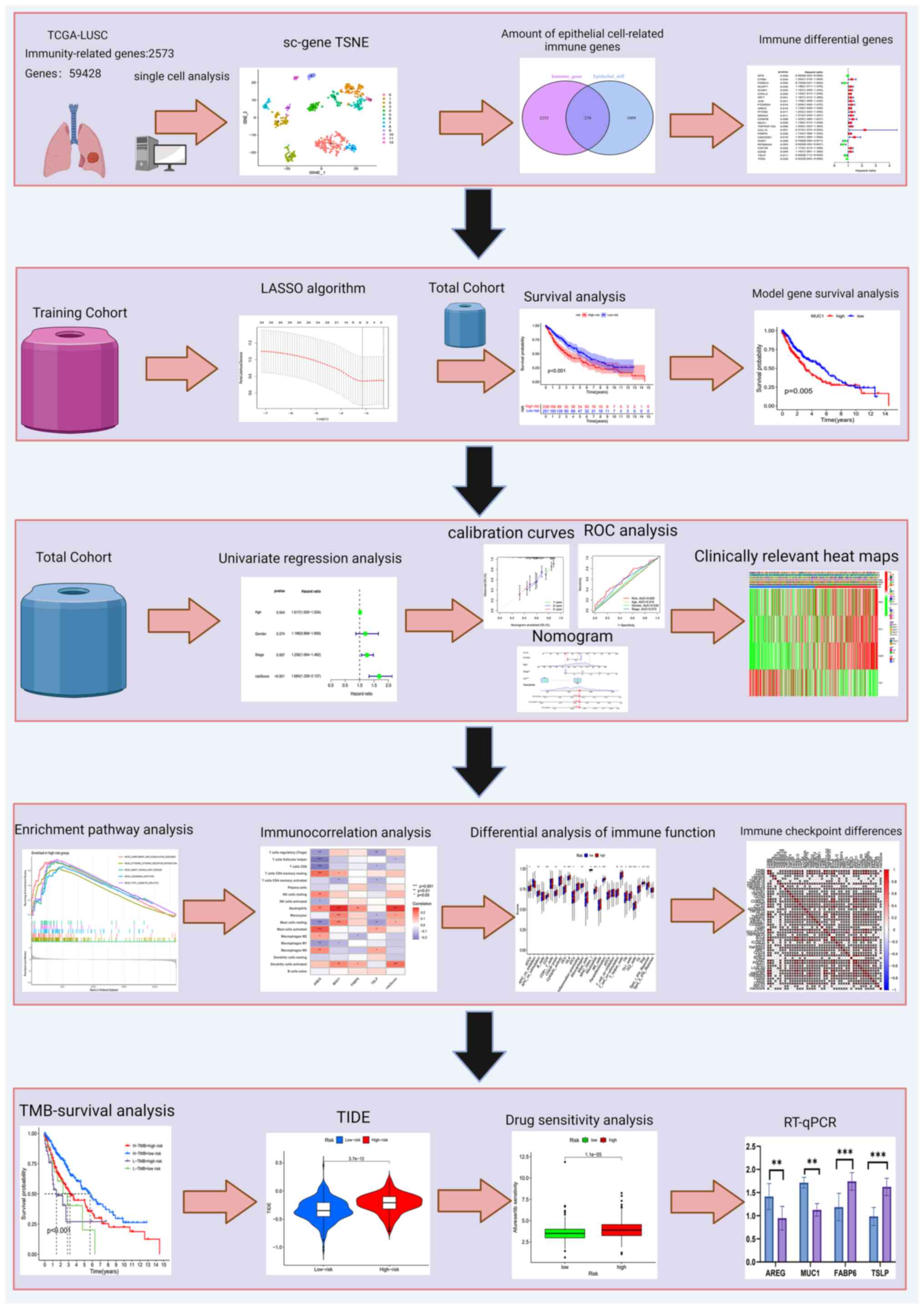
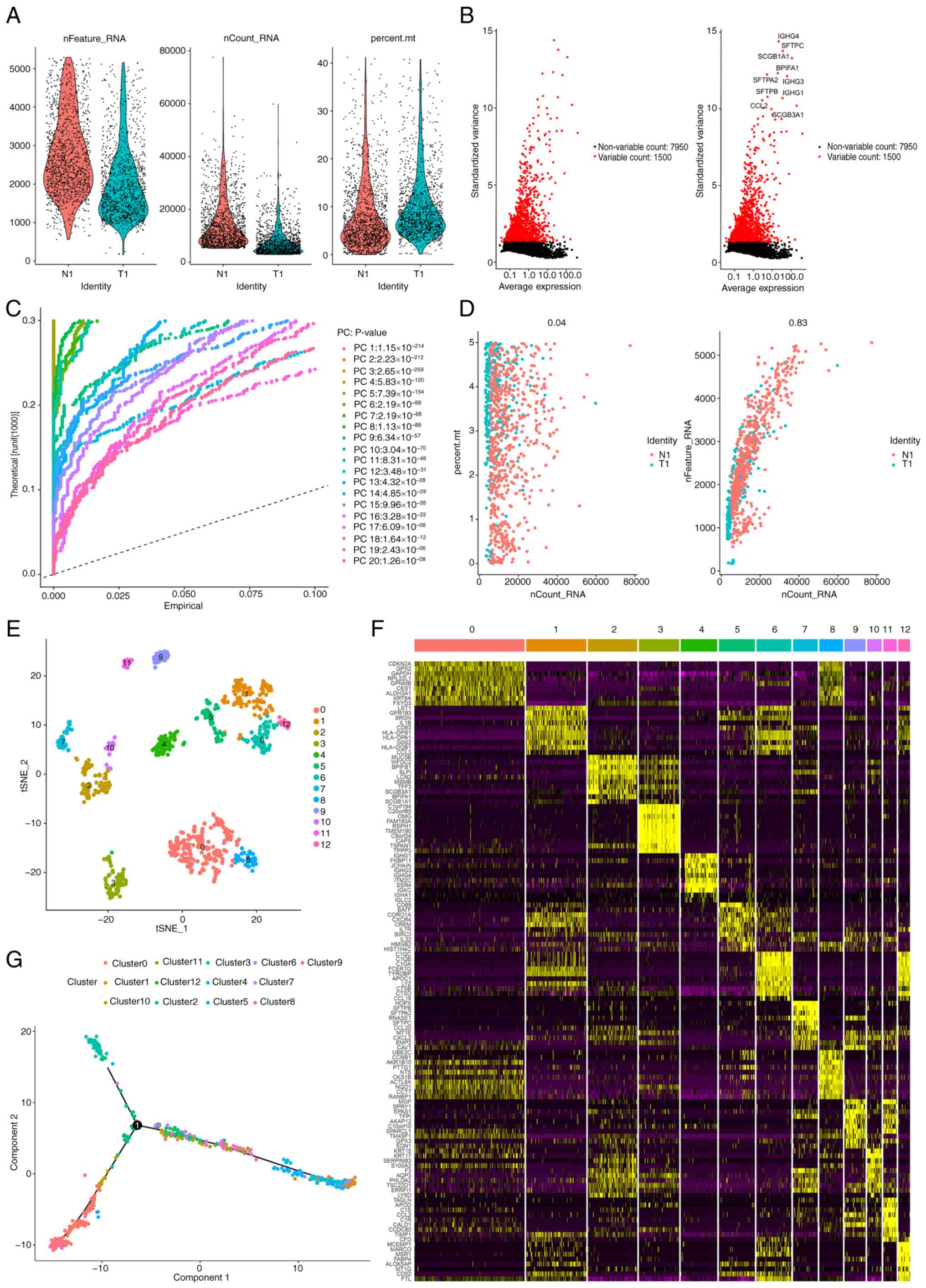
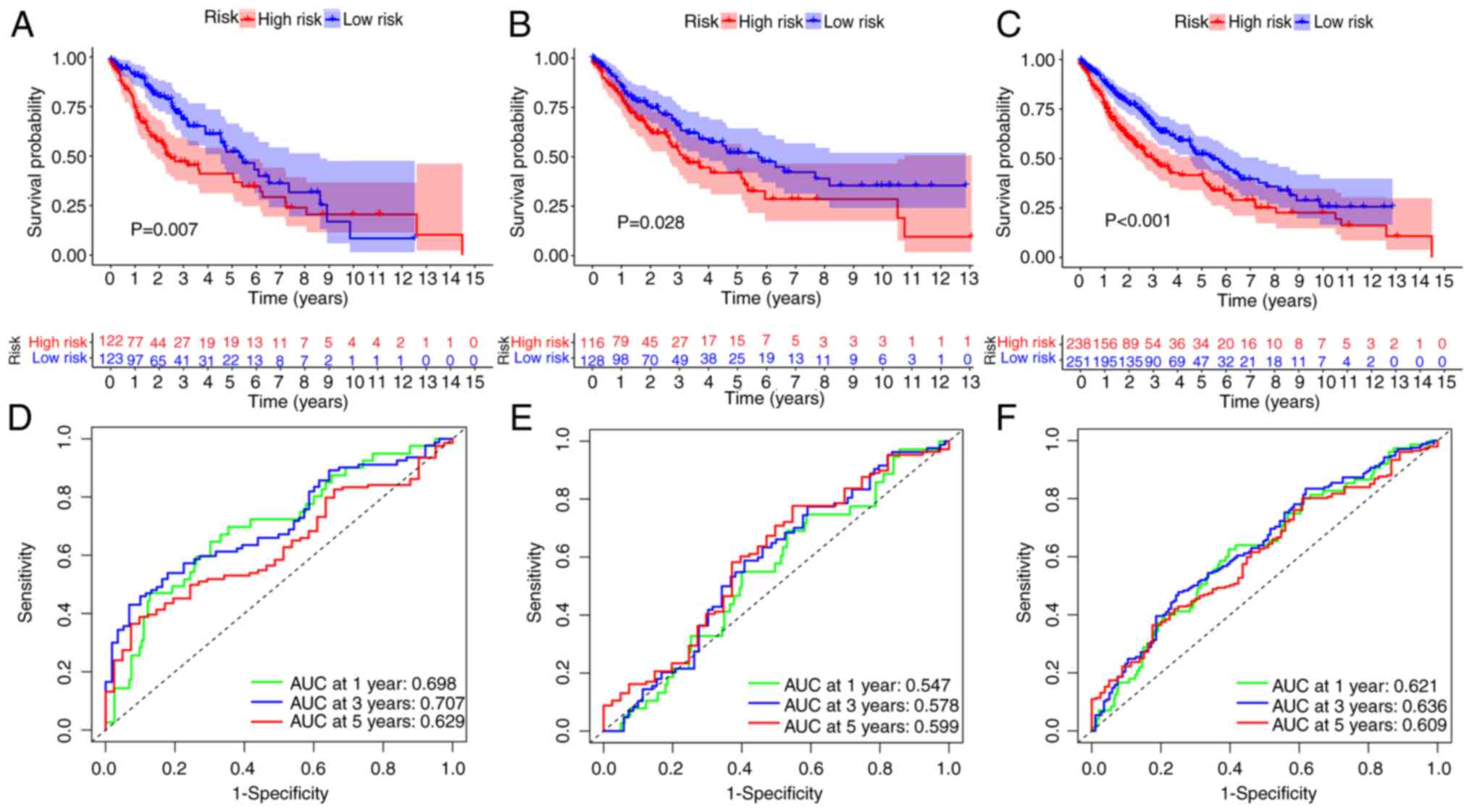
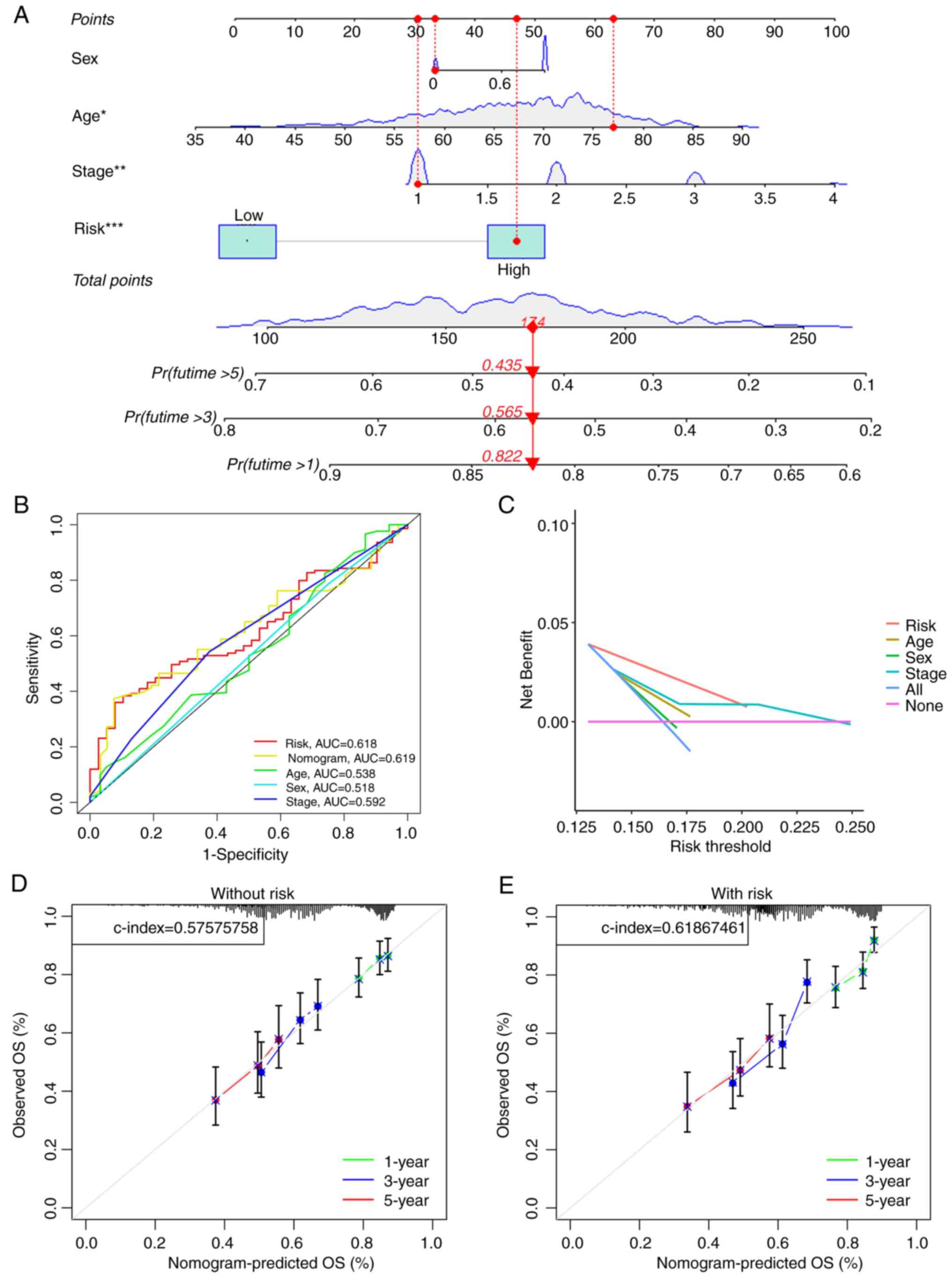
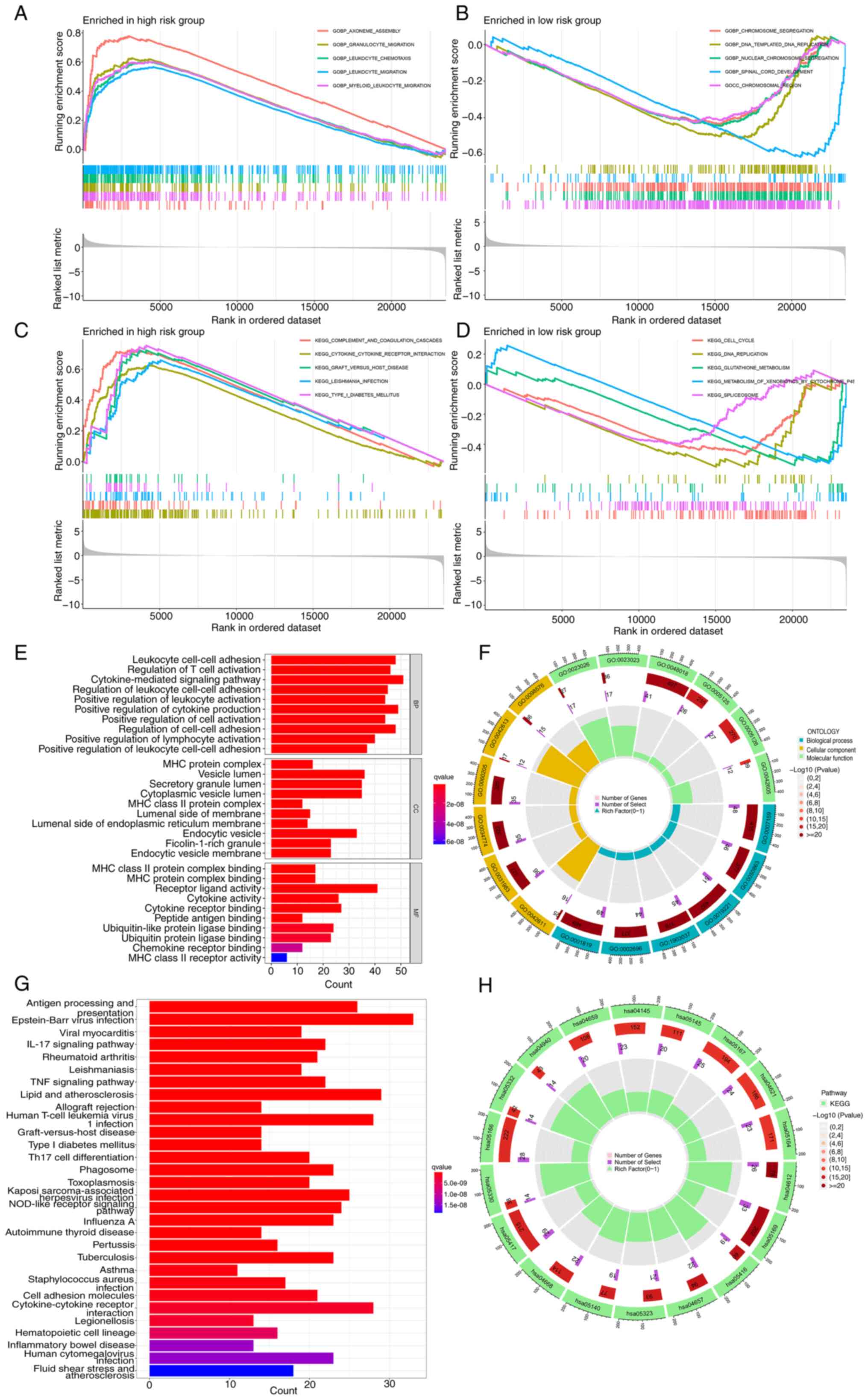
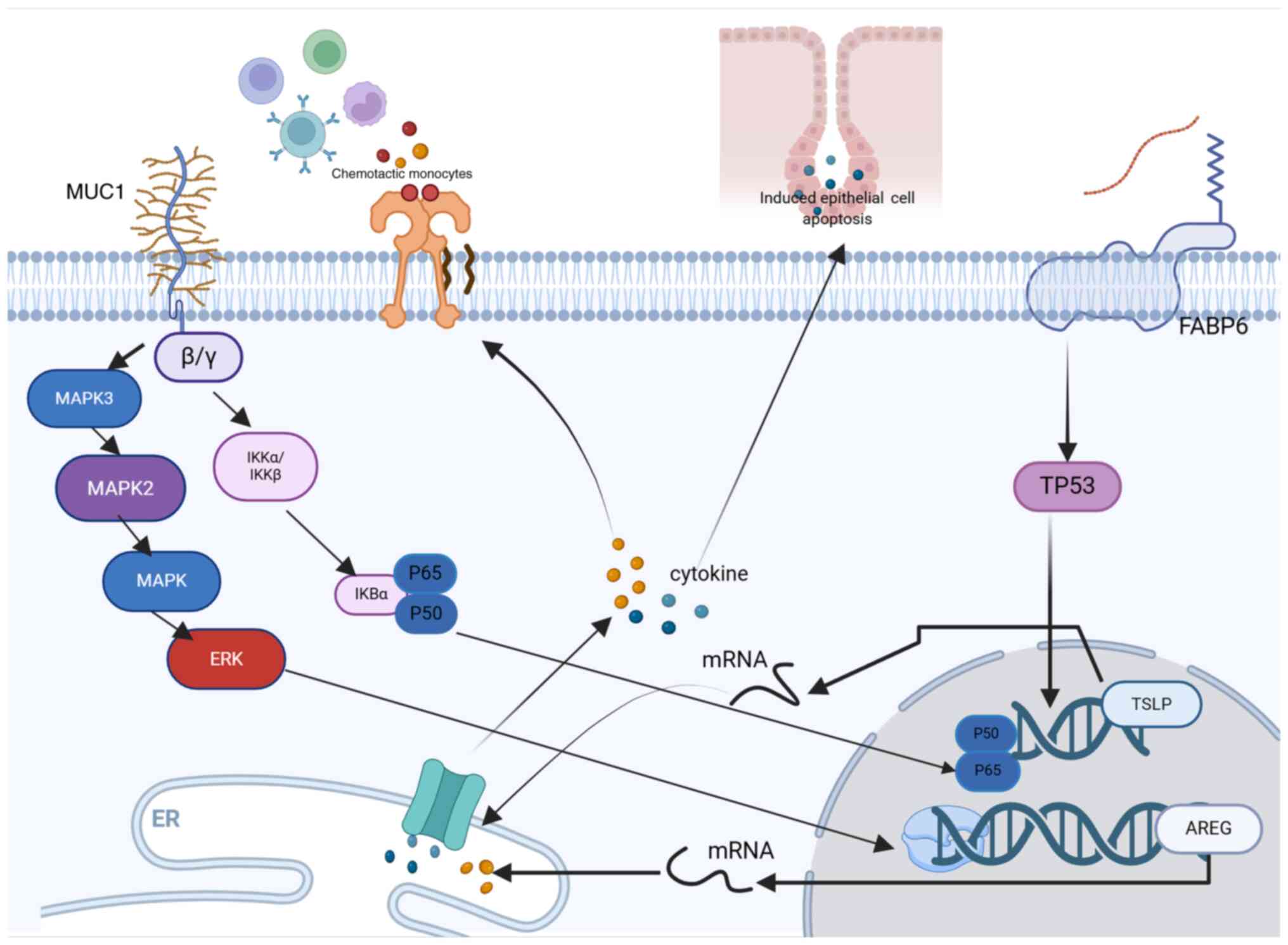
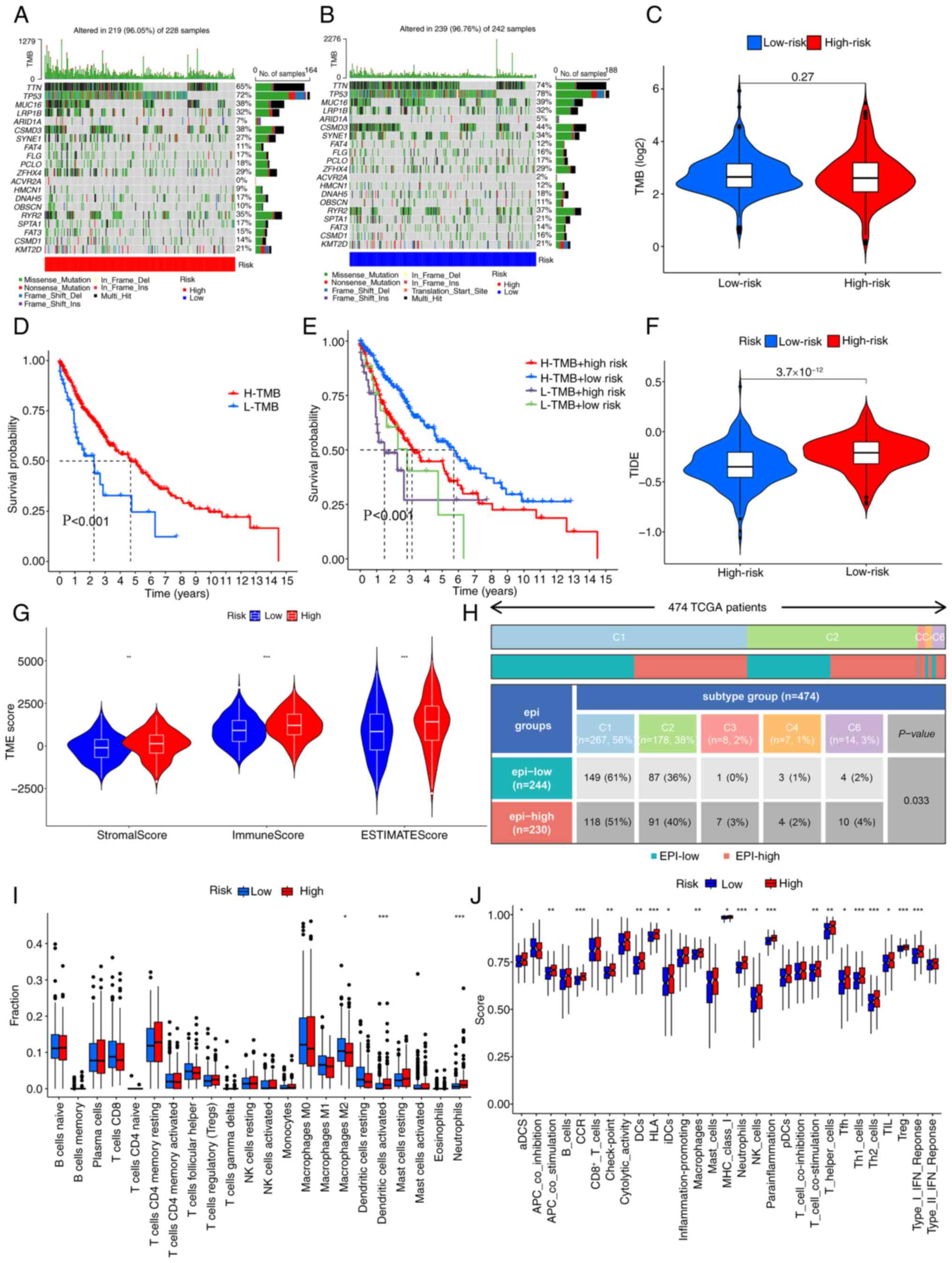
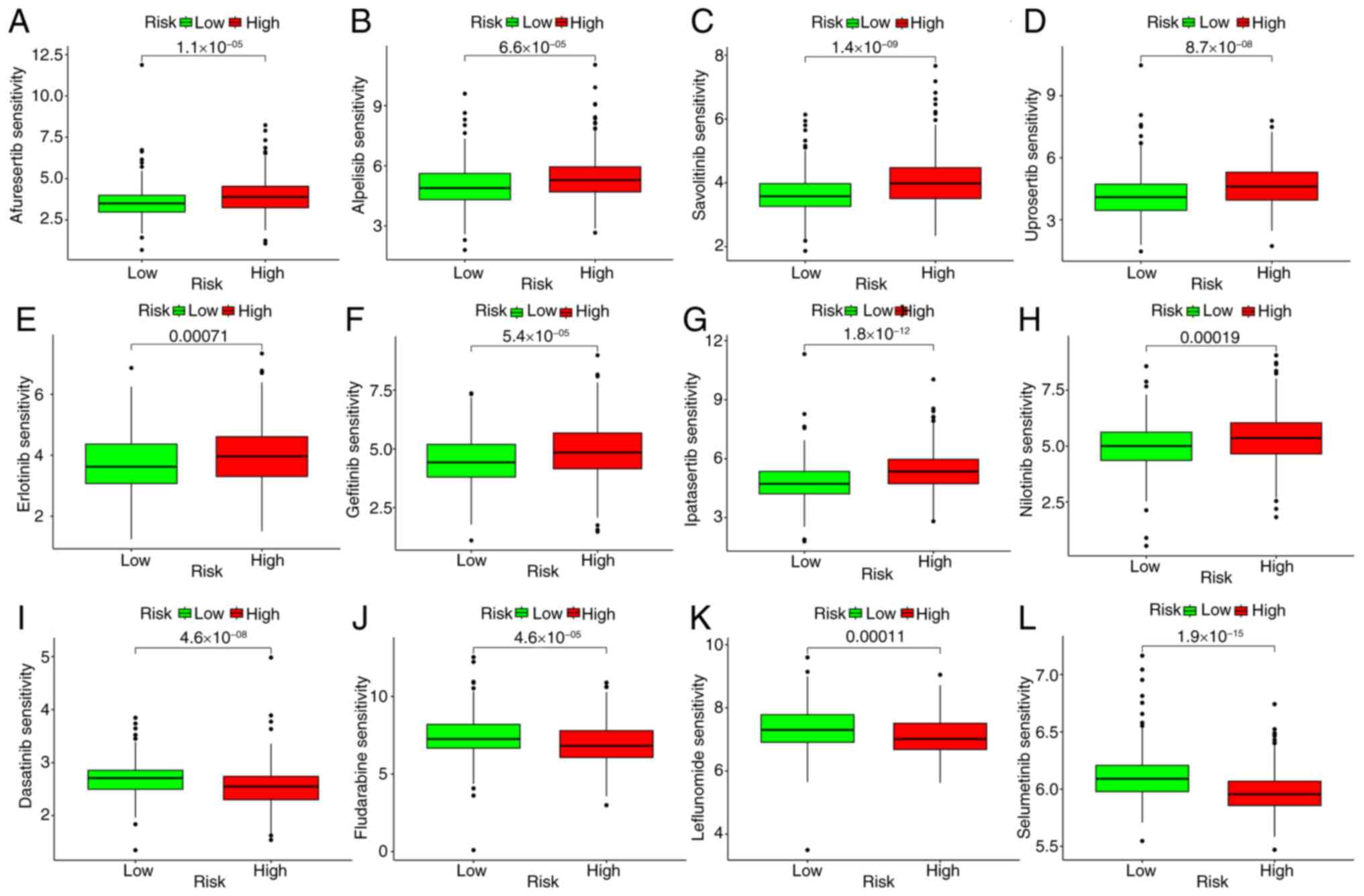
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rodak O, Peris-Díaz MD, Olbromski M,
Podhorska-Okołów M and Dzięgiel P: Current landscape of non-small
cell lung cancer: Epidemiology, histological classification,
targeted therapies, and immunotherapy. Cancers (Basel).
13:47052021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Piñeros M, Parkin DM, Ward K, Chokunonga
E, Ervik M, Farrugia H, Farrugia H, Gospodarowicz M, O'Sullivan B,
Soerjomataram I, et al: Essential TNM: A registry tool to reduce
gaps in cancer staging information. Lancet Oncol. 20:e103–e111.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruni D, Angell HK and Galon J: The immune
contexture and immunoscore in cancer prognosis and therapeutic
efficacy. Nat Rev Cancer. 20:662–680. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sands JM, Nguyen T, Shivdasani P, Sacher
AG, Cheng ML, Alden RS, Jänne PA, Kuo FC, Oxnard GR and Sholl LM:
Next-generation sequencing informs diagnosis and identifies
unexpected therapeutic targets in lung squamous cell carcinomas.
Lung Cancer. 140:35–41. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han X, Zhou Z, Fei L, Sun H, Wang R, Chen
Y, Chen H, Wang J, Tang H, Ge W, et al: Construction of a human
cell landscape at single-cell level. Nature. 581:303–309. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Joanito I, Wirapati P, Zhao N, Nawaz Z,
Yeo G, Lee F, Eng CLP, Macalinao DC, Kahraman M, Srinivasan H, et
al: Single-cell and bulk transcriptome sequencing identifies two
epithelial tumor cell states and refines the consensus molecular
classification of colorectal cancer. Nat Genet. 54:963–975. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen HT, Liu H, Mao MJ, Tan Y and Mo XQ:
Crosstalk between autophagy and epithelial-mesenchymal transition
and its application in cancer therapy. Mol Cancer. 18:1012019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang Z, Wu C, Liu X, Lu S, You L, Guo F,
Stalin A, Zhang J, Zhang F, Wu Z, et al: Single-cell and bulk RNA
sequencing reveal malignant epithelial cell heterogeneity and
prognosis signatures in gastric carcinoma. Cells. 11:25502022.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu K, Lin CJ, Hatcher A, Lozzi B, Kong K,
Huang-Hobbs E, Cheng YT, Beechar VB, Zhu W, Zhang Y, et al: PIK3CA
variants selectively initiate brain hyperactivity during
gliomagenesis. Nature. 578:166–171. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aran D, Looney AP, Liu L, Wu E, Fong V,
Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, et al:
Reference-based analysis of lung single-cell sequencing reveals a
transitional profibrotic macrophage. Nat Immunol. 20:163–172. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cao J, Spielmann M, Qiu X, Huang X,
Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers
FJ, et al: The single-cell transcriptional landscape of mammalian
organogenesis. Nature. 566:496–502. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Q, Qiao W, Zhang H, Liu B, Li J, Zang
C, Mei T, Zheng J and Zhang Y: Nomogram established on account of
Lasso-Cox regression for predicting recurrence in patients with
early-stage hepatocellular carcinoma. Front Immunol.
13:10196382022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H, Han D, Hou Y, Chen H and Chen Z:
Statistical inference methods for two crossing survival curves: A
comparison of methods. PLoS One. 10:e01167742015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang J, Zhang JL, Ang L, Li MC, Zhao M,
Wang Y and Wu Q: Proposing a novel molecular subtyping scheme for
predicting distant recurrence-free survival in breast cancer
post-neoadjuvant chemotherapy with close correlation to metabolism
and senescence. Front Endocrinol (Lausanne). 14:12655202023.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Petegrosso R, Li Z and Kuang R: Machine
learning and statistical methods for clustering single-cell
RNA-sequencing data. Brief Bioinform. 21:1209–1223. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smith-Miles K and Geng X: Revisiting
facial age estimation with new insights from instance space
analysis. IEEE Trans Pattern Anal Mach Intell. 44:2689–2697. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Singh S, Singh VK and Rai G:
Identification of differentially expressed hematopoiesis-associated
genes in term low birth weight newborns by systems genomics
approach. Curr Genomics. 20:469–482. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Postow MA, Sidlow R and Hellmann MD:
Immune-related adverse events associated with immune checkpoint
blockade. N Engl J Med. 378:158–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie T, Peng S, Liu S, Zheng M, Diao W,
Ding M, Fu Y, Guo H, Zhao W and Zhuang J: Multi-cohort validation
of ascore: An anoikis-based prognostic signature for predicting
disease progression and immunotherapy response in bladder cancer.
Mol Cancer. 23:302024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maeser D, Gruener RF and Huang RS:
oncoPredict: An R package for predicting in vivo or cancer patient
drug response and biomarkers from cell line screening data. Brief
Bioinform. 22:bbab2602021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–428. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guénin S, Mauriat M, Pelloux J, Van
Wuytswinkel O, Bellini C and Gutierrez L: Normalization of qRT-PCR
data: The necessity of adopting a systematic, experimental
conditions-specific, validation of references. J Exp Bot.
60:487–493. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zaiss DMW, Gause WC, Osborne LC and Artis
D: Emerging functions of amphiregulin in orchestrating immunity,
inflammation, and tissue repair. Immunity. 42:216–226. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun R, Zhao H, Gao DS, Ni A, Li H, Chen L,
Lu X, Chen K and Lu B: Amphiregulin couples IL1RL1+ regulatory T
cells and cancer-associated fibroblasts to impede antitumor
immunity. Sci Adv. 9:eadd73992023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang S, Deng T, Cheng H, Liu W, Shi D,
Yuan J, He Z, Wang W, Chen B, Ma L, et al: Macrophage-organoid
co-culture model for identifying treatment strategies against
macrophage-related gemcitabine resistance. J Exp Clin Cancer Res.
42:1992023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Murthy D, Attri KS, Suresh V, Rajacharya
GH, Valenzuela CA, Thakur R, Zhao J, Shukla SK, Chaika NV, LaBreck
D, et al: The MUC1-HIF-1α signaling axis regulates pancreatic
cancer pathogenesis through polyamine metabolism remodeling. Proc
Natl Acad Sci USA. 121:e23155091212024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li XX, Wang LJ, Hou J, Liu HY, Wang R,
Wang C and Xie WH: Identification of long noncoding RNAs as
predictors of survival in triple-negative breast cancer based on
network analysis. Biomed Res Int. 2020:89703402020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao T, Cen Q and Lei H: A review on
development of MUC1-based cancer vaccine. Biomed Pharmacother.
132:1108882020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hendrick AG, Müller I, Willems H, Leonard
PM, Irving S, Davenport R, Ito T, Reeves J, Wright S, Allen V, et
al: Identification and investigation of novel binding fragments in
the fatty acid binding protein 6. J Med Chem. 59:8094–8102. 2020.
View Article : Google Scholar
|
|
31
|
Braile M, Fiorelli A, Sorriento D, Di
Crescenzo RM, Galdiero MR, Marone G, Santini M, Varricchi G and
Loffredo S: Human lung-resident macrophages express and are targets
of thymic stromal lymphopoietin in the tumor microenvironment.
Cells. 10:20122021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Corren J and Ziegler SF: TSLP: From
allergy to cancer. Nat Immunol. 20:1603–1609. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Matsuzaki H, Asou N, Kawaguchi Y, Hata H,
Yoshinaga T, Kinuwaki E, Ishii T, Yamaguchi K and Takatsuki K:
Human T-cell leukemia virus type 1 associated with small cell lung
cancer. Cancer. 66:1763–1768. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mamtani R, Clark AS, Scott FI, Brensinger
CM, Boursi B, Chen L, Xie F, Yun H, Osterman MT, Curtis JR and
Lewis JD: Association between breast cancer recurrence and
immunosuppression in rheumatoid arthritis and inflammatory bowel
disease: A cohort study. Arthritis Rheumatol. 68:2403–2411. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shi X, Dong A, Jia X, Zheng G, Wang N,
Wang Y, Yang C, Lu J and Yang Y: Integrated analysis of single-cell
and bulk RNA-sequencing identifies a signature based on T-cell
marker genes to predict prognosis and therapeutic response in lung
squamous cell carcinoma. Front Immunol. 13:9929902022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lai X, Fu G, Du H, Xie Z, Lin S, Li Q and
Lin K: Identification of a cancer-associated fibroblast classifier
for predicting prognosis and therapeutic response in lung squamous
cell carcinoma. Medicine (Baltimore). 102:e350052023. View Article : Google Scholar : PubMed/NCBI
|