Introduction
Papillary thyroid carcinoma (PTC) is the most common
pathological subtype of thyroid cancer, followed by follicular and
medullary thyroid cancer (1). Due
to its increasing incidence rate, PTC is considered a serious
concern for society (2). Although
the majority of PTC cases have a relatively good prognosis, the
risk of regional recurrence and PTC-related deaths should not be
underestimated in patients with lymph node infiltration (3,4).
Unlike medullary thyroid cancer, which is characterized by the
secretion of calcitonin and carcinoembryonic antigen, there are no
significant molecular biomarkers for PTC (1). Therefore, more research on the
identification of novel biomarkers is needed for the early
diagnosis and individual treatment of patients with PTC.
Circular RNAs (circRNAs) are a class of endogenous
non-coding RNAs, which are usually generated by the back-cleavage
of exons in protein-encoding genes (5). Due to their tissue-specific expression
profile and highly conserved characteristics, circRNAs have become
targets for cancer treatment (6,7). Zhang
et al (8) revealed that
circ_TIAM1 may be involved in the development of PTC by acting as
an oncogene. Additionally, another study demonstrated that
circ_102171 could interact with CTNNBIP1 protein to activate the
Wnt/β-catenin pathway, thus promoting PTC progression (9). However, the role and mechanism of
circRNAs in PTC remain to be elucidated.
hsa_circ_0004846, also known as circ_SAMD4A (chr14:
55168779-55169298), is a circRNA with a length of 519 nucleotides
that is derived from the reverse cleavage of SAMD4A mRNA. Previous
studies have shown that hsa_circ_0004846 is involved in the
development of osteosarcoma (10,11).
For example, Zhao et al (11) revealed that hsa_circ_0004846 could
enhance the proliferation and the stemness features of osteosarcoma
cells. Furthermore, hsa_circ_0004846 silencing has been shown to
suppress the epithelial-mesenchymal transition and malignant
behavior of osteosarcoma cells (12). However, to the best of our
knowledge, there are currently no reports on the role of
hsa_circ_0004846 in other types of cancer, including PTC.
With the advancement of sequencing technology,
circRNAs have been shown to regulate the expression of their target
genes by acting as competing endogenous RNAs (ceRNAs) of microRNAs
(miRNAs/miRs) (13). miRNAs, a
class of endogenous and small noncoding RNAs, act as regulators of
cell-cell communication during tumor progression (14). For example, miR-221 and miR-222 have
been reported to be significantly upregulated in PTC compared with
in benign thyroid nodules (15).
Emerging evidence has suggested that serum miRNAs, such as
miR-146-b and miR-221, may be considered as novel biomarkers for
diagnosing PTC (16). Furthermore,
miR-142-3p may induce apoptosis, and inhibit cell proliferation and
metastasis in PTC (17). However,
the regulatory mechanism of miR-142-3p in PTC remains unclear.
The present study aimed to reveal the role of
hsa_circ_0004846 in PTC. Therefore, hsa_circ_0004846-overexpressing
and -depleted TPC-1 and IHH-4 cell lines were established. Based on
the ceRNA hypothesis, the current study further explored whether
the effect of hsa_circ_0004846 on PTC cells was mediated by the
miR-142-3p/Pellino E3 ubiquitin protein ligase 1 (PELI1) axis.
Materials and methods
Human tissue samples
A total of 34 pairs of frozen PTC tissues and
paracancerous tissues were obtained from patients (mean age, 45
years; age range, 20–65 years; 7 male patients and 27 female
patients) who underwent surgical resection at The First Affiliated
Hospital of Anhui Medical University (Hefei, China) between
February 2021 and June 2022. No patients received radiotherapy or
chemotherapy before surgery, and there was no history of neck
radiation exposure during adolescence or childhood.
Cell culture
The thyroid cancer cell line BCPAP, the PTC cell
line TPC-1 and the normal human thyroid cell line Nthy-ori 3–1 were
purchased from iCell Bioscience Inc., and were cultured in
RPMI-1640 medium (Beijing Solarbio Science & Technology Co.,
Ltd.) supplemented with 10% fetal bovine serum (FBS; Zhejiang
Tianhang Biotechnology Co., Ltd.). Another PTC cell line (IHH-4)
was obtained from Nanjing CoBioer Biotechnology Co., Ltd. and was
maintained in DMEM (Wuhan Servicebio Technology Co., Ltd.)
supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin
(100 mg/ml). All cells were incubated at 37°C in an incubator
containing 5% CO2. Profiling and authentication of
BCPAP, TPC-1, IHH-4 and Nthy-ori 3–1 cell lines was performed using
the short tandem repeat identification criteria developed by the
International Cell Line Authentication Committee (18).
Cell transduction and
transfection
The EGFP-expressing lentivirus vector
pLKO.1-EGFP-puro was purchased from Hunan Fenghui Biotechnology
Co., Ltd. (cat. no. FH1717), whereas the GFP-expressing pLC5-ciR
lentivirus vector was obtained from Guangzhou Geneseed Biotech Co.,
Ltd. The short hairpin RNA (shRNA) against hsa_circ_0004846
(sh-circRNA; sense, 5′-CAAGAATCATTAACCAATGGC-3′ and antisense,
5′-GCCATTGGTTAATGATTCTTG-3′) and the corresponding negative control
shRNA (sh-NC; sense, 5′-TTCTCCGAACGTGTCACGT-3′ and antisense,
5′-ACGTGACACGTTCGGAGAA-3′) were obtained from General Biology
(Anhui) Co., Ltd., and were sub-cloned into the pLKO.1-EGFP-puro
lentivirus vector. The full-length hsa_circ_0004846 sequence was
sub-cloned into the pLC5-ciR lentivirus vector (Lv-circRNA). The
empty pLC5-ciR was used as a negative control (Lv-Vector).
To obtain lentiviral particles, the 2nd lentiviral
generation system was used. Briefly, 293T cells (Cellverse Co.,
Ltd.) were transfected with 14 µg lentiviral plasmids, 10.5 µg
packaging plasmids and 3.5 µg envelope plasmids using
Lipofectamine™ 3000 (Invitrogen; Thermo Fisher Scientific, Inc.)
for 6 h at 37°C. After replacing the medium with fresh medium, the
lentiviral particles were collected after continued incubation for
72 h at 37°C. For stable transduction, the aforementioned
lentivirus particles (Lv-sh-NC, Lv-sh-circRNA, Lv-Vector and
Lv-circRNA) were used to infect TPC-1 (MOI=10) and IHH-4 (MOI=20)
cells. After 24 h, TPC-1 cells were selected with 2 µg/ml puromycin
and IHH-4 cells were selected with 3 µg/ml puromycin for 7 days.
The maintenance concentration of puromycin used for TPC-1 and IHH-4
cells was 1 and 1.5 µg/ml, respectively. Subsequently, a BX53
fluorescence microscope (Olympus Corporation) was used to confirm
the transduction efficiency. Once the stably transduced cells were
constructed, the cells underwent subsequent experiments according
to the experimental protocol.
miR-142-3p-mimic, NC-mimic, PELI1 small interfering
(si)RNA (si-PEL1) and the siRNA negative control (si-NC) were
purchased from General Biology (Anhui) Co., Ltd. The sequences were
as follows: miR-142-3p-mimic sense, 5′-UGUAGUGUUUCCUACUUUAUGGA-3′,
antisense, 5′-UCCAUAAAGUAGGAAACACUACA-3′; NC-mimic sense,
5′-UUCUCCGAACGUGUCACGUTT-3′, antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; si-PELI1 sense,
5′-CAGUCAGUACAAAGCACUAUATT-3′, antisense, UAUAGUGCUUUGUACUGACUGTT;
and si-NC sense, 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. TPC-1 cells were transfected with the
indicated mimics (75 pmol) and siRNAs (75 pmol) using Lipofectamine
3000 for 15 min at 37°C. according to the manufacturer's protocol.
A total of 48 h after transfection, the cells were collected for
subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells and tissues using
the TRIpure Reagent (BioTeke Corporation), and was then reverse
transcribed into the corresponding cDNA using the BeyoRT II M-MLV
reverse transcriptase (Beyotime Institute of Biotechnology) or the
miRNA First Strand cDNA Synthesis (Tailing Reaction) Kit (cat. no.
B532451; Sangon Biotech Co., Ltd.) according to the manufacturers'
protocols. qPCR was performed on the Exicycler™ 96 system (Bioneer
Corporation) using SYBR Green (Beijing Solarbio Science &
Technology Co., Ltd.) and 2X Taq PCR MasterMix kit (Beijing
Solarbio Science & Technology Co., Ltd.). The cycling
conditions comprised an initial denaturation step for 5 min at
95°C, followed by 40 cycles at 95°C for 10 sec (denaturation),
annealing at 60°C for 10 sec and elongation at 72°C for 15 sec. The
expression levels of the target mRNA and circRNA were normalized to
the housekeeping gene β-actin, whereas miR-142-3p expression was
normalized to U6. The relative quantification of target genes in
frozen clinical PTC/paracancerous tissues and cultured cells was
calculated using the 2−ΔCq and 2−ΔΔCq
methods, respectively (19). The
primer sequences were as follows: hsa_circ_0004846 forward,
5′-CGGGATTCTGGGATTTG-3′; reverse, 5′-TTGGGCAGCAGTTTCAT-3′; PELI1
forward, 5′-CTACCGTGAAGCATTTA-3′; reverse, 5′-ACAGAGGAACATAGGGA-3′;
β-actin forward, 5′-GGCACCCAGCACAATGAA-3′; reverse,
5′-TAGAAGCATTTGCGGTGG-3′; and miR-142-3p forward,
5′-TGTAGTGTTTCCTACTTTATGGA-3′. The U6 primers and the miR-142-3p
reverse primer were contained in the corresponding miRNA First
Strand cDNA Synthesis (Tailing Reaction) kit for RT.
Characterization of
hsa_circ_0004846
Total RNA was extracted from TPC-1 cells using the
TRIpure Reagent, and was then reverse transcribed into the
corresponding cDNA using the BeyoRT II M-MLV reverse transcriptase
according to manufacturer's protocol. Genomic DNA (gDNA) was
extracted from TPC-1 cells by using a gDNA extraction kit (BioTeke
Corporation). To validate the formation of hsa_circ_0004846, the
divergent primer (DP) sequences that amplify only circular
transcripts and the convergent primer (CP) sequences that detect
linear RNA molecules were designed to amplify linear mRNA (SAMD4A;
Fig. 1A) and hsa_circ_0004846 using
cDNA and gDNA as templates. β-actin was employed as the internal
control. The sequences of the primers were as follows:
hsa_circ_0004846 CP (forward, 5′-GGGATTCTGGGATTTGC-3′; reverse,
5′-CGTATTGATTGTGGTGGG-3′); hsa_circ_0004846 DP (forward,
5′-CGGGATTCTGGGATTTG-3′; reverse, 5′-TTGGGCAGCAGTTTCAT-3′); β-actin
CP (forward, 5′-GGCACCCAGCACAATGAA-3′; reverse,
5′-TAGAAGCATTTGCGGTGG-3′); and the DP for β-actin (forward,
5′-GCCTCGCTGTCCACCTT-3′; reverse, 5′-TCTGACCCATGCCCACC-3′), which
was used as a negative control. Semi-quantitative PCR was carried
out on the Exicycler™ 96 machine (Bioneer Corporation) in a 20-µl
reaction system containing 1 µl cDNA or gDNA, 10 µl 2X Taq PCR
MasterMix and 1 µl forward/reverse primer for divergent primers
(DPs) and convergent primers (CPs), respectively. The thermocycling
condition were as follows: 2 min at 94°C for pre-denaturation; 40
cycles at 94°C for 10 sec for denaturation, 54°C for 20 sec for
annealing and 72°C for 30 sec for extension. β-actin was used as a
loading control. Subsequently, 1.5% agarose gel (Biowest) was used
for electrophoresis of PCR products, and Gold View dye (Beijing
Solarbio Science & Technology Co., Ltd.) was used for
visualization.
Western blot analysis
Total protein extracts were isolated from cells
using the RIPA lysis buffer (Beijing Solarbio Science &
Technology Co., Ltd.) and were quantified using the BCA Protein
Assay Kit (Beijing Solarbio Science & Technology Co., Ltd.).
Western blot analysis was performed as previously described
(12). The primary antibodies used
were as follows: Anti-phosphorylated (p)-AKT (cat. no. AP0140;
dilution, 1:500), anti-AKT (cat. no. A17909; dilution, 1:1,000),
anti-p-glycogen synthase kinase-3β (GSK-3β; cat. no. AP0039;
dilution, 1:500) and anti-GSK-3β (cat. no. A2081; dilution,
1:1,000) (all from ABclonal Biotech Co., Ltd). The following
secondary antibodies were purchased from Beijing Solarbio Science
& Technology Co., Ltd.: Goat anti-rabbit IgG/HRP (cat. no.
SE134; dilution, 1:3,000) and goat anti-mouse IgG/HRP (cat. no.
SE131; dilution, 1:3,000). GAPDH (cat. no. 60004-1-Ig; dilution,
1:20,000; Wuhan Sanying Biotechnology) was used as the internal
control.
Cell Counting Kit 8 (CCK-8) assay
Cell proliferation and viability were assessed using
a CCK-8 Assay Kit (Nanjing KeyGen Biotech Co., Ltd.), according to
the manufacturer's instructions. Briefly, after being transfected
for 48 h, 5×103 cells/well were seeded into 96-well
plates and incubated for 24, 48 or 72 h. Subsequently, each well
was supplemented with 10 µl CCK-8 solution and incubated for an
additional 2 h. Finally, the absorbance in each well was measured
at a wavelength of 450 nm using an 800TS Microplate Reader (Biotek;
Agilent Technologies, Inc.).
Colony formation assay
The colony formation assay was performed as
previously described (20).
Briefly, TPC-1 and IHH-4 cells transduced with the corresponding
lentiviral particles were seeded into culture dishes (400
cells/dish) and maintained in an incubator for 14 days. After
washing with PBS, cells were fixed with 4% paraformaldehyde for 1
min at room temperature and then stained with Wright-Giemsa
solution (Nanjing Jiancheng Bioengineering Institute) for 5 min at
room temperature. The visible colonies containing ≥50 cells were
manually counted under an IX53 light microscope (Olympus
Corporation). The colony formation rate was calculated using the
following formula: Colony formation rate (%)=(number of
colonies/400) ×100.
5-ethynyl-2′-deoxyuridine (EdU)
staining
The EdU Imaging Detection Kit (Nanjing KeyGen
Biotech Co., Ltd.) was utilized to evaluate cell proliferation,
according to the manufacturer's instructions. Briefly, transduced
TPC-1 and IHH-4 cells were treated with 10 µM EdU staining solution
at 37°C for 2 h, followed by fixing with 4% paraformaldehyde for 15
min at room temperature and staining with the Click-iT reaction
solution for 30 min at room temperature. The cell nuclei were
stained with DAPI for 5 min at room temperature. Finally, images of
the cells were captured under an IX53 light microscope (Olympus
Corporation).
Immunofluorescence (IF) staining of
vimentin
To detect the protein expression levels of vimentin
in PTC cells, IF staining was conducted. Specifically, TPC-1 and
IHH-4 cells were fixed with 4% paraformaldehyde for 15 min at room
temperature. After washing with PBS, cells were permeabilized with
0.1% Triton X-100 (Beyotime Institute of Biotechnology) for 30 min
at room temperature. Subsequently, the slides were blocked with 1%
bovine serum albumin (Sangon Biotech Co., Ltd.) for 15 min at room
temperature and incubated with a primary antibody against vimentin
(cat. no. A19607; dilution, 1:100; ABclonal Biotech Co., Ltd)
overnight at 4°C, followed by incubation with Goat anti-Rabbit IgG
(Heavy chain) Superclonal™ Secondary Antibody, Alexa Fluor™ 555
(cat. no. A27039; dilution, 1:200; Invitrogen; Thermo Fisher
Scientific, Inc.) for 1 h at room temperature. Subsequently, the
cell nuclei were stained with DAPI for 5 min at room temperature
and images were captured at magnification of ×400 under a BX53
fluorescence microscope (Olympus Corporation).
Cell migration and invasion
assays
For the cell migration assay, 300 µl cell suspension
containing 5×103 PTC cells was added to the upper
Transwell chambers (24 wells; pore size, 8.0 µm). For the cell
invasion assay, 5×104 PTC cells in 300 µl were added to
the upper chambers, which were precoated with Matrigel for 2 h at
37°C. The lower chamber was supplemented with 700 µl culture medium
containing 10% FBS. Subsequently, the chambers were incubated in a
37°C incubator with 5% CO2. Following incubation for 24
h, the Transwell chambers were washed with PBS, fixed in 4%
paraformaldehyde for 20 min at room temperature and stained with
0.5% crystal violet solution (Amresco, LLC) for 5 min at room
temperature. Images of the migratory and invasive cells were
captured under a light microscope (IX53; Olympus Corporation).
Dual-luciferase reporter assay
For the dual-luciferase reporter assay, the
wild-type (WT) and mutant (Mut) sequences of the 3′untranslated
region (3′UTR) of PELI1 (WT-PELI1 3′UTR or Mut-PELI1 3′UTR), and
WT-hsa_circ_0004846 and Mut-hsa_circ_0004846 fragments were
sub-cloned into the pmirGLO vector [General Biology (Anhui) Co.,
Ltd.]. 293T cells were co-transfected with pmirGLO vectors
(pmirGLO-WT-PELI1-3′UTR, pmirGLO-Mut-PELI1-3′UTR,
pmirGLO-WT-hsa_circ_0004846 or pmirGLO-hsa_circ_0004846) and miRNA
mimics (miR-142-3p-mimic or NC-mimic) using Lipofectamine 3000.
After 48 h, firefly and Renilla luciferase activities were
detected using a dual-luciferase reporter gene assay kit (Nanjing
KeyGen Biotech Co., Ltd.). The relative luciferase activity was
normalized to Renilla luciferase activity.
Bioinformatics analysis
The circRNA Interactome database (https://circinteractome.nia.nih.gov/)
was used to predict the downstream targets for hsa_circ_0004846,
while an online target prediction (http://www.mirdb.org/) was performed to discover the
downstream targets for miR-142-3p.
Statistical analysis
Data are presented as the mean ± SD. The in
vitro experiments were repeated at last three times.
Statistical analysis was performed using GraphPad Prism 9.0
software (Dotmatics). The paired Student's t-test was used to
assess the differences between the paired PTC tumor and
paracancerous tissues. Unless otherwise specified, the differences
between two groups were compared by unpaired Student's t-test
(two-tailed), while those among multiple groups were compared using
one-way ANOVA followed by Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
hsa_circ_0004846 is upregulated in PTC
tissues and thyroid cancer cell lines
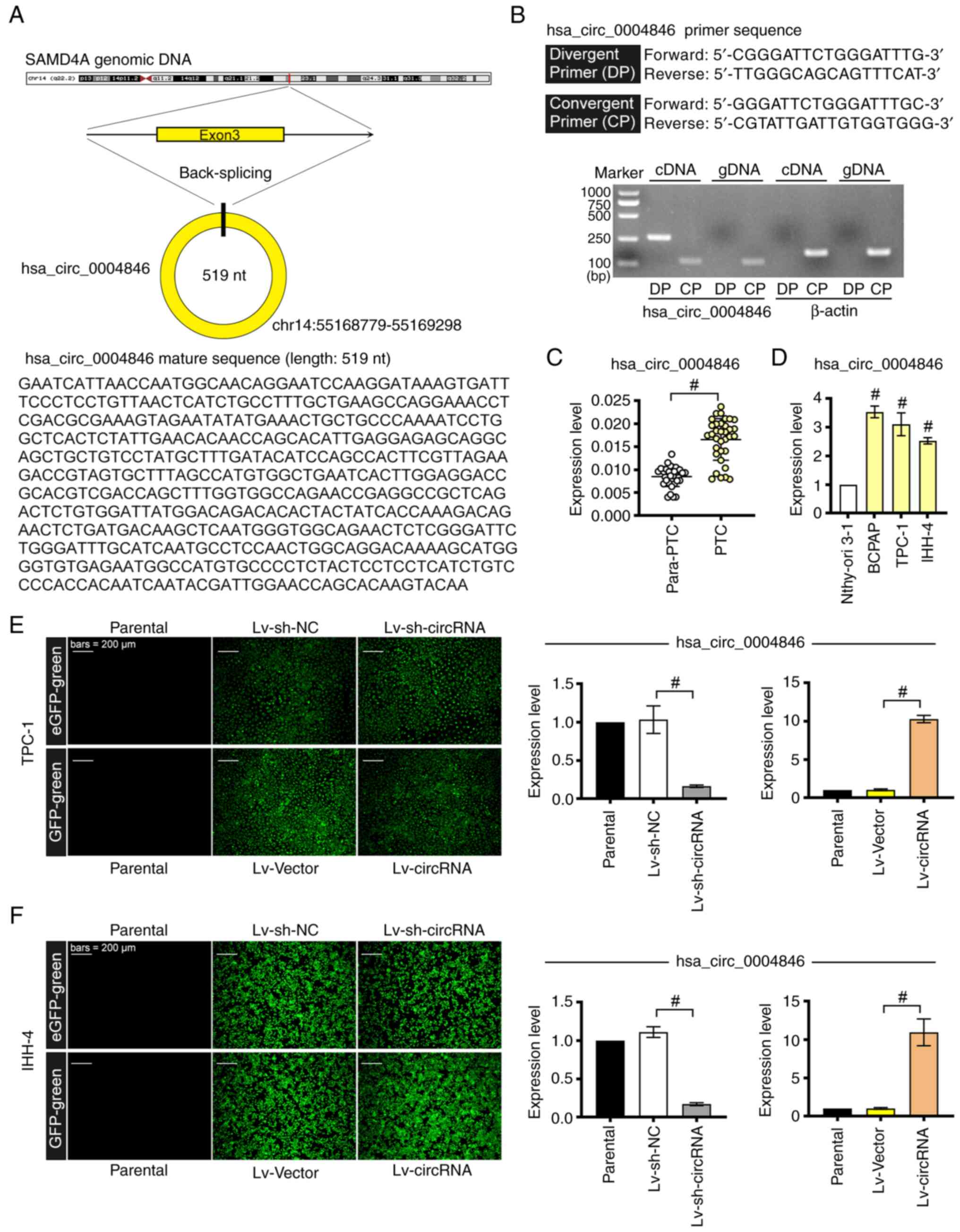
hsa_circ_0004846 (length, 519 nucleotides; chr14:
55168779-55169298) is derived from back-splicing of SAMD4A mRNA
(Fig. 1A). Firstly, the specificity
of the primers for hsa_circ_0004846 were verified by RT-qPCR. As
shown in Fig. 1B, convergent
primers amplified both cDNA and gDNA, whereas divergent primers
detected hsa_circ_0004846 in cDNA but not in gDNA. Subsequently,
the analysis revealed that hsa_circ_0004846 was more highly
expressed in PTC tissues compared with that in paracancerous
tissues (Fig. 1C). Consistently,
hsa_circ_0004846 was also upregulated in the thyroid cancer cell
line BCPAP and the two PTC cell lines, TPC-1 and IHH-4, compared
with that in the Nthy-ori 3–1 normal human thyroid cell line
(Fig. 1D). These findings
encouraged further investigation of the role of hsa_circ_0004846 in
PTC. Therefore, hsa_circ_0004846-overexpressing (Lv-circRNA) and
-depleted (Lv-sh-circRNA) TPC-1 and IHH-4 cell lines were
established via lentiviral infection. The transduction efficiency
was confirmed by fluorescence detection and RT-qPCR analysis
(Fig. 1E and F).
hsa_circ_0004846 promotes PTC cell
proliferation, migration and invasion
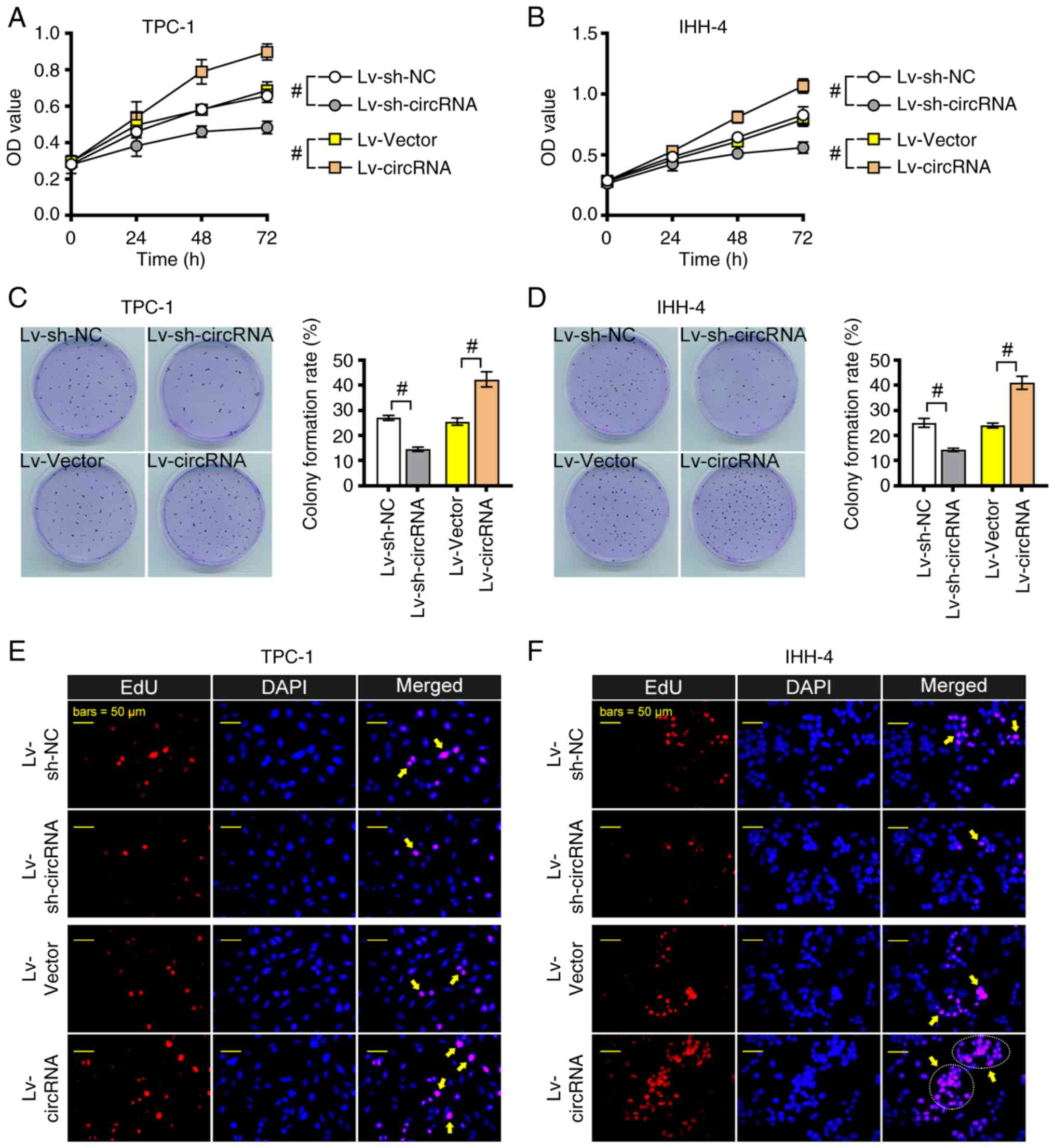
As shown in Fig. 2A and
B, TPC-1 and IHH-4 cell proliferation was significantly
enhanced following hsa_circ_0004846 overexpression; however,
proliferation was markedly reduced in hsa_circ_0004846-depleted PTC
cells. Similar results were observed using the colony formation
assay (Fig. 2C and D). EdU staining
further verified that hsa_circ_0004846 overexpression enhanced PTC
cell proliferation, whereas hsa_circ_0004846 knockdown mitigated
this process (Fig. 2E and F).
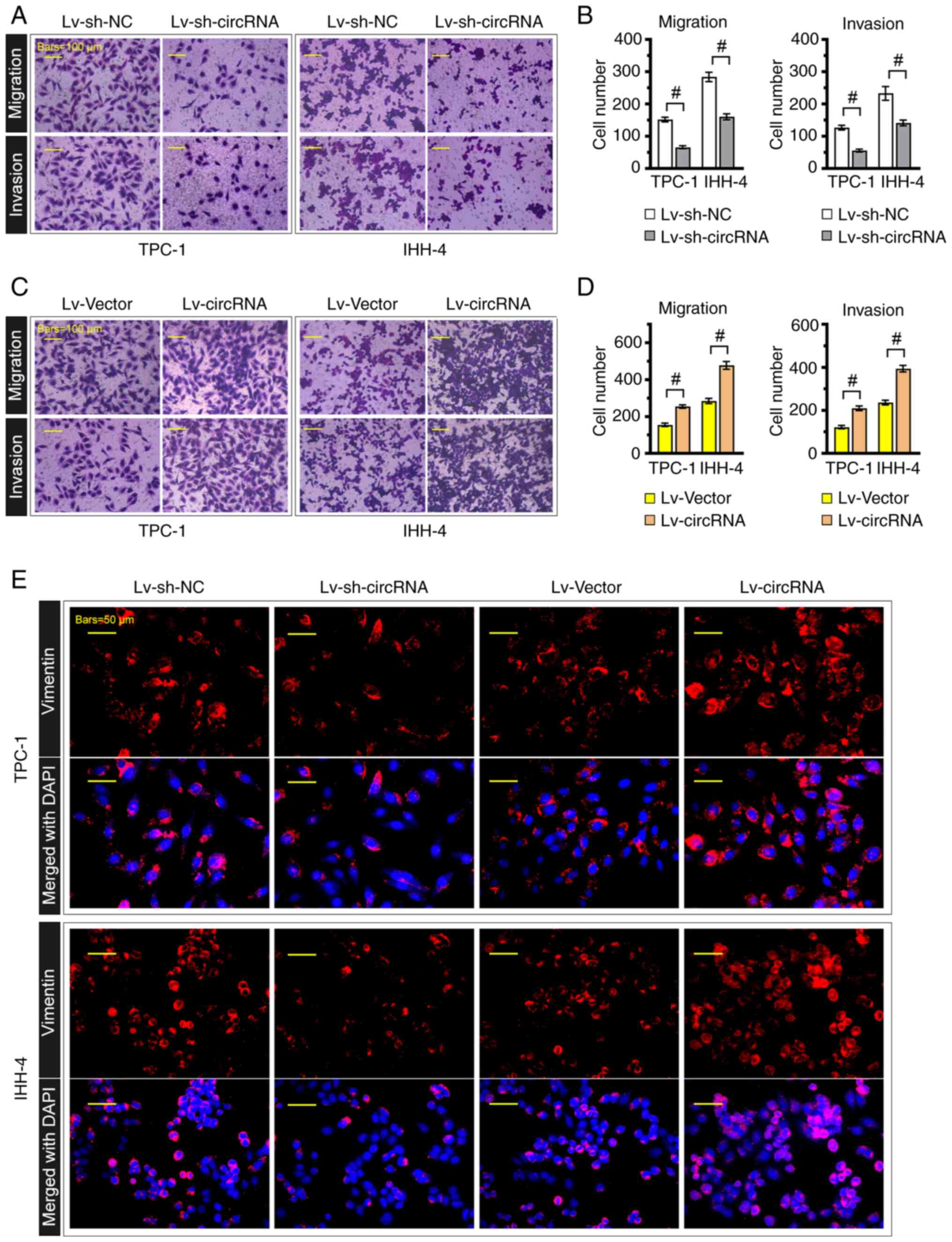
Furthermore, Transwell assays showed that hsa_circ_0004846
knockdown significantly attenuated PTC cell migration and invasion
(Fig. 3A and B); however, TPC-1 and
IHH-4 cells transduced with the hsa_circ_0004846 overexpression
lentivirus displayed stronger migratory and invasive capabilities
(Fig. 3C and D). Consistently, as
shown in Fig. 3E, vimentin loss was
observed in hsa_circ_0004846-silenced cells, whereas
hsa_circ_0004846-overexpressing cells displayed the opposite
effect.
hsa_circ_0004846 activates the
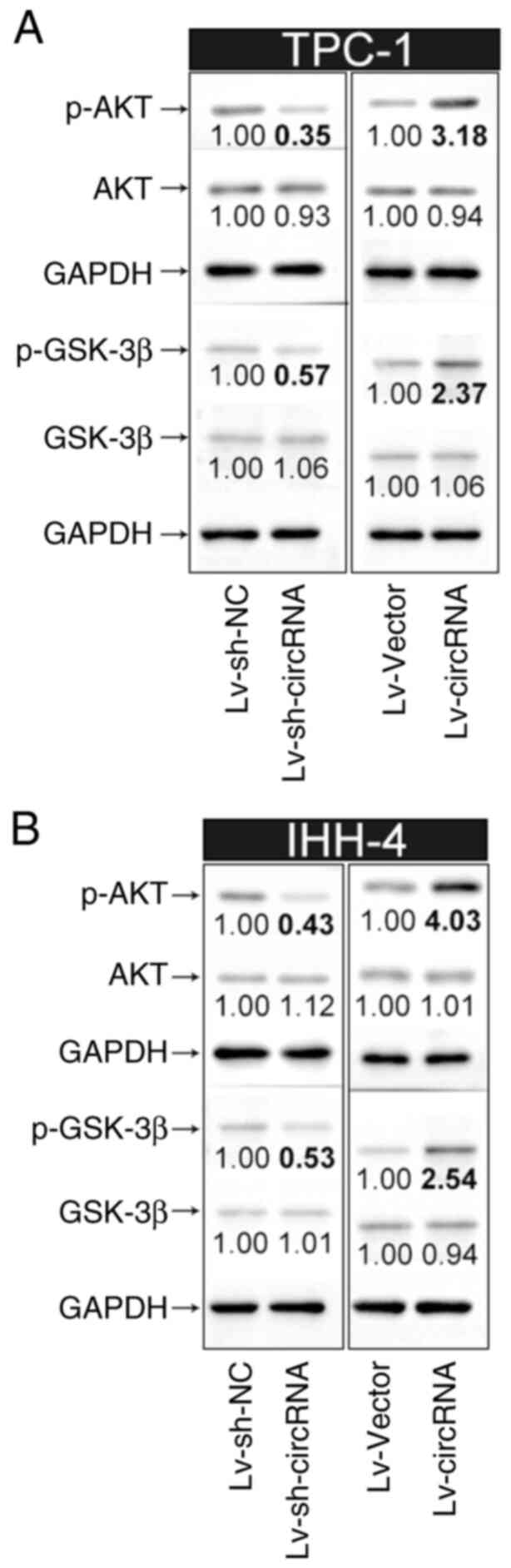
PI3K/AKT pathway in PTC cells
Given that the PI3K/AKT pathway serves a notable
role in the pathogenesis of PTC (21), the current study further explored
whether hsa_circ_0004846-induced PTC cell proliferation was
mediated by activation of the PI3K/AKT pathway. As shown in
Fig. 4A and B, hsa_circ_0004846
overexpression in TPC-1 and IHH-4 cells upregulated p-AKT and
p-GSK-3β. By contrast, hsa_circ_0004846 knockdown notably
suppressed the expression levels of p-AKT and p-GSK-3β. However,
hsa_circ_0004846 overexpression or silencing had no effect on the
protein expression levels of total AKT and GSK-3β.
hsa_circ_0004846 serves as a sponge
for miR-142-3p, which in turn targets PELI1 in PTC cells
Bioinformatics analysis was performed using the
circRNA Interactome database. The analysis predicted that
hsa_circ_0004846 could bind to miR-142-3p (Fig. 5A-a). Dual-luciferase reporter assays
verified that the luciferase activity was significantly decreased
in cells co-transfected with miR-142-3p-mimic and WT-circRNA
compared with that in the NC-mimic + WT-circRNA group (Fig. 5A-b). Furthermore, RT-qPCR analysis
indicated that hsa_circ_0004846 knockdown could upregulate
miR-142-3p and downregulate PELI1 in TPC-1 cells (Fig. 5B and C). By contrast, downregulated
miR-142-3p and upregulated PELI1 expression were observed in
hsa_circ_0004846-overexpressing cells. In addition, bioinformatics
analysis using an online target prediction suggested that PELI1
could be a downstream target for miR-142-3p (Fig. 5D-a). Dual-luciferase reporter assays
verified that co-transfection of cells with WT-PELI1-3′UTR and
miR-142-3p-mimic inhibited luciferase activity compared with that
in the corresponding control group (WT-PELI1-3′UTR + NC-mimic)
(Fig. 5D-b). No significant changes
were observed between the Mut-PELI1-3′UTR + miR-142-3p-mimic and
Mut-PELI1-3′UTR + NC-mimic groups. Furthermore, TPC-1 cells were
transfected with miR-142-3p mimics to overexpress miR-142-3p
(Fig. 5E). The subsequent RT-qPCR
analysis showed that miR-142-3p overexpression significantly
inhibited PELI1 expression (Fig.
5F). Considering that circRNAs often act as ceRNAs of miRNAs to
regulate the expression of their target genes, these results
indicated that hsa_circ_0004846 could promote PELI1 expression by
inhibiting miR-142-3p expression.
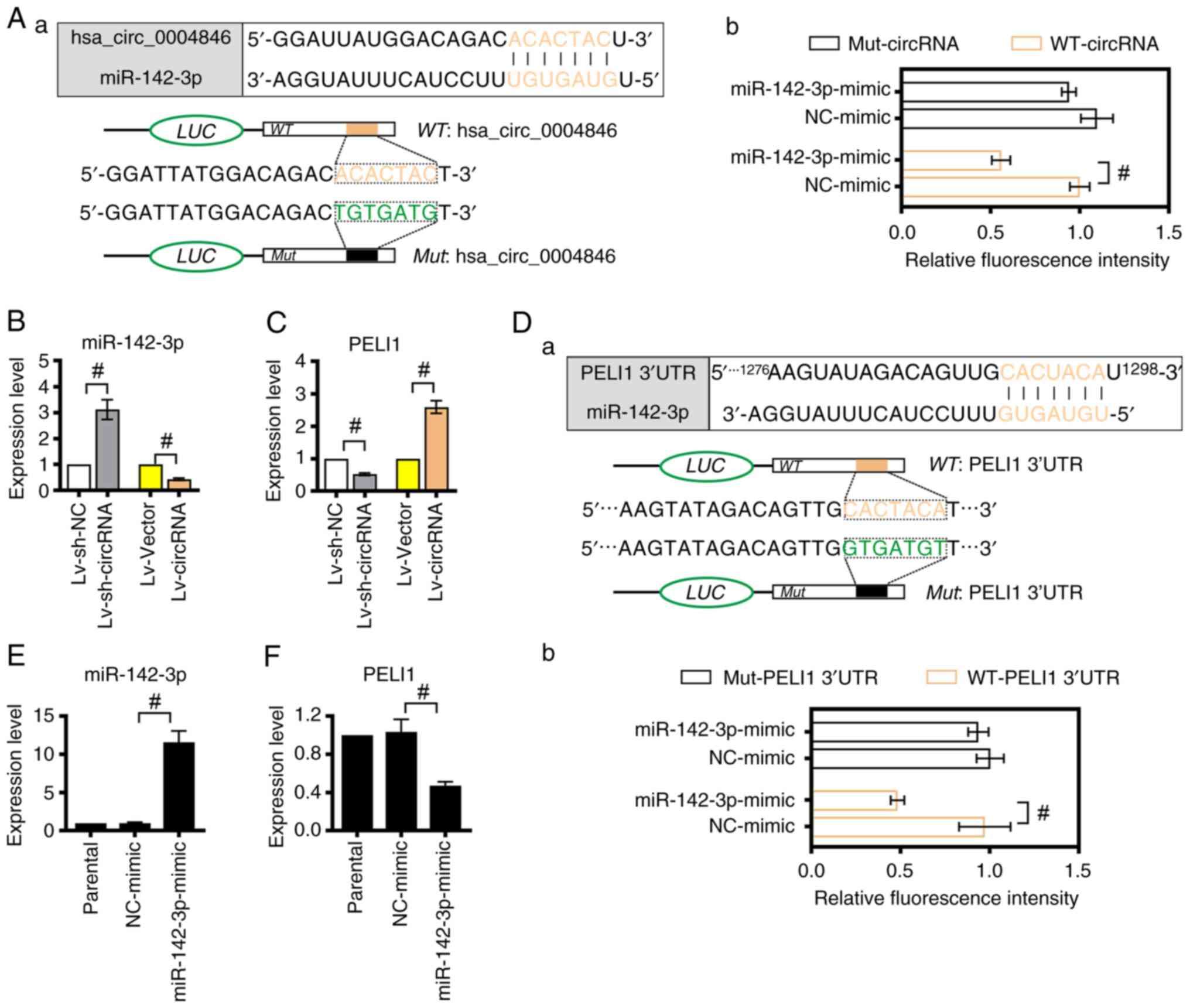 | Figure 5.hsa_circ_0004846 upregulates PELI1 by
sponging miR-142-3p. (A-a) Schematic diagram shows the binding site
of miR-142-3p on hsa_circ_0004846 (https://circinteractome.nia.nih.gov/). (A-b) A
dual-luciferase reporter assay was performed to verify the
interaction between miR-142-3p and hsa_circ_0004846. Expression
levels of (B) miR-142-3p and (C) PELI1 in
hsa_circ_0004846-overexpressing or -depleted TPC-1 cells were
detected by RT-qPCR. (D-a) Schematic diagram shows the binding site
between miR-142-3p and the 3′UTR of PELI1, which was predicted
using the miRDB online database (http://www.mirdb.org/). (D-b) A dual-luciferase
reporter assay was carried out to verify the interaction between
miR-142-3p and 3′-UTR PELI1. Expression levels of (E) miR-142-3p
and (F) PELI1 in TPC-1 cells transfected with miR-142-3p-mimic or
NC-mimic were measured by RT-qPCR analysis. #P<0.05.
circ, circular; PELI1, Pellino E3 ubiquitin protein ligase 1; Lv,
lentivirus; miR, microRNA; Mut, mutant; NC, negative control;
RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin;
WT, wild-type; 3′UTR, 3′ untranslated region. |
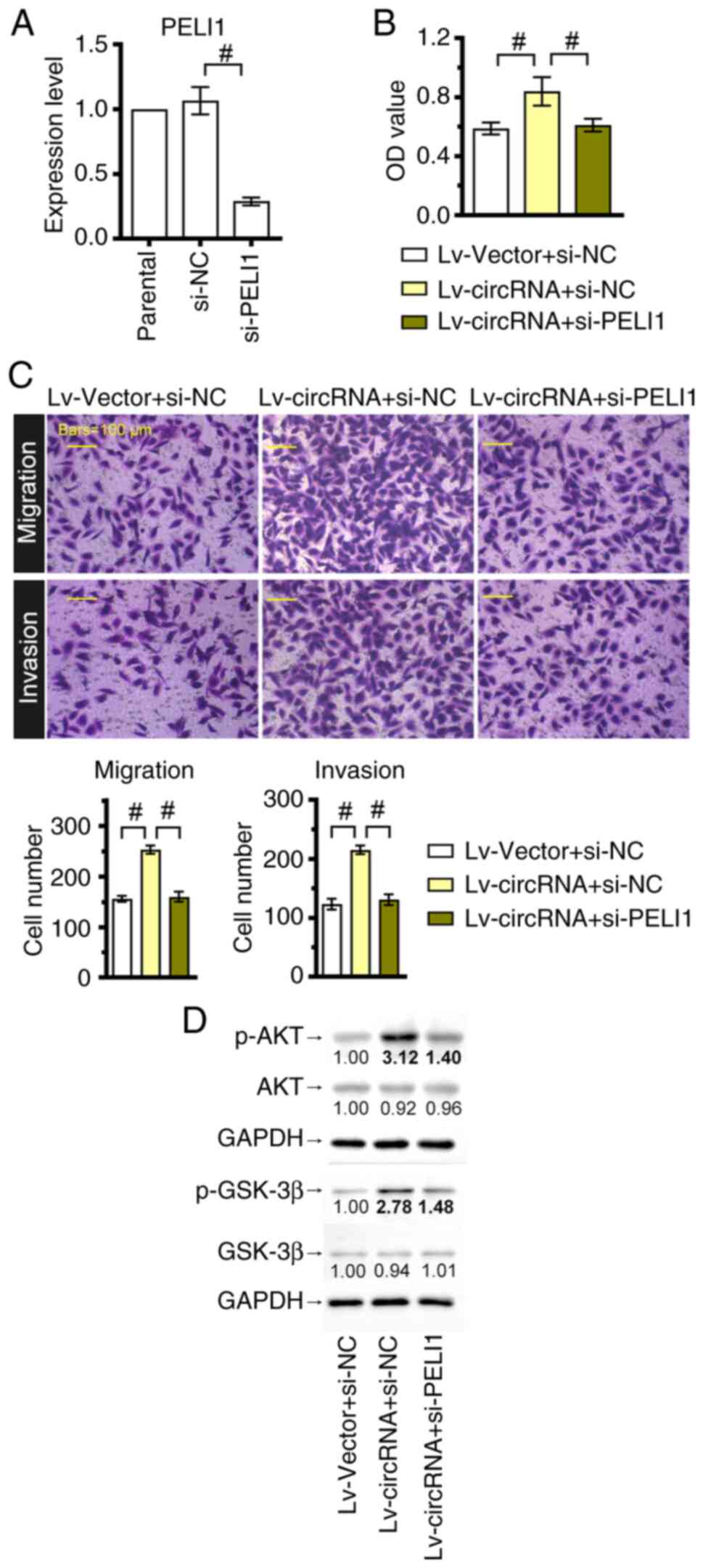
PELI1 silencing reverses the
cancer-promoting effects of hsa_circ_0004846
Since the molecular mechanism of hsa_circ_0004846 in
the miR-142-3p/PELI1 axis was uncovered, a PELI1-silenced TPC-1
cell line was constructed to further explore the role of this axis
in regulating tumorigenesis. The efficiency of cell transfection
with PELI1-siRNA was verified by RT-qPCR analysis (Fig. 6A). Furthermore, the CCK-8 assay
demonstrated that PELI1 silencing significantly inhibited cell
viability in hsa_circ_0004846-overexpressing cells (Fig. 6B). Consistently, PELI1 silencing
also reversed the enhanced invasion and migration of
hsa_circ_0004846-overexpressing TPC-1 cells (Fig. 6C). In addition, western blotting
revealed that the increased expression levels of p-AKT and p-GSK-3β
induced by hsa_circ_0004846 were reversed after PELI1 knockdown
(Fig. 6D).
Discussion
Currently, surgery and iodine 131 radiation therapy
are the most common treatment strategies for PTC (22). According to the American Thyroid
Association guidelines, prophylactic lymph node dissection is
encouraged for high-risk patients with advanced primary tumors
(23); however, the range of lymph
node dissection and its auxiliary role in postoperative radioactive
iodine are still a subject of controversy, as it is beneficial for
preventing further recurrence, but is associated with a higher
incidence of complications (22,24).
Therefore, more effective treatment strategies are needed for
patients with advanced and iodine-resistant PTC.
circRNA-related disorders have been well proven in
several human diseases, including cancer (25,26).
Emerging evidence has indicated that dysregulated circRNAs are
associated with PTC progression. For example, circ_0011373 has been
shown to be upregulated in PTC tissues and to promote PTC cell
proliferation via the miR-1271/low-density lipoprotein
receptor-related protein 6 axis (27). Jiang et al (28) suggested that circLDLR knockdown
could induce PTC cell apoptosis via the miR-637/LIM domain only 4
axis. These findings indicated the role of circRNAs in PTC
progression. To the best of our knowledge, the present study is the
first to demonstrate that hsa_circ_0004846 is significantly
upregulated in PTC tissues and cell lines. Notably,
hsa_circ_0004846 is produced by reverse cleavage of the exon 3 of
SAMD4A. To date, the expression and function of hsa_circ_0004846
have only been reported in osteosarcoma. Zhao et al
(11) indicated that the high
expression of hsa_circ_0004846 was associated with the metastasis
and low overall survival rate of osteosarcoma cells. Their
functional experiments also showed that hsa_circ_0004846 enhanced
the malignant phenotypes of osteosarcoma cells in vivo and
vitro. In the present study, the in vitro cell
experiments revealed that hsa_circ_0004846 overexpression could
markedly upregulate vimentin. A previous study has shown that
vimentin is involved in cell attachment and migration in several
types of cancer, including PTC (29). Furthermore, herein, hsa_circ_0004846
could activate the PI3K/AKT pathway, as evidenced by the increased
expression levels of p-AKT and p-GSK-3β in
hsa_circ_0004846-overexpressing PTC cells. GSK-3β, a major member
of the destruction complex (APC, GSK-3β and Axin-2), is an
important substrate of the PI3K/AKT pathway (30). In addition, the phosphorylation of
AKT could promote the phosphorylation of GSK-3β to promote cancer
cell metastasis (31). These
findings indicated that hsa_circ_0004846 may serve an important
role in the proliferation, migration and invasion of PTC cells.
Mechanistically, circRNAs have been widely reported
to act as ‘miRNA sponges’ that can regulate the expression levels
of miRNAs. In the current study, bioinformatics analysis using an
online database (circRNA Interactome) predicted that miR-142-3p may
be a downstream factor of hsa_circ_0004846. This hypothesis was
further verified by a dual-luciferase reporter assay. Previous
studies have verified that miR-142-3p expression is reduced in
follicular thyroid adenoma tissues, while the forced expression of
miR-142-3p could suppress thyroid cancer WRO and FTC133 cell
proliferation, thus supporting the tumor suppressor role of
miR-142-3p in thyroid cancer (32).
In the present study, miR-142-3p was revealed to be downregulated
in hsa_circ_0004846-overexpressing PTC cells. In addition, miRNAs
primarily act as the fine-tuners of target genes by binding to
their 3′UTR (33). Based on the
preliminary bioinformatics analysis results (miRDB), it was
hypothesized that miR-142-3p might serve a role in regulating PELI1
expression. PELI1 is a novel cancer-related E3 ubiquitin ligase,
which is involved in the progression of breast cancer (34), lung cancer (35) and lymphoma (36). Thus far, to the best of our
knowledge, there are no studies reporting the association between
miR-142-3p and PELI1. The current study was therefore the first to
verify that miR-142-3p could bind to the 3′UTR of PELI1 and
inversely regulate PELI1 mRNA abundance in PTC cells. Taken
together, these results suggested that hsa_circ_0004846 could
sponge miR-142-3p to regulate PELI1 expression.
Although the majority of studies have shown that
PELI1 is a carcinogenic gene (37,38), a
previous study revealed that PELI1 may act as a tumor suppressor
gene in esophageal squamous cancer (ESC), since PELI1
overexpression could promote the radiotherapy sensitivity of ESC
cells by inhibiting the non-canonical nuclear factor κB pathway
(39). However, Zheng et al
(40) proposed that PELI1 was
upregulated in PTC tissues, and PELI1 overexpression could promote
the diffusion and migration of PTC cells by activating the PI3K/AKT
pathway. In line with the aforementioned findings, the results of
the present study indicated that PELI1 silencing could reverse
activation of the PI3K/AKT pathway mediated by hsa_circ_0004846,
thus suggesting that PELI1 could mediate the cancer-promoting
effects of hsa_circ_0004846 on PTC cells.
There are some limitations in the present study.
This work did not exclude the fact that other miRNAs could be
involved in the regulation of PELI1 by hsa_circ_0004846, since a
downstream target gene could be simultaneously targeted by several
miRNAs. Therefore, this could be a focus of our future research.
Furthermore, since the current study primarily focused on in
vitro experiments, in vivo validation experiments should
be performed in future studies.
In conclusion, the present study suggested that
hsa_circ_0004846 overexpression promoted PTC cell proliferation,
migration and invasion by activating the PI3K/AKT pathway.
Mechanistically, hsa_circ_0004846 acted as a sponge of miR-142-3p
to upregulate PELI1. Overall, the aforementioned findings indicated
that hsa_circ_0004846 could serve a carcinogenic role in PTC by
regulating the miR-142-3p/PELI1 axis.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XD and YW conceptualized and designed the study. XD,
RL and JX performed the experiments. JX, GH, WW and QL contributed
to data acquisition. XD, RL and YW analyzed the data. XD, RL, JX,
GH, WW and QL interpreted the data. XD and RL confirm the
authenticity of all the raw data. XD edited the original draft. XD
and YW reviewed and edited the manuscript. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Anhui Medical
University (approval no. Quick-PJ 2023-01-27) and was in accordance
with the ethical standards formulated in The Declaration of
Helsinki. All patients provided written informed.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gambardella C, Offi C, Clarizia G, Romano
RM, Cozzolino I, Montella M, Di Crescenzo RM, Mascolo M, Cangiano
A, Di Martino S, et al: Medullary thyroid carcinoma with double
negative calcitonin and CEA: A case report and update of literature
review. BMC Endocr Disord. 19:1032019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jiang C, Cheng T, Zheng X, Hong S, Liu S,
Liu J, Wang J and Wang S: Clinical behaviors of rare variants of
papillary thyroid carcinoma are associated with survival: A
population-level analysis. Cancer Manag Res. 10:465–472. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fröhlich E and Wahl R: The current role of
targeted therapies to induce radioiodine uptake in thyroid cancer.
Cancer Treat Rev. 40:665–674. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ryu YJ, Lim SY, Na YM, Park MH, Kwon SY
and Lee JS: Prostate-specific membrane antigen expression predicts
recurrence of papillary thyroid carcinoma after total
thyroidectomy. BMC Cancer. 22:12782022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Q, Wang W, Zhou Q, Chen C, Yuan W,
Liu J, Li X and Sun Z: Roles of circRNAs in the tumour
microenvironment. Mol Cancer. 19:142020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang D, Tao L, Xu N, Lu X, Wang J, He G,
Tang Q, Huang K, Shen S and Chu J: CircRNA circTIAM1 promotes
papillary thyroid cancer progression through the miR-646/HNRNPA1
signaling pathway. Cell Death Discov. 8:212022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bi W, Huang J, Nie C, Liu B, He G, Han J,
Pang R, Ding Z, Xu J and Zhang J: CircRNA circRNA_102171 promotes
papillary thyroid cancer progression through modulating
CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin
Cancer Res. 37:2752018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei W, Ji L, Duan W and Zhu J: CircSAMD4A
contributes to cell doxorubicin resistance in osteosarcoma by
regulating the miR-218-5p/KLF8 axis. Open Life Sci. 15:848–859.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Y and Zhang J: CircSAMD4A accelerates
cell proliferation of osteosarcoma by sponging miR-1244 and
regulating MDM2 mRNA expression. Biochem Biophys Res Commun.
516:102–111. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie C, Chen B, Wu B, Guo J, Shi Y and Cao
Y: CircSAMD4A regulates cell progression and epithelial-mesenchymal
transition by sponging miR-342-3p via the regulation of FZD7
expression in osteosarcoma. Int J Mol Med. 46:107–118.
2020.PubMed/NCBI
|
|
13
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
White WL: Erratum to: Why I hate the index
finger. Hand (N Y). 6:2332011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Y, Dong B, Huang L and Huang H: Serum
microRNAs as biomarkers for the diagnosis of papillary thyroid
carcinoma: A meta-analysis. Bosn J Basic Med Sci. 22:862–871. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang Y, Liu Y, Zhang Y, Ouyang J, Feng Y,
Li S, Wang J, Zhang C, Tan L, Zhong J and Zou L: MicroRNA-142-3P
suppresses the progression of papillary thyroid carcinoma by
targeting FN1 and inactivating FAK/ERK/PI3K signaling. Cell Signal.
109:1107922023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Almeida JL and Korch CT: Authentication of
human and mouse cell lines by short tandem repeat (STR. DNA
genotype analysis. In Assay Guidance Manual. Markossian S, Grossman
A, Arkin M, Auld D, Austin C, Baell J, Brimacombe K, Chung TDY,
Coussens NP, Dahlin JL, et al: Eli Lilly & Company and the
National Center for Advancing Translational Sciences; 2004
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T). method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao X, Fu J, Hu B, Chen L, Wang J, Fang
J, Ge C, Lin H, Pan K, Fu L, et al: Serine metabolism regulates YAP
activity through USP7 in colon cancer. Front Cell Dev Biol.
9:6391112021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Petrulea MS, Plantinga TS, Smit JW,
Georgescu CE and Netea-Maier RT: PI3K/Akt/mTOR: A promising
therapeutic target for non-medullary thyroid carcinoma. Cancer
Treat Rev. 41:707–713. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Conzo G, Mauriello C, Docimo G,
Gambardella C, Thomas G, Cavallo F, Tartaglia E, Napolitano S,
Varriale R, Rossetti G, et al: Clinicopathological pattern of lymph
node recurrence of papillary thyroid cancer. Implications for
surgery. Int J Surg. 12 (Suppl 1):S194–S197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cooper DS, Doherty GM, Haugen BR, Kloos
RT, Lee SL, Mandel SJ, Mazzaferri EL, McIver B, Pacini F,
Schlumberger M, et al: Revised American Thyroid Association
management guidelines for patients with thyroid nodules and
differentiated thyroid cancer. Thyroid. 19:1167–1214. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Conzo G, Docimo G, Mauriello C,
Gambardella C, Esposito D, Cavallo F, Tartaglia E, Napolitano S and
Santini L: The current status of lymph node dissection in the
treatment of papillary thyroid cancer. A literature review. Clin
Ter. 164:e343–e346. 2013.PubMed/NCBI
|
|
25
|
Wang L, Wang P, Su X and Zhao B:
Circ_0001658 promotes the proliferation and metastasis of
osteosarcoma cells via regulating miR-382-5p/YB-1 axis. Cell
Biochem Funct. 38:77–86. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ding B, Fan W and Lou W: hsa_circ_0001955
Enhances in vitro proliferation, migration, and invasion of HCC
cells through miR-145-5p/NRAS axis. Mol Ther Nucleic Acids.
22:445–455. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang G, Mao L and Hu X: Circ_0011373
promotes papillary thyroid carcinoma progression by regulating
miR-1271/LRP6 axis. Hormones (Athens). 22:375–387. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang YM, Liu W, Jiang L and Chang H:
CircLDLR promotes papillary thyroid carcinoma tumorigenicity by
regulating miR-637/LMO4 axis. Dis Markers.
2021.39771892021.PubMed/NCBI
|
|
29
|
Chong ST, Tan KM, Kok CYL, Guan SP, Lai
SH, Lim C, Hu J, Sturgis C, Eng C, Lam PYP and Ngeow J: IL13RA2 is
differentially regulated in papillary thyroid carcinoma vs
follicular thyroid carcinoma. J Clin Endocrinol Metab.
104:5573–5584. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jere SW, Houreld NN and Abrahamse H: Role
of the PI3K/AKT mTOR and GSK3β. signalling pathway and
photobiomodulation in diabetic wound healing. Cytokine Growth
Factor Rev. 50:52–59. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han J, Zhang M, Nie C, Jia J, Wang F, Yu
J, Bi W, Liu B, Sheng R, He G, et al: miR-215 suppresses papillary
thyroid cancer proliferation, migration, and invasion through the
AKT/GSK-3β/Snail signaling by targeting ARFGEF1. Cell Death Dis.
10:1952019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Colamaio M, Puca F, Ragozzino E, Gemei M,
Decaussin-Petrucci M, Aiello C, Bastos AU, Federico A, Chiappetta
G, Del Vecchio L, et al: miR-142-3p down-regulation contributes to
thyroid follicular tumorigenesis by targeting ASH1L and MLL1. J
Clin Endocrinol Metab. 100:E59–E69. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu SS, Qi J, Teng ZD, Tian FT, Lv XX, Li
K, Song YJ, Xie WD, Hu ZW and Li X: Resistomycin attenuates
triple-negative breast cancer progression by inhibiting E3 ligase
Pellino-1 and inducing SNAIL/SLUG degradation. Signal Transduct
Target Ther. 5:1332020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jeon YK, Kim CK, Hwang KR, Park HY, Koh J,
Chung DH, Lee CW and Ha GH: Pellino-1 promotes lung carcinogenesis
via the stabilization of Slug and Snail through K63-mediated
polyubiquitination. Cell Death Differ. 24:469–480. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park HY, Go H, Song HR, Kim S, Ha GH, Jeon
YK, Kim JE, Lee H, Cho H, Kang HC, et al: Pellino 1 promotes
lymphomagenesis by deregulating BCL6 polyubiquitination. J Clin
Invest. 124:4976–4988. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fei X, Zhu C, Liu P, Liu S, Ren L, Lu R,
Hou J, Gao Y, Wang X and Pan Y: PELI1: Key players in the oncogenic
characteristics of pancreatic Cancer. J Exp Clin Cancer Res.
43:912024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou W, Hu Y, Wang B, Yuan L, Ma J and
Meng X: Aberrant expression of PELI1 caused by Jagged1 accelerates
the malignant phenotype of pancreatic cancer. Cell Signal.
111:1108772023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ko CJ, Zhang L, Jie Z, Zhu L, Zhou X, Xie
X, Gao T, Yang JY, Cheng X and Sun SC: The E3 ubiquitin ligase
Peli1 regulates the metabolic actions of mTORC1 to suppress
antitumor T cell responses. EMBO J. 40:e1045322021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zheng T, Zhou Y, Xu X, Qi X, Liu J, Pu Y,
Zhang S, Gao X, Luo X, Li M, et al: MiR-30c-5p loss-induced PELI1
accumulation regulates cell proliferation and migration via
activating PI3K/AKT pathway in papillary thyroid carcinoma. J
Transl Med. 20:202022. View Article : Google Scholar : PubMed/NCBI
|