS, Jin B, Hersey
P, et al: Dual processing of FAT1 cadherin protein by human
melanoma cells generates distinct protein products. J Biol Chem.
286:28181–28191. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cao LL, Riascos-Bernal DF, Chinnasamy P,
Dunaway CM, Hou R, Pujato MA, O'Rourke BP, Miskolci V, Guo L,
Hodgson L, et al: Control of mitochondrial function and cell growth
by the atypical cadherin Fat1. Nature. 539:575–578. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Magg T, Schreiner D, Solis GP, Bade EG and
Hofer HW: Processing of the human protocadherin Fat1 and
translocation of its cytoplasmic domain to the nucleus. Exp Cell
Res. 307:100–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Riascos-Bernal DF, Maira A and Sibinga
NES: The atypical cadherin FAT1 limits mitochondrial respiration
and proliferation of vascular smooth muscle cells. Front Cardiovasc
Med. 9:9057172022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hou R, Liu L, Anees S, Hiroyasu S and
Sibinga NE: The Fat1 cadherin integrates vascular smooth muscle
cell growth and migration signals. J Cell Biol. 173:417–429. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Riascos-Bernal DF, Ressa G, Korrapati A
and Sibinga NES: The FAT1 cadherin drives vascular smooth muscle
cell migration. Cells. 12:16212023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tanoue T and Takeichi M: Mammalian Fat1
cadherin regulates actin dynamics and cell-cell contact. J Cell
Biol. 165:517–528. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Saburi S, Hester I, Goodrich L and McNeil
H: Functional interactions between Fat family cadherins in tissue
morphogenesis and planar polarity. Development. 139:1806–1820.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ahmed AF, de Bock CE, Sontag E,
Hondermarck H, Lincz LF and Thorne R: FAT1 cadherin controls
neuritogenesis during NTera2 cell differentiation. Biochem Biophys
Res Commun. 514:625–631. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Braun GS, Kuszka A, Dau C, Kriz W and
Moeller MJ: Interaction of atypical cadherin Fat1 with SoHo adaptor
proteins CAP/ponsin and ArgBP2. Biochem Biophys Res Commun.
472:88–94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peng Z, Gong Y and Liang X: Role of FAT1
in health and disease. Oncol Lett. 21:3982021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li M, Zhong Y and Wang M: Fat1 suppresses
the tumor-initiating ability of nonsmall cell lung cancer cells by
promoting Yes-associated protein 1 nuclear-cytoplasmic
translocation. Environ Toxicol. 36:2333–2341. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Katoh M: Function and cancer genomics of
FAT family genes (review). Int J Oncol. 41:1913–1918. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Morris LG, Ramaswami D and Chan TA: The
FAT epidemic: A gene family frequently mutated across multiple
human cancer types. Cell Cycle. 12:1011–1012. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Morris LG, Kaufman AM, Gong Y, Ramaswami
D, Walsh LA, Turcan Ş, Eng S, Kannan K, Zou Y, Peng L, et al:
Recurrent somatic mutation of FAT1 in multiple human cancers leads
to aberrant Wnt activation. Nat Genet. 45:253–261. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He Z, Li R and Jiang H: Mutations and copy
number abnormalities of hippo pathway components in human cancers.
Front Cell Dev Biol. 9:6617182021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Faraji F, Ramirez SI, Quiroz PY,
Mendez-Molina AN and Gutkind JS: Genomic hippo pathway alterations
and persistent YAP/TAZ activation: New hallmarks in head and neck
cancer. Cells. 11:13702022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen M, Sun X, Wang Y, Ling K, Chen C, Cai
X, Liang X and Liang Z: FAT1 inhibits the proliferation and
metastasis of cervical cancer cells by binding β-catenin. Int J
Clin Exp Pathol. 12:3807–3818. 2019.PubMed/NCBI
|
|
22
|
Ma W, Niu Z, Han D, Wang B and Wang X:
Circ-FAT1 up-regulates FOSL2 expression by sponging miR-619-5p to
facilitate colorectal cancer progression. Biochem Genet.
60:1362–1379. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jia L, Wang Y and Wang CY: circFAT1
promotes cancer stemness and immune evasion by promoting STAT3
activation. Adv Sci (Weinh). 8:20033762021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang TL, Miao XJ, Shuai YR, Sun HP, Wang
X, Yang M and Zhang N: FAT1 inhibits the proliferation of DLBCL
cells via increasing the m(6)A modification of YAP1 mRNA. Sci Rep.
14:118362024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pastushenko I, Mauri F, Song Y, de Cock F,
Meeusen B, Swedlund B, Impens F, Van Haver D, Opitz M, Thery M, et
al: Fat1 deletion promotes hybrid EMT state, tumour stemness and
metastasis. Nature. 589:448–455. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang X, Liu J, Liang X, Chen J, Hong J,
Li L, He Q and Cai X: History and progression of Fat cadherins in
health and disease. Onco Targets Ther. 9:7337–7343. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Katoh Y and Katoh M: Comparative
integromics on FAT1, FAT2, FAT3 and FAT4. Int J Mol Med.
18:523–528. 2006.PubMed/NCBI
|
|
28
|
Chen ZG, Saba NF and Teng Y: The diverse
functions of FAT1 in cancer progression: Good, bad, or ugly? J Exp
Clin Cancer Res. 41:2482022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Z, Lin K and Xiao H: A pan-cancer
analysis of the FAT1 in human tumors. Sci Rep. 12:215982022.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu W, Yang L, Gao Y, Zhou Y, Shi Y, Liu
K, Yu R, Shao Y, Zhang W, Wu G and He J: Clinical value of FAT1
mutations to indicate the immune response in colorectal cancer
patients. Genomics. 116:1108082024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ding C, Huang H, Wu D, Chen C, Hua Y, Liu
J, Li Y, Liu H and Chen J: Pan-cancer analysis predict that FAT1 is
a therapeutic target and immunotherapy biomarker for multiple
cancer types including non-small cell lung cancer. Front Immunol.
15:13690732024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Valletta D, Czech B, Spruss T, Ikenberg K,
Wild P, Hartmann A, Weiss TS, Oefner PJ, Müller M, Bosserhoff AK
and Hellerbrand C: Regulation and function of the atypical cadherin
FAT1 in hepatocellular carcinoma. Carcinogenesis. 35:1407–1415.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun
ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, et al: Genetic landscape of
esophageal squamous cell carcinoma. Nat Genet. 46:1097–1102. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cui Y, Chen H, Xi R, Cui H, Zhao Y, Xu E,
Yan T, Lu X, Huang F, Kong P, et al: Whole-genome sequencing of 508
patients identifies key molecular features associated with poor
prognosis in esophageal squamous cell carcinoma. Cell Res.
30:902–913. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang N, Shi J, Shi X, Chen W and Liu J:
Mutational characterization and potential prognostic biomarkers of
Chinese patients with esophageal squamous cell carcinoma. Onco
Targets Ther. 13:12797–12809. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang L, Zhou Y, Cheng C, Cui H, Cheng L,
Kong P, Wang J, Li Y, Chen W, Song B, et al: Genomic analyses
reveal mutational signatures and frequently altered genes in
esophageal squamous cell carcinoma. Am J Hum Genet. 107:3752020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hu X, Zhai Y, Shi R, Qian Y, Cui H, Yang
J, Bi Y, Yan T, Yang J, Ma Y, et al: FAT1 inhibits cell migration
and invasion by affecting cellular mechanical properties in
esophageal squamous cell carcinoma. Oncol Rep. 39:2136–2146.
2018.PubMed/NCBI
|
|
40
|
Mishra R, Nikoo MZ, Veeraballi S and Singh
A: Venetoclax and hypomethylating agent combination in myeloid
malignancies: Mechanisms of synergy and challenges of resistance.
Int J Mol Sci. 25:4842023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Wang G, Ma Y, Teng J, Wang Y, Cui
Y, Dong Y, Shao S, Zhan Q and Liu X: FAT1, a direct transcriptional
target of E2F1, suppresses cell proliferation, migration and
invasion in esophageal squamous cell carcinoma. Chin J Cancer Res.
31:609–619. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hu X, Zhai Y, Kong P, Cui H, Yan T, Yang
J, Qian Y, Ma Y, Wang F, Li H, et al: FAT1 prevents epithelial
mesenchymal transition (EMT) via MAPK/ERK signaling pathway in
esophageal squamous cell cancer. Cancer Lett. 397:83–93. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ashrafizadeh M, Zarrabi A, Hushmandi K,
Kalantari M, Mohammadinejad R, Javaheri T and Sethi G: Association
of the epithelial-mesenchymal transition (EMT) with cisplatin
resistance. Int J Mol Sci. 21:40022020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hu X, Zhai Y, Kong P, Cui H, Yan T, Yang
J, Qian Y, Ma Y, Wang F, Li H, et al: Corrigendum to ‘FAT1 prevents
epithelial mesenchymal transition (EMT) via MAPK/ERK signaling
pathway in esophageal squamous cell cancer’ [(Canc. Lett. 397
(2017) 83–93)]. Cancer Lett. 494:1–2. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhai Y, Shan C, Zhang H, Kong P, Zhang L,
Wang Y, Hu X and Cheng X: FAT1 downregulation enhances stemness and
cisplatin resistance in esophageal squamous cell carcinoma. Mol
Cell Biochem. 477:2689–2702. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mai Z, Yuan J, Yang H, Fang S, Xie X, Wang
X, Xie J, Wen J and Fu J: Inactivation of Hippo pathway
characterizes a poor-prognosis subtype of esophageal cancer. JCI
Insight. 7:e1552182022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lu Y, Wang Z, Zhou L, Ma Z, Zhang J, Wu Y,
Shao Y and Yang Y: FAT1 and PTPN14 regulate the malignant
progression and chemotherapy resistance of esophageal cancer
through the hippo signaling pathway. Anal Cell Pathol (Amst).
2021:92903722021.PubMed/NCBI
|
|
48
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cramer JD, Burtness B, Le QT and Ferris R:
The changing therapeutic landscape of head and neck cancer. Nat Rev
Clin Oncol. 16:669–683. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Leemans CR, Snijders PJF and Brakenhoff
RH: The molecular landscape of head and neck cancer. Nat Rev
Cancer. 18:269–282. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cancer Genome Atlas Network, .
Comprehensive genomic characterization of head and neck squamous
cell carcinomas. Nature. 517:576–582. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Campbell JD, Yau C, Bowlby R, Liu Y,
Brennan K, Fan H, Taylor AM, Wang C, Walter V, Akbani R, et al:
Genomic, pathway network, and immunologic features distinguishing
squamous carcinomas. Cell Rep. 23:194–212.e6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Alamoud KA and Kukuruzinska MA: Emerging
insights into Wnt/β-catenin signaling in head and neck cancer. J
Dent Res. 97:665–673. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zwirner K, Hilke FJ, Demidov G, Fernandez
JS, Ossowski S, Gani C, Thorwarth D, Riess O, Zips D, Schroeder C
and Welz S: Radiogenomics in head and neck cancer: Correlation of
radiomic heterogeneity and somatic mutations in TP53, FAT1 and
KMT2D. Strahlenther Onkol. 195:771–779. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Moreira A, Poulet A, Masliah-Planchon J,
Lecerf C, Vacher S, Chérif LL, Dupain C, Marret G, Girard E, Syx L,
et al: Prognostic value of tumor mutational burden in patients with
oral cavity squamous cell carcinoma treated with upfront surgery.
ESMO Open. 6:1001782021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kim S, Lee C, Kim H and Yoon SO: Genetic
characteristics of advanced oral tongue squamous cell carcinoma in
young patients. Oral Oncol. 144:1064662023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Su SC, Lin CW, Liu YF, Fan WL, Chen MK, Yu
CP, Yang WE, Su CW, Chuang CY, Li WH, et al: Exome sequencing of
oral squamous cell carcinoma reveals molecular subgroups and novel
therapeutic opportunities. Theranostics. 7:1088–1099. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chai AWY, Lim KP and Cheong SC:
Translational genomics and recent advances in oral squamous cell
carcinoma. Semin Cancer Biol. 61:71–83. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kim KT, Kim BS and Kim JH: Association
between FAT1 mutation and overall survival in patients with human
papillomavirus-negative head and neck squamous cell carcinoma. Head
Neck. 38 (Suppl 1):E2021–E2029. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xi Y, Negrao MV, Akagi K, Xiao W, Jiang B,
Warner SC, Dunn JD, Wang J, Symer DE and Gillison ML: Noninvasive
genomic profiling of somatic mutations in oral cavity cancers. Oral
Oncol. 140:1063722023. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Inchanalkar M, Srivatsa S, Ambatipudi S,
Bhosale PG, Patil A, Schäffer AA, Beerenwinkel N and Mahimkar MB:
Genome-wide DNA methylation profiling of HPV-negative leukoplakia
and gingivobuccal complex cancers. Clin Epigenetics. 15:932023.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chaudhary S, Dam V, Ganguly K, Sharma S,
Atri P, Chirravuri-Venkata R, Cox JL, Sayed Z, Jones DT, Ganti AK,
et al: Differential mutation spectrum and immune landscape in
African Americans versus Whites: A possible determinant to health
disparity in head and neck cancer. Cancer Lett. 492:44–53. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Santos-de-Frutos K, Segrelles C and Lorz
C: Hippo pathway and yap signaling alterations in squamous cancer
of the head and neck. J Clin Med. 8:21312019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Martin D, Degese MS, Vitale-Cross L,
Iglesias-Bartolome R, Valera JLC, Wang Z, Feng X, Yeerna H, Vadmal
V, Moroishi T, et al: Assembly and activation of the Hippo
signalome by FAT1 tumor suppressor. Nat Commun. 9:23722018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen N, Golczer G, Ghose S, Lin B,
Langenbucher A, Webb J, Bhanot H, Abt NB, Lin D, Varvares M, et al:
YAP1 maintains active chromatin state in head and neck squamous
cell carcinomas that promotes tumorigenesis through cooperation
with BRD4. Cell Rep. 39:1109702022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Alonso-Juarranz M, Sen O, Pérez P,
González-Corchón MA, Cabezas-Camarero S, Saiz-Pardo M, Viñas-Lopez
J, Recio-Poveda L, Botella LM and Falahat F: The distinctive
features behind the aggressiveness of oral and cutaneous squamous
cell carcinomas. Cancers (Basel). 15:32272023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lin SC, Lin LH, Yu SY, Kao SY, Chang KW,
Cheng HW and Liu CJ: FAT1 somatic mutations in head and neck
carcinoma are associated with tumor progression and survival.
Carcinogenesis. 39:1320–1330. 2018.PubMed/NCBI
|
|
68
|
Wu MH, Lu RY, Yu SJ, Tsai YZ, Lin YC, Bai
ZY, Liao RY, Hsu YC, Chen CC and Cai BH: PTC124 rescues nonsense
mutation of two tumor suppressor genes NOTCH1 and FAT1 to repress
HNSCC cell proliferation. Biomedicines. 10:29482022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lan T, Ge Q, Zheng K, Huang L, Yan Y,
Zheng L, Lu Y and Zheng D: FAT1 Upregulates in oral squamous cell
carcinoma and promotes cell proliferation via cell cycle and DNA
repair. Front Oncol. 12:8700552022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kim SI, Woo SR, Noh JK, Lee MK, Lee YC,
Lee JW, Ko SG and Eun YG: Clinical significance of FAT1 gene
mutation and mRNA expression in patients with head and neck
squamous cell carcinoma. Mol Oncol. 16:1661–1679. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Hsu TN, Huang CM, Huang CS, Huang MS, Yeh
CT, Chao TY and Bamodu OA: Targeting FAT1 inhibits carcinogenesis,
induces oxidative stress and enhances cisplatin sensitivity through
deregulation of LRP5/WNT2/GSS signaling axis in oral squamous cell
carcinoma. Cancers (Basel). 11:18832019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chen Z, Zhang C, Chen J, Wang D, Tu J, Van
Waes C, Saba NF, Chen ZG and Chen Z: The proteomic landscape of
growth factor signaling networks associated with FAT1 mutations in
head and neck cancers. Cancer Res. 81:4402–4416. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xu F, Cui WQ, Liu C, Feng F, Liu R, Zhang
J and Sun CG: Prognostic biomarkers correlated with immune
infiltration in non-small cell lung cancer. FEBS Open Bio.
13:72–88. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu Q, Zhang J, Guo C, Wang M, Wang C, Yan
Y, Sun L, Wang D, Zhang L, Yu H, et al: Proteogenomic
characterization of small cell lung cancer identifies biological
insights and subtype-specific therapeutic strategies. Cell.
187:184–203.e28. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Peng J, Xiao L, Zou D and Han L: A somatic
mutation signature predicts the best overall response to
anti-programmed cell death protein-1 treatment in epidermal growth
factor receptor/anaplastic lymphoma kinase-negative non-squamous
non-small cell lung cancer. Front Med (Lausanne). 9:8083782022.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Hao F, Ma Q and Zhong D: Potential
predictive value of comutant LRP1B and FAT for immune response in
non-small cell lung cancer: LRP1B and FAT comutation enhance immune
response. Transl Oncol. 24:1014932022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Fang W, Ma Y, Yin JC, Hong S, Zhou H, Wang
A, Wang F, Bao H, Wu X, Yang Y, et al: comprehensive genomic
profiling identifies novel genetic predictors of response to
anti-PD-(L)1 therapies in non-small cell lung cancer. Clin Cancer
Res. 25:5015–5026. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhang W, Tang Y, Guo Y, Kong Y, Shi F,
Sheng C, Wang S and Wang Q: Favorable immune checkpoint inhibitor
outcome of patients with melanoma and NSCLC harboring FAT1
mutations. NPJ Precis Oncol. 6:462022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang L, Wang Y, Wang L, Wang M, Li S, He
J, Ji J, Li K and Cao L: Identifying survival of pan-cancer
patients under immunotherapy using genomic mutation signature with
large sample cohorts. J Mol Med (Berl). 102:69–79. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Fu Y, Yang Z, Hu Z, Yang Z, Pan Y, Chen J,
Wang J, Hu D, Zhou Z, Xu L, et al: Preoperative serum ctDNA
predicts early hepatocellular carcinoma recurrence and response to
systemic therapies. Hepatol Int. 16:868–878. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhu HY, Cao GY, Wang SP, Chen Y, Liu GD,
Gao YJ and Hu JP: POU2F1 promotes growth and metastasis of
hepatocellular carcinoma through the FAT1 signaling pathway. Am J
Cancer Res. 7:1665–1679. 2017.PubMed/NCBI
|
|
82
|
Xu J, Wang B, Liu ZT, Lai MC, Zhang ML and
Zheng SS: miR-223-3p regulating the occurrence and development of
liver cancer cells by targeting FAT1 gene. Math Biosci Eng.
17:1534–1547. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Huang ZL, Zhang PB, Zhang JT, Li F, Li TT
and Huang XY: Comprehensive genomic profiling identifies FAT1 as a
negative regulator of EMT, CTCs, and metastasis of hepatocellular
carcinoma. J Hepatocell Carcinoma. 10:369–382. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li X, Jiang J, Zhao X, Wang J, Han H, Zhao
Y, Peng B, Zhong R, Ying W and Qian X: N-glycoproteome analysis of
the secretome of human metastatic hepatocellular carcinoma cell
lines combining hydrazide chemistry, HILIC enrichment and mass
spectrometry. PLoS One. 8:e819212013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Meng P, Zhang YF, Zhang W, Chen X, Xu T,
Hu S, Liang X, Feng M, Yang X and Ho M: Identification of the
atypical cadherin FAT1 as a novel glypican-3 interacting protein in
liver cancer cells. Sci Rep. 11:402021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Malik N, Kundu A, Gupta Y, Irshad K, Arora
M, Goswami S, Mahajan S, Sarkar C, Suri V, Suri A, et al:
Protumorigenic role of the atypical cadherin FAT1 by the
suppression of PDCD10 via RelA/miR221-3p/222-3p axis in
glioblastoma. Mol Carcinog. 62:1817–1831. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Li LC, Zhang M, Feng YK and Wang XJ:
IDH1-R132H suppresses glioblastoma malignancy through
FAT1-ROS-HIF-1α signaling. Neurol India. 68:1050–1058. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Yu J, Gao H, Su Z, Yue F and Tian X:
Effect of FAT1 gene expression on the prognosis of medulloblastoma
in children: A protocol for systematic review and meta-analysis.
Medicine (Baltimore). 99:e230202020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Madan E, Dikshit B, Gowda SH, Srivastava
C, Sarkar C, Chattopadhyay P, Sinha S and Chosdol K: FAT1 is a
novel upstream regulator of HIF1α and invasion of high grade
glioma. Int J Cancer. 139:2570–2582. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Srivastava C, Irshad K, Dikshit B,
Chattopadhyay P, Sarkar C, Gupta DK, Sinha S and Chosdol K: FAT1
modulates EMT and stemness genes expression in hypoxic
glioblastoma. Int J Cancer. 142:805–812. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Dikshit B, Irshad K, Madan E, Aggarwal N,
Sarkar C, Chandra PS, Gupta DK, Chattopadhyay P, Sinha S and
Chosdol K: FAT1 acts as an upstream regulator of oncogenic and
inflammatory pathways, via PDCD4, in glioma cells. Oncogene.
32:3798–3808. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Irshad K, Srivastava C, Malik N, Arora M,
Gupta Y, Goswami S, Sarkar C, Suri V, Mahajan S, Gupta DK, et al:
Upregulation of atypical cadherin FAT1 promotes an
immunosuppressive tumor microenvironment via TGF-β. Front Immunol.
13:8138882022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Wang L, Lyu S, Wang S, Shen H, Niu F, Liu
X, Liu J and Niu Y: Loss of FAT1 during the progression from DCIS
to IDC and predict poor clinical outcome in breast cancer. Exp Mol
Pathol. 100:177–183. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhao F, Miyashita M, Hattori M, Yoshimatsu
T, Howard F, Kaneva K, Jones R, Bell JSK, Fleming GF, Jaskowiak N,
et al: Racial disparities in pathological complete response among
patients receiving neoadjuvant chemotherapy for early-stage breast
cancer. JAMA Netw Open. 6:e2333292023. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Li Z, Razavi P, Li Q, Toy W, Liu B, Ping
C, Hsieh W, Sanchez-Vega F, Brown DN, Da Cruz Paula AF, et al: Loss
of the FAT1 tumor suppressor promotes resistance to CDK4/6
inhibitors via the hippo pathway. Cancer Cell. 34:893–905. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Xi J and Ma CX: Sequencing endocrine
therapy for metastatic breast cancer: What do we do after disease
progression on a CDK4/6 inhibitor? Curr Oncol Rep. 22:572020.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Bu J, Zhang Y, Wu S, Li H, Sun L, Liu Y,
Zhu X, Qiao X, Ma Q, Liu C, et al: KK-LC-1 as a therapeutic target
to eliminate ALDH(+) stem cells in triple negative breast cancer.
Nat Commun. 14:26022023. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wong K, Abascal F, Ludwig L,
Aupperle-Lellbach H, Grassinger J, Wright CW, Allison SJ, Pinder E,
Phillips RM, Romero LP, et al: Cross-species oncogenomics offers
insight into human muscle-invasive bladder cancer. Genome Biol.
24:1912023. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Wang F, Liu P, An H and Zhang Y:
Sulforaphane suppresses the viability and metastasis, and promotes
the apoptosis of bladder cancer cells by inhibiting the expression
of FAT-1. Int J Mol Med. 46:1085–1095. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Cazier JB, Rao SR, McLean CM, Walker AK,
Wright BJ, Jaeger EE, Kartsonaki C, Marsden L, Yau C, Camps C, et
al: Whole-genome sequencing of bladder cancers reveals somatic
CDKN1A mutations and clinicopathological associations with mutation
burden. Nat Commun. 5:37562014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Jiang S, Zhu Y, Chen Z, Huang Z, Liu B, Xu
Y, Li Z, Lin Z and Li M: S100A14 inhibits cell growth and
epithelial-mesenchymal transition (EMT) in prostate cancer through
FAT1-mediated Hippo signaling pathway. Hum Cell. 34:1215–1226.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Kang MH, Jeong GS, Smoot DT, Ashktorab H,
Hwang CM, Kim BS, Kim HS and Park YY: Verteporfin inhibits gastric
cancer cell growth by suppressing adhesion molecule FAT1.
Oncotarget. 8:98887–98897. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhang W, Ji K, Min C, Zhang C, Yang L,
Zhang Q, Tian Z, Zhang M, Wang X and Li X: Oncogenic LINC00857
recruits TFAP2C to elevate FAT1 expression in gastric cancer.
Cancer Sci. 114:63–74. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Holowatyj AN, Wen W, Gibbs T, Seagle HM,
Keller SR, Edwards DRV, Washington MK, Eng C, Perea J, Zheng W and
Guo X: Racial/Ethnic and sex differences in somatic cancer gene
mutations among patients with early-onset colorectal cancer. Cancer
Discov. 13:570–579. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Li P, Meng Q, Xue Y, Teng Z, Chen H, Zhang
J, Xu Y, Wang S, Yu R, Ou Q, et al: Comprehensive genomic profiling
of colorectal cancer patients reveals differences in mutational
landscapes among clinical and pathological subgroups. Front Oncol.
12:10001462022. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Tang J, Peng W, Tian C, Zhang Y, Ji D,
Wang L, Jin K, Wang F, Shao Y, Wang X and Sun Y: Molecular
characteristics of early-onset compared with late-onset colorectal
cancer: A case controlled study. Int J Surg. 110:4559–4570.
2024.PubMed/NCBI
|
|
107
|
Jiang NN, Yue GGL, Li P, Ye YS, Gomes AJ,
Kwok FHF, Lee JKM, Gao S, Lau CB and Xu G: Discovery of
dearomatized isoprenylated acylphloroglucinols with colon tumor
suppressive activities in mice via inhibiting NFκB-FAT1-PDCD4
signaling activation. Eur J Med Chem. 239:1145322022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Yang J, Zhao S, Su J, Liu S, Wu Z, Ma W,
Tang M, Wu J, Mao E, Han L, et al: Comprehensive genomic profiling
reveals prognostic signatures and insights into the molecular
landscape of colorectal cancer. Front Oncol. 13:12855082023.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Grifantini R, Taranta M, Gherardini L,
Naldi I, Parri M, Grandi A, Giannetti A, Tombelli S, Lucarini G,
Ricotti L, et al: Magnetically driven drug delivery systems
improving targeted immunotherapy for colon-rectal cancer. J Control
Release. 280:76–86. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ardjmand A, de Bock CE, Shahrokhi S, Lincz
LF, Boyd AW, Burns GF and Thorne RF: Fat1 cadherin provides a novel
minimal residual disease marker in acute lymphoblastic leukemia.
Hematology. 18:315–322. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Zhou H, Xiao M, Zhou X, Hao Y, Xin C, Tang
Y, Liang Y, Zhang Y and Li S: Aplastic anemia preceding acute
lymphoblastic leukemia in an adult with FAT1 mutation. Minerva Med.
110:593–594. 2019.PubMed/NCBI
|
|
112
|
Feng J, Li Y, Jia Y, Fang Q, Gong X, Dong
X, Ru K, Li Q, Zhao X, Liu K, et al: Spectrum of somatic mutations
detected by targeted next-generation sequencing and their
prognostic significance in adult patients with acute lymphoblastic
leukemia. J Hematol Oncol. 10:612017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Sun X, Liu X, Li Y, Shi X, Li Y, Tan R,
Jiang Y, Sui X, Ge X, Xu H, et al: Characteristics of molecular
genetic mutations and their correlation with prognosis in
adolescent and adult patients with acute lymphoblastic leukemia.
Oncology. 102:85–98. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Chang YH, Yu CH, Jou ST, Lin CY, Lin KH,
Lu MY, Wu KH, Chang HH, Lin DT, Lin SW, et al: Targeted sequencing
to identify genetic alterations and prognostic markers in pediatric
T-cell acute lymphoblastic leukemia. Sci Rep. 11:7692021.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
de Bock CE, Ardjmand A, Molloy TJ, Bone
SM, Johnstone D, Campbell DM, Shipman KL, Yeadon TM, Holst J,
Spanevello MD, et al: The Fat1 cadherin is overexpressed and an
independent prognostic factor for survival in paired
diagnosis-relapse samples of precursor B-cell acute lymphoblastic
leukemia. Leukemia. 26:918–926. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Neumann M, Seehawer M, Schlee C, Vosberg
S, Heesch S, von der Heide EK, Graf A, Krebs S, Blum H, Gökbuget N,
et al: FAT1 expression and mutations in adult acute lymphoblastic
leukemia. Blood Cancer J. 4:e2242014. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Liu Y, Easton J, Shao Y, Maciaszek J, Wang
Z, Wilkinson MR, McCastlain K, Edmonson M, Pounds SB, Shi L, et al:
The genomic landscape of pediatric and young adult T-lineage acute
lymphoblastic leukemia. Nat Genet. 49:1211–1218. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Liebig S, Neumann M, Silva P,
Ortiz-Tanchez J, Schulze V, Isaakidis K, Schlee C, Schroeder MP,
Beder T, Morris LGT, et al: FAT1 expression in T-cell acute
lymphoblastic leukemia (T-ALL) modulates proliferation and WNT
signaling. Sci Rep. 13:9722023. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
de Bock CE, Down M, Baidya K, Sweron B,
Boyd AW, Fiers M, Burns GF, Molloy TJ, Lock RB, Soulier J, et al:
T-cell acute lymphoblastic leukemias express a unique truncated
FAT1 isoform that cooperates with NOTCH1 in leukemia development.
Haematologica. 104:e204–e207. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Garg M, Nagata Y, Kanojia D, Mayakonda A,
Yoshida K, Keloth SH, Zang ZJ, Okuno Y, Shiraishi Y, Chiba K, et
al: Profiling of somatic mutations in acute myeloid leukemia with
FLT3-ITD at diagnosis and relapse. Blood. 126:2491–2501. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Sethi S, Madden B, Moura MC, Nasr SH,
Klomjit N, Gross L, Negron V, Charlesworth MC, Alexander MP, Leung
N, et al: Hematopoietic stem cell transplant-membranous nephropathy
is associated with protocadherin FAT1. J Am Soc Nephrol.
33:1033–1044. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ahn JS, Kim HJ, Kim YK, Lee SS, Jung SH,
Yang DH, Lee JJ, Kim NY, Choi SH, Jung CW, et al: DNMT3A R882
mutation with FLT3-ITD positivity is an extremely poor prognostic
factor in patients with normal-karyotype acute myeloid leukemia
after allogeneic hematopoietic cell transplantation. Biol Blood
Marrow Transplant. 22:61–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zeng X, Zhang Y, Zhao K, Zhou L, Zhou Y,
Xuan L, Cao R, Xu J, Dai M and Liu Q: Somatic mutations predict
prognosis in myelodysplastic syndrome patients with normal
karyotypes. Signal Transduct Target Ther. 6:2742021. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zhong WJ, Liu XD, Zhong LY, Li KB, Sun QX,
Xu X, Wei T, Li QS and Zhu ZG: Comparison of gene mutation spectra
in younger and older Chinese acute myeloid leukemia patients and
its prognostic value. Gene. 770:1453442021. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Huang X, Li Y, Zhang J, Yan L, Zhao H,
Ding L, Bhatara S, Yang X, Yoshimura S, Yang W, et al: Single-cell
systems pharmacology identifies development-driven drug response
and combination therapy in B cell acute lymphoblastic leukemia.
Cancer Cell. 42:552–567. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Laginestra MA, Cascione L, Motta G,
Fuligni F, Agostinelli C, Rossi M, Sapienza MR, Righi S, Broccoli
A, Indio V, et al: Whole exome sequencing reveals mutations in FAT1
tumor suppressor gene clinically impacting on peripheral T-cell
lymphoma not otherwise specified. Mod Pathol. 33:179–187. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Laginestra MA, Cascione L, Motta G,
Fuligni F, Agostinelli C, Rossi M, Sapienza MR, Righi S, Broccoli
A, Indio V, et al: Correction: Whole exome sequencing reveals
mutations in FAT1 tumor suppressor gene clinically impacting on
peripheral T-cell lymphoma not otherwise specified. Mod Pathol.
33:3192020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhang C, Mou B, Xu J, Wang J, Liu Q, Yang
Y, Tang W, Zhong X and Xu C: Angioimmunoblastic T-cell lymphoma:
Novel recurrent mutations and prognostic biomarkers by cell-free
DNA profiling. Br J Haematol. 203:807–819. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Hansen SV, Hansen MH, Cédile O, Møller MB,
Haaber J, Abildgaard N and Nyvold CG: Detailed characterization of
the transcriptome of single B cells in mantle cell lymphoma
suggesting a potential use for SOX4. Sci Rep. 11:190922021.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Zhao A, Wu F, Wang Y, Li J, Xu W and Liu
H: Analysis of genetic alterations in ocular adnexal
mucosa-associated lymphoid tissue lymphoma with whole-exome
sequencing. Front Oncol. 12:8176352022. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Yang P, Liu SZ, Li CY, Zhang WL, Wang J,
Chen YT, Li S, Liu CL, Liu H, Cai QQ, et al: Genetic and prognostic
analysis of blastoid and pleomorphic mantle cell lymphoma: A
multicenter analysis in China. Ann Hematol. 103:2381–2391. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Kortüm KM, Langer C, Monge J, Bruins L,
Zhu YX, Shi CX, Jedlowski P, Egan JB, Ojha J, Bullinger L, et al:
Longitudinal analysis of 25 sequential sample-pairs using a custom
multiple myeloma mutation sequencing panel (M(3)P). Ann Hematol.
94:1205–1211. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Sethi S, Madden B, Moura MC, Nasr SH,
Alexander MP, Debiec H, Torrel N, Gross L, Negron V, Specks U, et
al: FAT1 is a target antigen in a subset of de novo allograft
membranous nephropathy associated with antibody mediated rejection.
Kidney Int. 106:985–990. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Zhang Q, Li MK, Hu XY, Wu YX, Wang YY,
Zhao PP, Cheng LN, Yu RH, Zhang XD, Chen S, et al: The tumor
suppressor Fat1 is dispensable for normal murine hematopoiesis. J
Leukoc Biol. 116:909–914. 2024. View Article : Google Scholar : PubMed/NCBI
|