Introduction
Despite the application of surgery and chemotherapy
and/or radiotherapy, the prognosis of patients with malignant
glioma is still extremely poor. Glioblastoma multiforme (GBM) is
the most malignant glioma, the median survival of which is
approximately 1 year (1–3). The invasive growth pattern of glioma
cells is partially responsible for its poor response to treatment
(4). Some characteristic oncogenes
may take responsibility for the infiltration and invasion.
Epigenetic mechanisms are important factors in the
regulation of gene expression in tumors. To date, research has
discovered many epigenetic mechanisms, for example promoter
hypermethylation/hypomethylation, histone modifications, or
non-coding RNA expression (5).
Moreover, DNA methylation is typically a more stable and
inheritable epigenetic pattern, and is an important hallmark of
tumor development and progression (6–8).
Although the important role of hypermethylation in the silencing of
tumor-suppressor genes is now well-documented (9), a decrease in the level of methylation
also contributes to numerous types of human cancer (10,11).
The RAS superfamily of small GTPases includes 5
groups: Ras, Rho, Rab, Ran and Arf. Rab family proteins are
important in regulating signal transduction and cellular processes,
such as differentiation, proliferation, vesicle transport, nuclear
assembly and cytoskeleton formation. In addition, various Rab
proteins are important for the adhesion and migration of cancer
cells. Rab27b can control vesicle exocytosis and release important
growth regulators into the tumor microenvironment, regulating
invasive growth and metastasis in human breast tumors (12). Yet, the role of Rab27b in glioma
remains unclear.
In the present study, the promoter methylation
status of Rab27b was evaluated in our methylation microarray (119
samples) and 52 independent samples. Correlation of Rab27b
methylation with clinical data was assessed. Rab27b expression was
also detected to determine whether promoter hypomethylation was
correlated with the high expression level in gliomas. Furthermore,
we explore the probable mechanistic link through experiments in
vitro and in vivo.
Materials and methods
Ethics
Written informed consent from each donor or the next
of kin was obtained. This study was performed following approval of
the Ethics Committee of the Beijing Tiantan Hospital and is in
compliance with the Helsinki Declaration.
Datasets used in this study
All patients were from the Chinese Glioma Genome
Atlas (CGGA). All of the patients selected in the present study
underwent surgical resection between January 2006 and December 2010
and subsequently received concomitant and adjuvant temozolomide and
radiotherapy. Data concerning overall survival (OS), defined as the
period from operation to death, were collected mainly when patients
visited the clinics or by phone interview with patients and/or
their relatives. Patients who died of non-primary diseases were
excluded (13). Tumor tissue
samples were obtained by surgical resection before treatment with
radiation and chemotherapy. Respected specimens were snap-frozen
and stored in liquid nitrogen until nucleic acid extraction. Only
samples with >80% tumor cells were selected. The microarray data
set was deposited in the Gene Expression Omnibus (GEO) (accession
no. GSE53227 and GSE53228) according to 'minimum information about
a microarray experiment' (MIAME) guidelines.
DNA extraction
All of the tissue samples were immediately
snap-frozen in liquid nitrogen after surgery. A hematoxylin and
eosin-stained frozen section was prepared for assessment of the
percentage of tumor cells before DNA extraction. Genomic DNA was
isolated from frozen tumor tissues using the QIAamp DNA Mini kit
(Qiagen) according to the manufacturer's instructions. DNA
concentration and quality were measured using the NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies, Houston, TX, USA)
(8).
Genome-wide DNA methylation
profiling
A series of 119 glioma samples (63 low-grade
gliomas, 33 anaplastic gliomas and 23 glioblastomas) were assessed
by methylation microarray. We used the Illumina Infinium Human
Methylation 27 BeadChips (Illumina Inc.) as previously described
(14). The BeadChip contains 27,578
highly informative CpG sites covering >14,000 human RefSeq
genes. This allows researchers to interrogate all of these sites
per sample at a single nucleotide resolution. Bisulfite
modification of DNA, chip processing and data analysis were
performed following the manufacturer's manual at the Wellcome Trust
Centre for Human Genetics Genomics Laboratory (Oxford, UK). The
array results were analyzed with the BeadStudio software (Illumina
Inc.).
Bisulfite sequencing PCR (BSP)
Fifty-six glioma samples (4 normal brain tissues and
20 low-grade and 32 high-grade gliomas) were measured by BSP and
immunohistochemistry (IHC). Purified DNA was treated with sodium
bisulfite (Sigma, Phoenix, AZ, USA). The PCR products were
confirmed by agarose gel electrophoresis and visualized using
ethidium bromide staining. The primers used were:
5′-AGTAAGATTTGTTTGAGGTGAGTTT-3′ and
3′-AAAACAACTACTTATCTCCTCAACC-5′. There were 11 CPG sites in the
amplified region (18q21.2, 54715778-54715993). The specific
experimental steps were in accordance with the test method from the
EZ DNA Methylation-Direct kit (Zymo Research).
Immunohistochemistry
Ninety-one glioma samples included 30 low-grade and
20 anaplastic gliomas and 41 glioblastomas. IHC was performed as
previously described (17). The
degree of immunostaining of the sections was viewed and scored
separately by two independent investigators. The scores were
determined by combining the proportion of positively stained tumor
cells and the intensity of staining. Binary decision criterion of
Rab27b expression was performed as previously described (8,16).
Cell lines and culture
Human glioblastoma cell lines U87 and LN229 were
obtained from the Institute of Biochemistry and Cell Biology,
Chinese Academy of Science (Shanghai, China). The cells were
maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco,
Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS)
and incubated at 37°C in 5% CO2. Upon 80% confluency,
the cells were starved in DMEM with 1% FBS for 24 h and maintained
in this low serum condition for the course of treatment (8).
Rab27b gene knockdown by siRNA
Specific oligos targeting the Rab27b gene were
selected (OriGene Technologies, Inc.). siRNA1, the most efficient
one, screened from 3 siRNAs, was used to knockdown Rab27b (5′-AAA
GGT GTG GTT TAT AAT GCA-3′), and a RNA interference negative
control was purchased from Qiagen (venlo, The Netherlands).
Logarithmically growing cells were seeded at a density of
105 cells/6-cm dish and transfected with 5 µmol
Rab27b siRNA using Lipofectamine 2000 (Invitrogen) according to the
manufacturer's instructions. Forty-eight hours later, the cells
were used for in vitro functional assays as described
below.
Transwell invasion assay
The Transwell invasion assay was conducted in
24-well cell culture chambers using Transwell inserts (Corning Life
Sciences, Corning, NY, USA; BD Biosciences, San Jose, CA, USA) with
an 8-µm pore membrane precoated with Matrigel (BD
Biosciences). U87 and LN229 cells were plated at the density of
1×104/upper well in 200 µl culture medium (DMEM,
no FBS), as well as control group and siRNA group, respectively.
The lower chamber was filled with 500 µl medium (DMEM, 12%
FBS). The cells were allowed to invade for 24 h, after which, the
non-invading cells with Matrigel matrix were removed from the upper
surface of the membrane by scrubbing with a cotton-tipped swab.
Cells on the lower surface of the filter were fixed for 30 min in
methanol and glacial acetic acid mixture (3:1), air-dried briefly,
and stained with crystal violet. The mean number of invaded cells
was counted from five preselected microscopic fields at ×200
magnification; all experiments were performed in triplicate.
Western blot analysis
After cell treatment, cell lysates were prepared via
lysis buffer, electro phoresed onto SDS-polyacrylamide gels, and
transferred to polyvinylidene difluoride membranes (4). Membranes were probed with antibodies
against Rab27b (Proteintech) and glyceraldehyde-3-phosphate
dehydrogenase (GAPDH, A-3; Santa Cruz Biotechnology) at dilutions
of 1:1,000. Blots were detected with horseradish peroxidase-labeled
anti-rabbit antibodies (1:5,000 dilution) developed using enhanced
chemiluminescence (ECL) reagents (Amersham Pharmacia, UK). The
primary antibody against MMP-9 (Oncogene, Boston, MA, USA), was
used to examine protein expression.
Determination of gelatinase activity
Cells (1×106/35-mm dish) undergoing
Rab27b siRNA treatments or the negative control were subsequently
incubated in 2 ml serum-free medium for 24 h. The supernatants were
centrifuged at 10,000 rpm for 5 min to remove cell debris and their
protein contents were determined using Bradford reagent (Bio-Rad,
USA). Gelatinolytic activity of MMP-9 and MMP-2 was examined by
gelatin zymography. Briefly, equal amounts (10 µg) of
protein were mixed with SDS gel-loading buffer and then loaded
without reduction or heating onto 10% polyacrylamide gels
containing 0.1% gelatin (Sigma). To detect specific activity,
SDS-PAGE was carried out. After running, the gels were renatured
for 1 h in 0.25% Triton X-100 at room temperature with gentle
agitation, and then incubated in developing buffer (50 mM Tris-HCl,
100 mM NaCl, 10 mM CaCl2 and 0.02% NaN3, pH
7.5) for 24 h at 37°C. After briefly washing in water, the gels
were stained with Coomassie blue R-250 for 1 h to reveal the
gelatinolytic activity. Gels were destained with 40% methanol and
5% acetic acid until clear white bands against a blue background
were visible.
Nude mouse tumor xenograft model and
treatment with siRNA of Rab27b
LN229 glioma cells were subcutaneously injected into
5-week-old female nude mice (Cancer Institute of the Chinese
Academy of Medical Science). When the tumor volume reached 50
mm3, the mice were randomly divided into two groups (6
mice/group). Each group was treated with siRNA of Rab27b or
negative control oligo in 10 µl Lipofectamine through local
injection of the xenograft tumor at multiple sites. The treatment
was performed once every 3 days for 21 days. The tumor volume was
measured with a caliper twice a week, using the following formula:
volume = length × width2/2.
Statistical analysis
Significant Analysis of Microarrays (SAM) was used
for the genes significantly differentially methylated between high-
and low-grade gliomas. The t-test or Chi-square test was applied
for statistical analysis of the correlation between two independent
variables. Survival curves were estimated using the Kaplan-Meier
method, and statistical differences were evaluated using the
two-sided log-rank test. A p<0.05 was considered
significant.
Results
Rab27b methylation status is correlated
with tumor grade progression and prognosis
In a microarray of 119 glioma samples, we found that
Rab27b was significantly hypomethylated in high-grade gliomas
compared with low-grade gliomas (p=0.02, Fig. 1A). The methylation status of Rab27b
was correlated with OS in the high-grade glioma samples through
Kaplan-Meier survival curve analysis (p<0.01, Fig. 1B). There was a big CpG island
located in nearly 2,000 bp upstream of the Rab27b promoter region
(Fig. 1C). These CG sites in the
CpG island were detected by BSP method, and glioma cell lines (U87
and LN229) harbored a low methylation level than the level in the
glio cell line (HA cells). Similar to these results, the
methylation levels in these sites from the high-grade glioma
tissues were lower than levels in the low-grade glioma and normal
brain tissues.
 | Figure 1Rab27b hypomethylation is associated
with tumor grade progression and poor prognosis as determined in
the CGGA data. (A) The methylation level of the Rab27b promoter was
significantly lower in high-grade glioma than that in low-grade
gliomas. (B) In high-grade gliomas, patients with a low methylation
level of Rab27b had shorter OS than those with high methylation.
(C) There was a big CpG island located in upstream of the Rab27b
promoter region. Methylation analysis using BSP method showed
differential methylation levels in different cell lines and glioma
tissues. CGGA, Chinese Glioma Genome Atlas; OS, overall survival;
BSP, bisulfite sequencing PCR. Note: Hypermethylation, HA cells
(normal brain cells), normal brain tissue, low-grade glioma (OA,
oligoastrocytoma; A, astrocytoma); Hypomethylation, glioma cell
lines (U87 and LN229); high-grade glioma (AOA, anaplastic
oligoastrocytoma; AA, anaplastic astrocytoma; GBM, glioblastoma
multiforme). |
The methylation status of Rab27b was significantly
lower in high-grade gliomas (68.8%) compared with the status in
low-grade gliomas (31.2%) (Table I,
p<0.01), which was significantly associated with glioma grade
progression. In 32 high-grade glioma samples, 80% showed low Rab27b
promoter methylation accompanied by high Rab27b protein expression
detected by the IHC method, and the remaining samples showed high
Rab27b promoter methylation along with low protein expression.
 | Table IMethylation status of Rab27b was
negatively correlated with grade and Rab27b expression. |
Table I
Methylation status of Rab27b was
negatively correlated with grade and Rab27b expression.
| Characteristics | Methylation status of
52 glioma samples
|
|---|
| Low n/total (%) | High n/total (%) |
|---|
| Low grade | 6/20
(30) | 14/20 (70) |
| High gradea | 22/32 (68.8) | 10/32 (31.2) |
| Characteristics | Methylation status of
32 high-grade samples
|
|---|
| Low n/total (%) | High n/total (%) |
|---|
| Rab27b low
expression | 5/22 (22.7) | 17/22 (77.3) |
| Rab27b high
expressionb | 8/10 (80) | 2/10
(20) |
Rab27b expression is associated with
glioma grade progression and confers a poor prognosis to high-grade
glioma patients with high Rab27b expression
We performed IHC of 91 glioma samples from mainland
Han Chinese patients. We analyzed the correlation between Rab27b
protein expression and the histological stage of the gliomas.
Rab27b expression ranged from low to high along with the grade of
progression of the gliomas (p<0.01, Fig. 2A and C). Survival analysis showed
that patients with high Rab27b expression had significantly shorter
OS (p=0.01) than those with low expression in the high-grade glioma
patients (Fig. 2B).
Rab27b promotes glioma invasion via
activation of MMP-9 but not MMP-2
Western blot analysis identified that Rab27b was
significantly knocked down by siRNA compared with the control group
in the U87 cells (Fig. 3A). To
confirm the effect of Rab27b on the regulation of intracellular
molecules involved in aggressive cell behavior, we examined MMP-9
expression. MMP-9 expression was downregulated following Rab27b
knockdown when compared to the expression levels in the control
group. The gelatinase activity assay showed that MMP-9 activation
was weakened but not MMP-2 following Rab27b knockdown (Fig. 3B). After siRNA knockdown of Rab27b
in the glioma cell lines, the U87 and LN229 cell lines were used
for Transwell invasion assay. siRNA significantly attenuated the
effect of Rab27b on cell invasion of the U87 cells (p<0.01,
Fig. 3C) as well as the LN229 cells
(p<0.01), respectively.
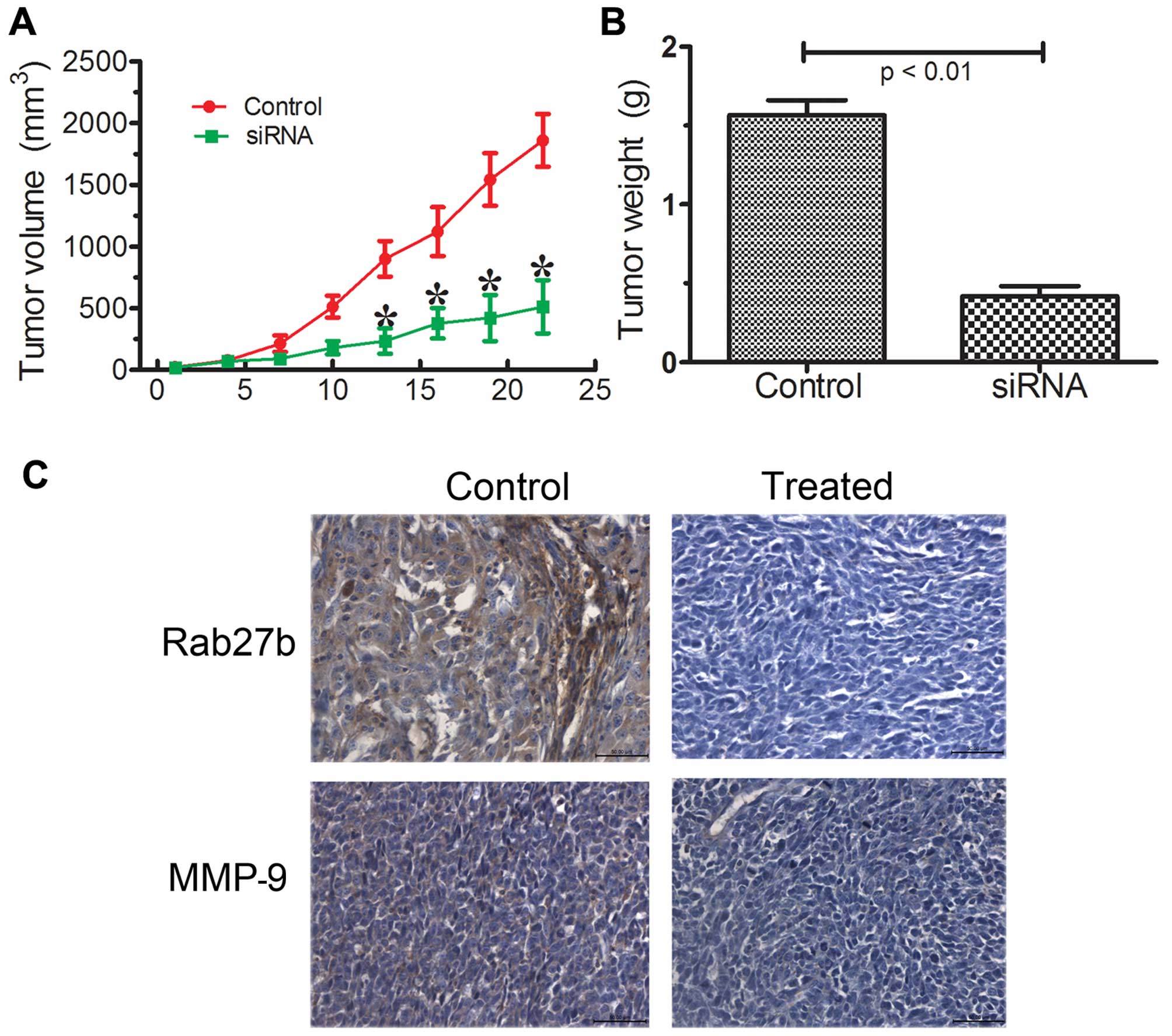
Rab27b promotes glioma growth in
vivo
To investigate the potential impact of Rab27b
expression in vivo, a LN229 cell xenograft model was
utilized. The Rab27b-treated group displayed a significant growth
reduction, whereas tumor growth was not impacted by the negative
control (Fig. 4A). Tumor mass
(Fig. 4B) was significantly higher
in the control tumors when compared with those treated with Rab27b.
IHC analysis confirmed downregulated expression of Rab27b in the
treated group (Fig. 4C).
Rab27b-treated glioma model displayed decreased expression of MMP-9
compared with the control. These data indicated that Rab27b
knockdown in vivo functioned similarly as the effect in
vitro.
Discussion
In the present study, we investigated the
methylation and expression level of Rab27b in 3 independent
cohorts, including 262 glioma patients. The methylation level of
the Rab27b promoter was associated with tumor grade progression and
survival in malignant gliomas. We further detected the difference
in expression of Rab27b in different grade gliomas, and observed a
significant increase in Rab27b expression from low- to high-grade
gliomas. Rab27b was an independent unfavorable prognostic factor
for the patients with high-grade glioma. In vitro analysis
revealed that Rab27b promoted the invasion of glioma cells, at
least in part, by activating MMP-9. As far as we know, this is the
first study on the methylation status, expression difference, and
function of Rab27b in glioma.
A large body of evidence demonstrates that DNA
methylation plays a role in gene regulation in animal cells.
Hypomethylation of oncogenes and hypermethylation of suppressor
gene promoters are pivotal alterations in tumor development
(17). Genome-wide DNA methylation
profiling has revealed a glioma-CpG island methylator phenotype
(G-CIMP+) subtype that has a strong positive correlation
with survival prognosis in GBM patients (18), however the role of promoter
demethylation remains obscure. In the present study, a genome-wide
DNA methylation microarray including 119 gliomas was performed. We
found that the methylation level of Rab27b in high-grade gliomas
was significantly lower than that in low-grade ones. Kaplan-Meier
survival curve analysis showed that patients with a low methylation
level of Rab27b had shorter OS compared to the high methylation
group in patients with high-grade glioma. To identify whether the
expression of Rab27b was correlated with the status of methylation
in glioma, we detected both the Rab27b expression and methylated
level of the promoter region in 52 glioma tissues. As shown in
Table I, the Rab27b expression was
negatively associated with the promoter methylation status
(p<0.01). In another independent cohort including 91 glioma
samples, we found that patients with high-grade glioma had
significant higher Rab27b expression than those patients with
low-grade glioma. In 61 high-grade glioma patients, high Rab27b
expression was associated with adverse outcomes. The results
indicated that glioma with Rab27b hypomethylation and higher
protein expression may be a more biologically aggressive phenotype,
and that elevated expression of Rab27b resulting in a shorter
survival was at least partially due to hypomethylation of the
Rab27b promoter region.
To date, no information has been provided concerning
the Rab27b gene in gliomas, and its clinical effect on survival
remains unknown. Ras-associated binding (Rab)-GTPases are members
of the Ras family of small GTPases. Many Rab proteins are
abnormally expressed in different cancer tissues. Rab25 has been
shown to decrease apoptosis, as well as to increase both the
proliferation and aggressiveness of ovarian and breast cancer
(16). Rab5 is involved in EGFR
signaling and was found to promote proliferation in lung
adenocarcinoma cell lines (20).
Rab27b was reported to control vesicle exocytosis and release
important growth regulators into the tumor microenvironment,
leading to regulation of invasive growth and metastasis in human
breast tumors (12). We performed
functional assays in malignant glioma U87 and LN229 cell lines and
confirmed that Rab27b promoted the invasion of glioma cells.
Furthermore, Rab27b regulated the expression and activation status
of MMP-9 which characteristically participates in tumor invasion.
Thus, these functional assays confirmed our hypothesis.
In summary, Rab27b promoter hypomethylation and
expression are significantly correlated with the tumor grade
progression of glioma. The methylation status of Rab27b was
negatively associated with its expression. Patients harboring
Rab27b hypomethylation and high expression had reduced OS in
high-grade glioma. Rab27b promoted glioma invasion, at least in
part, via regulation of MMP-9 expression and activation.
Acknowledgments
This research was supported by grants from the
Health and Family Planning Commission Research Project of
Heilongjiang province (2014-329), the Government Spending
Postdoctoral Fellowship of Heilongjiang province (LRB14-422), the
National High Technology Research and Development Program
(2012AA02A508), the National Natural Science Foundation of China
(nos. 81402052, 81201993, 81372700 and 81302200) and the
International Science and Technology Cooperation Program
(2012DFA30470).
References
|
1
|
Zhang W, Yan W, You G, Bao Z, Wang Y, Liu
Y, You Y and Jiang T: Genome-wide DNA methylation profiling
identifies ALDH1A3 promoter methylation as a prognostic predictor
in G-CIMP- primary glioblastoma. Cancer Lett. 328:120–125. 2013.
View Article : Google Scholar
|
|
2
|
Yan W, Zhang W, You G, Zhang J, Han L, Bao
Z, Wang Y, Liu Y, Jiang C, Kang C, et al: Molecular classification
of gliomas based on whole genome gene expression: A systematic
report of 225 samples from the Chinese Glioma Cooperative Group.
Neuro Oncol. 14:1432–1440. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Y, Li S, Chen L, You G, Bao Z, Yan W,
Shi Z, Chen Y, Yao K, Zhang W, et al: Glioblastoma with an
oligodendroglioma component: Distinct clinical behavior, genetic
alterations, and outcome. Neuro Oncol. 14:518–525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang H, Wang Y and Jiang C: Stromal
protein periostin identified as a progression associated and
prognostic biomarker in glioma via inducing an invasive and
proliferative phenotype. Int J Oncol. 42:1716–1724. 2013.PubMed/NCBI
|
|
5
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gaspar-Maia A, Alajem A, Meshorer E and
Ramalho-Santos M: Open chromatin in pluripotency and reprogramming.
Nat Rev Mol Cell Biol. 12:36–47. 2011. View
Article : Google Scholar
|
|
7
|
Feinberg AP and Tycko B: The history of
cancer epigenetics. Nat Rev Cancer. 4:143–153. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang H, Feng Y, Bao Z, Jiang C, Yan W,
Wang Y, Zhang C, Liu Y, Zhang Q, Zhang W, et al: Epigenetic
silencing of KAZALD1 confers a better prognosis and is associated
with malignant transformation/progression in glioma. Oncol Rep.
30:2089–2096. 2013.PubMed/NCBI
|
|
9
|
Baylin SB: DNA methylation and gene
silencing in cancer. Nat Clin Pract Oncol. 2(Suppl 1): S4–S11.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ehrlich M: DNA methylation in cancer: Too
much, but also too little. Oncogene. 21:5400–5413. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cadieux B, Ching TT, van den Berg SR and
Costello JF: Genome-wide hypomethylation in human glioblastomas
associated with specific copy number alteration,
methylenetetrahydrofolate reductase allele status, and increased
proliferation. Cancer Res. 66:8469–8476. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hendrix A, Maynard D, Pauwels P, Braems G,
Denys H, van den Broecke R, Lambert J, van Belle S, Cocquyt V,
Gespach C, et al: Effect of the secretory small GTPase Rab27B on
breast cancer growth, invasion, and metastasis. J Natl Cancer Inst.
102:866–880. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li S, Yan C, Huang L, Qiu X, Wang Z and
Jiang T: Molecular prognostic factors of anaplastic
oligodendroglial tumors and its relationship: A single
institutional review of 77 patients from China. Neuro Oncol.
14:109–116. 2012. View Article : Google Scholar :
|
|
14
|
Hill VK, Ricketts C, Bieche I, vacher S,
Gentle D, Lewis C, Maher ER and Latif F: Genome-wide DNA
methylation profiling of CpG islands in breast cancer identifies
novel genes associated with tumorigenicity. Cancer Res.
71:2988–2999. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang W, Qiu XG, Chen BS, Li SW, Cui Y,
Ren H and Jiang T: Antiangiogenic therapy with bevacizumab in
recurrent malignant gliomas: Analysis of the response and core
pathway aberrations. Chin Med J (Engl). 122:1250–1254. 2009.
|
|
16
|
Wang F, Yang C, Song Y, Jiang Y and Ding
Z: Periostin gene polymorphisms, protein levels and risk of
incident coronary artery disease. Mol Biol Rep. 39:359–367. 2012.
View Article : Google Scholar
|
|
17
|
Iacob G and Dinca EB: Current data and
strategy in glioblastoma multiforme. J Med Life. 2:386–393.
2009.
|
|
18
|
Laffaire J, Everhard S, Idbaih A, Crinière
E, Marie Y, de Reyniès A, Schiappa R, Mokhtari K, Hoang-Xuan K,
Sanson M, et al: Methylation profiling identifies 2 groups of
gliomas according to their tumorigenesis. Neuro Oncol. 13:84–98.
2011. View Article : Google Scholar :
|
|
19
|
Lebrand C, Corti M, Goodson H, Cosson P,
Cavalli V, Mayran N, Fauré J and Gruenberg J: Late endosome
motility depends on lipids via the small GTPase Rab7. EMBO J.
21:1289–1300. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tomshine JC, Severson SR, Wigle DA, Sun Z,
Beleford DA, Shridhar V and Horazdovsky BF: Cell proliferation and
epidermal growth factor signaling in non-small cell lung
adenocarcinoma cell lines are dependent on Rin1. J Biol Chem.
284:26331–26339. 2009. View Article : Google Scholar : PubMed/NCBI
|