Introduction
The incidence of malignant melanoma has increased
greatly in recent decades presenting a high mortality rate despite
intensive efforts in this area of research.
The chemokine receptor 4 (CXCR4) is a transmembrane
receptor that belongs to the chemokine receptor family CXC which
was initially reported to mediate homing of leukocytes into tissues
that produce its ligand stromal cell-derived factor 1 (SDF-1), also
known as CXCL12 (1,2).
A growing body of evidence indicates that CXCR4
plays a critical role in cancer since CXCL12 binding to CXCR4
initiates various downstream signaling pathways that result in a
plethora of responses involved in cell proliferation and metastasis
(3,4). It was reported that human melanoma
cells express a high level of the chemokine receptor CXCR4 when
compared with non-transformed melanocytes (5). The CXCR4 expression by malignant
melanoma is predictive of poor survival rate and metastasis since
its ligand CXCL12 is also increased in lungs which would explain
the high frequency of pulmonary metastasis (6).
Considering the critical role of the CXCR4/CXCL12
axis in many types of cancer, including human melanoma (7,8), there
is currently significant interest in the discovery and development
of CXCR4 antagonists (9).
Therefore, there is a need of experimental models of cancer to
reach this objective. One of the experimental models of melanoma
research currently used is the B16 melanoma, which spontaneously
originated in C57BL/6 mice. The primary tumor of B16 melanoma
contains subclones which differ in their ability to form
metastasis. The melanoma B16 was adapted to growth in vitro
in order to compare the metastatic properties of several clones.
Thus, the murine melanoma B16, F10 clone, is derived from
spontaneous melanoma B16 and adapted in vitro after ten lung
colonization cycles (10).
Recently, we demonstrated that the knockdown of chemokine receptor
CXCR4 by RNA interference (RNAi) significantly reduced the number
of pulmonary metastatic nodules (11). However, to use this experimental
model of cancer to discovery new CXCR4 antagonists, it is necessary
to demonstrate that CXCR4 silencing also results in the inhibition
of B16-F10 melanoma growth.
The potential therapeutic of RNAi technology has
also been explored in cancer since RNAi is able to selectively
knockdown critical genes involved in cell proliferation (12). The RNAi technology holds promise for
cancer treatment, but many hurdles need be overcome before the
clinical use of RNAi. It should be emphasized that the biggest
challenge is to develop methods to specifically delivery RNAi to
tumor cells. In the last few years, several strategies have been
used to surmount this barrier (13–15)
and the intratumoral injection of RNAi emerges as promising
approach in the case of solid tumors (16). We have previously used this approach
to elucidate the role played by the RNA-dependent protein kinase
(PKR) in the growth and metastasis of B16-F10 melanoma (17).
In the present study, we investigated the effect of
the intratumoral CXCR4 short hairpin (shRNA) expressing plasmid on
the growth of the B16-F10 melanoma in C57BL/6 mice. The strategy of
using a plasmid transient expression of shRNA anti-CXR4 has shown
that CXCR4 plays a critical role in the initial stages of
development of murine melanoma B16-F10 melanoma, suggesting that
this experimental model may be useful for the discovery and
development of CXCR4 antagonists.
Materials and methods
Animals
All the protocols involving animals were reviewed
and approved by our Institutional Animal Care Committee. We used
C57BL/6 mice, weighing 20–25 g, that were raised at the Central
Animal Laboratory of the School of Medicine of Ribeirão Preto, SP,
Brazil.
Culture of tumor cells
B16-F10 melanoma cells were maintained in RPMI-1640
medium supplemented with 10% inactivated fetal calf serum, 2 mM
L-glutamine all from (Invitrogen, Carlsbad, CA, USA) and 1%
penicillin/streptomycin (100 U/ml; Gibco Life Technologies,
Carlsbad, CA, USA) in a humidified atmosphere at 37°C and 5%
CO2.
Target sequence selection of CXCR4 mRNA
and plasmid vector construction
Two shRNA target sequences were selected from
different positions of mouse CXCR4 cDNA sequence (GenBank,
accession no. BC031665) corresponding to nucleotides 85-103
(CXCR4-1 shRNA) and 409-427 (CXCR4-2 shRNA). Each shRNA contains a
sense strand of 19 nucleotides followed by a short spacer
(AAGTTCTCT), the antisense strand and a stop signal (TTTTT) for RNA
polymerase III.
The selection of shRNA sequences was based on the
shRNA Target Finder and Design Tool available at Dharmacon website.
These target sequences were submitted to a BLAST search to ensure
that only the CXCR4 gene was the target. The target sequence of the
negative control group named control shRNA has no homology with
that of human or mice. The targeting sequence and location of each
shRNA in CXCR4 cDNA are shown in Table
I.
 | Table ISequences of shRNA
oligonucleotides. |
Table I
Sequences of shRNA
oligonucleotides.
| shRNA | Target
Positiona | Sequence |
|---|
| CXCR4-1 | 85–103 | F:
5′-ACCGCGATCAGTGTGAGTATATAAAGTTCTCTTATATACTCACGATCGCTTTTTC-3′ |
| R:
5′-GCAGAAAAAGCGATCAGTGTGAGTATATAAGAGAACTTTATATACTCACACTGATCG-3′ |
| CXCR4-2 | 409–427 | F:
5′-ACCGGTAAGGCTGTCCATATCATAAGTTCTCTATGATATGGACAGCCTTACCTTTTTC-3′ |
| R:
5′-GCAGAAAAAGGTAAGGCTGTCCATATCATAGAGAACTTATGATATGGACAGCCTTACCGGT-3′ |
| Control | | F:
5′-ACCGAAGCGCTGCCGCGACGTTGAAGTTCTCTCAACGTCGCGGCAGCGCTTCTTTTTC-3′ |
| R:
5′-TGCAGAAAAAGAAGCGCTGCCGCGACGTTGAGAGAACTTCAACGTCGCGGCAGCGCTTCGGT-3′ |
Two micrograms of sense and antisense
oligonucleotides were mixed and diluted in annealing buffer to a
final concentration of 40 ng/µl. The mixture was then heated
to 90°C for 3 min and then transferred to a water bath at 37°C and
incubated for 15 min. The annealed oligonucleotides were diluted in
nuclease-free water to a final concentration of 4 ng/µl.
Each paired oligonucleotide was ligated overnight by enzyme T4 DNA
ligase (3 U/µl) to the plasmid psiTRIKE (50 ng/µl).
The DH5α E. coli strain was used for cloning. Transformation
of plasmid DNA into competent E. coli was performed using
the heat shock method. After a short incubation in ice, a mixture
of chemically competent bacteria and plasmid DNA was placed at 42°C
for 45 sec and then placed back in ice. LB media were added and the
transformed cells were incubated at 37°C for 30 min with agitation.
The E. coli was plated and transformed bacteria was selected
based on resistance to ampicillin. Plasmid DNA was extracted of
transformed E. coli with FlexiPrep kit (Amersham
Biosciences, Little Chalfont, UK). The method employs a standard
alkaline lysis procedure, including treatment with RNase and
precipitation with isopropanol. Plasmid DNA was subsequently
purified by Sephaglas PF resin (Amersham Biosciences) according to
the manufacturer's instructions. Screening for the two inserts was
performed by digestion of one microgram of plasmid DNA with
PstI overnight for 37°C.
In vitro transfection
In vitro transfection of B16-F10 melanoma
cells were plated on tissue culture flasks at a density of
7×105 cells. After an overnight incubation and at a
confluence ~70–80%, these cells were transfected with 30 µg
of each CXCR4 shRNA and 30 µl of Lipofectamine 2000
(Invitrogen). The plasmid and Lipofectamine 2000 were diluted in
serum-free medium left at room temperature for 5 min, mixed
immediately and incubated for 20 min at room temperature at a v/w
ratio of liposomes to shRNA of 1:1. The culture medium was removed
and the shRNA-lipid complex (1.5 ml total volume) was added. The
transfection efficiency (~75–80%) was evaluated by using the green
fluorescent protein (GFP) expressing plasmid. Prior to the in
vivo study, we examined whether the two plasmid-based
CXCR4-specific shRNAs (CXCR4-1 shRNA and CXCR4-2 shRNA) were
effective in reducing the CXCR4 expression in cultured B16-F10
cells. Thus, tumor cells were transfected with CXCR4-1, CXCR4-2 or
control shRNA for 5 h and thereafter the cells were washed,
suspended in medium and maintained in culture for 24 or 48 h. To
determine the amount of mRNA CXCR4 and CXCR4 protein, lysates of
the B16-F10 melanoma cells were used for RNA isolation and western
blot analysis.
RNA isolation
Total cellular RNA was extracted using TRIzol-LS
Reagent (Invitrogen). The integrity of RNA was assessed using the
Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Analysis of CXCR4 expression
Reverse transcription-PCR was performed with 1.2
µg of the isolated total RNA and synthesized to cDNA in a 25
µl reaction system using reverse transcriptase (Promega,
Madison, WI, USA). RT conditions were 5 min denaturation at 65°C,
60 min at 37°C and 5 min at 75°C in a thermocycler (Abgene, Epsom,
UK). Reverse transcription was carried out with 0.5 µg of
the oligodT primer, 1 unit of reverse transcriptase, 1 unit of
RNase inhibitor (all from Invitrogen), 5 µl of 5X buffer and
4 µl MgCl2. The β-actin mRNA was used as a
loading control. PCR conditions for β-actin were 4 min denaturation
at 94°C, 40 cycles of 1 min at 94°C, 1 min at 52°C and 2 min at
72°C and 10 min elongation at 72°C in a thermocycler (Abgene). PCR
conditions for CXCR4 were 5 min denaturation at 94°C, 35 cycles of
1 min at 94°C, 1 min at 51°C and 1 min at 72°C, and 10 min
elongation at 72°C in a thermocycler (Abgene). The primers
sequences and GenBank Accession number are shown in Table II. PCR products of β-actin (364 bp)
and CXCR4 (291 bp) were analysed by electrophoresis in a 1.5%
agarose gel and visualized using UV fluorescence after staining
with ethidium bromide. Quantification of CXCR4 bands was performed
by using ImageQuant software, version 3.3 (Molecular Dynamics,
Inc., Sunnyvale, CA, USA) and the results were expressed in terms
of percentage.
 | Table IIPolymerase chain reaction primer
sequences. |
Table II
Polymerase chain reaction primer
sequences.
| Genes | Primer sequences | GenBank acession
no. |
|---|
| CXCR4 | F:
5′-ACAGGTACATCTGTGACCGCCTTT-3′ | BC031665 |
| R:
5′-TGCTCTCGAAGTCACATCCTTGCT-3′ |
| β-actin | F:
5′-TGGAATCCTGTGGCATCCATGAAAC-3′ | BC014861 |
| R:
5′-TAAAACGCAGCTCAGTAACAGTCCG-3′ |
Western blot analysis
B16-F10 adherent cells were detached using EDTA with
RPMI without fetal bovine serum and centrifuged at 4,000 rpm for 15
min. The cell pellet was resuspended in 300 µl PBS plus the
proteases inhibitors 0.1% aprotinin, 0.1% leupepsin and 1% Triton.
The sample was incubated under agitation on ice for 20 min and
after centrifuged at 12,000 rpm for 15 min at 4°C and the protein
concentration determined by Cadman method. Total cellular protein
(30 µg) was separated by electrophoresis through a 10%
SDS-PAGE resolving gel with an SDS-PAGE stacking gel. After
electrophoresis, proteins were transferred onto a Hybond-C
supported nitrocellulose membrane (Amersham Biosciences) by
electroblotting for 4 h at 45 V, 25°C, in transfer buffer (3.94 g
Tris-HCl, 18.80 g glycine, 240 ml methanol, 10% SDS). The membrane
was then blocked with 10% dried milk in TBS (20 mM Tris, 500 mM
NaCl) at room temperature overnight, after washed twice followed by
incubation at room temperature with 1:250 off rabbit anti-CXCR4
polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA)
in TBS-Tween-20 buffer for 90 min. The membrane was washed in TBS
1X Tween-20 for 30 min and secondary anti-rabbit antibodies labeled
with horseradish peroxidase (Amersham Biosciences) was added and
the membrane incubated at room temperature for 60 min under
agitation. Membranes were washed twice in TBS-T for 20 min and in
TBS for 5 min. Antibody labeled protein bands were visualized with
ECL detection reagents (Amersham Biosciences) applied following the
manufacturer's protocol. To use β-actin as a loading control, a
second gel was loaded with identical volume of the experimental
sample followed by blotting with the anti-β-actin antibody and the
detection was performed as described for CXCR4. Quantification of
bands was performed by using Image Quant software, version 3.3
(Molecular Dynamics Inc.) and the results were expressed in terms
of percentage.
Tumorigenic assay
Based on the in vitro findings, CXCR4-1 shRNA
was selected for in vivo experiments. B16-F10 melanoma cells
were transfected with CXCR4-1 shRNA or control shRNA for 5 h and
thereafter the cells were washed, suspended in medium and
maintained in culture for 24 h to inject into mice. After this
treatment, tumor cells were detached with EDTA, washed twice in PBS
and finally resuspended in RPMI. The viability of cells was
assessed by trypan blue staining and was >95%. Tumor cells
transfected were then inoculated subcutaneously (2×105
cells/animal) into the right flank of mice (n=10 per group),
animals were euthanized 14 days after treatment according to Kumar
et al (18) and tumors were
excised and weighted on a microbalance Sartorius Supermicro (model
S4; Sartorius, Goettingen, Germany).
Intratumoral injection of CXCR4-1
shRNA
Each animal received 2×105 tumor cells
subcutaneously into the right flank (n=10 per group). The tumors
were monitored, and treatment began when the average tumor diameter
reached 5–7 mm, typically 7 days after of tumor cell inoculation.
This tumor size allows the intratumoral injection to be performed
with safety. Thus, after 7 days of tumor inoculation mice received
a single intratumoral injection of 2 µg of CXCR4-1 shRNA
complexed with 2 µl of Lipofectamine 2000 dissolved in 50
µl of RPMI. Some mice received an intratumoral injection of
2 µg control shRNA-expressing plasmid complexed with 2
µl of Lipofectamine 2000 as a negative control. The mice
were sacrificed 7 days after the injection and the tumors
weighed.
Statistical analysis
One-way analysis of variance (ANOVA) was used to
analyze the significance between groups. All data represent mean ±
SD. P<0.05 was considered to indicate a statistically
significant difference.
Results
Analysis of CXCR4 expression by B16-F10
melanoma cells
Several studies have demonstrated the involvement of
chemokines and their receptors in tumor growth and metastasis. In
order to investigate the role of CXCR4 chemokine receptor in a
model of murine melanoma, we first examined whether B16-F10
melanoma cells express this chemokine receptor. Our results
indicate that these tumor cells constitutively express CXCR4 as
shown in Fig. 1.
Knockdown of CXCR4 mRNA by transfection
of B16-F10 melanoma cells with CXCR4 shRNA
To reduce the expression of CXCR4 mRNA in B16-F10
melanoma cells, two CXCR4 shRNAs were designed. Each CXCR4 shRNA
was annealed and ligated into the psiSTRIKE vector controlled by
Pol III U6 promoter. The tumor cells were transfected for 24 and 48
h with the plasmid-based CXCR4-1 shRNA, CXCR4-2 shRNA or control
shRNA expressing plasmids with Lipofectamine 2000. After
transfection, CXCR4 mRNA degradation was monitored by RT-PCR.
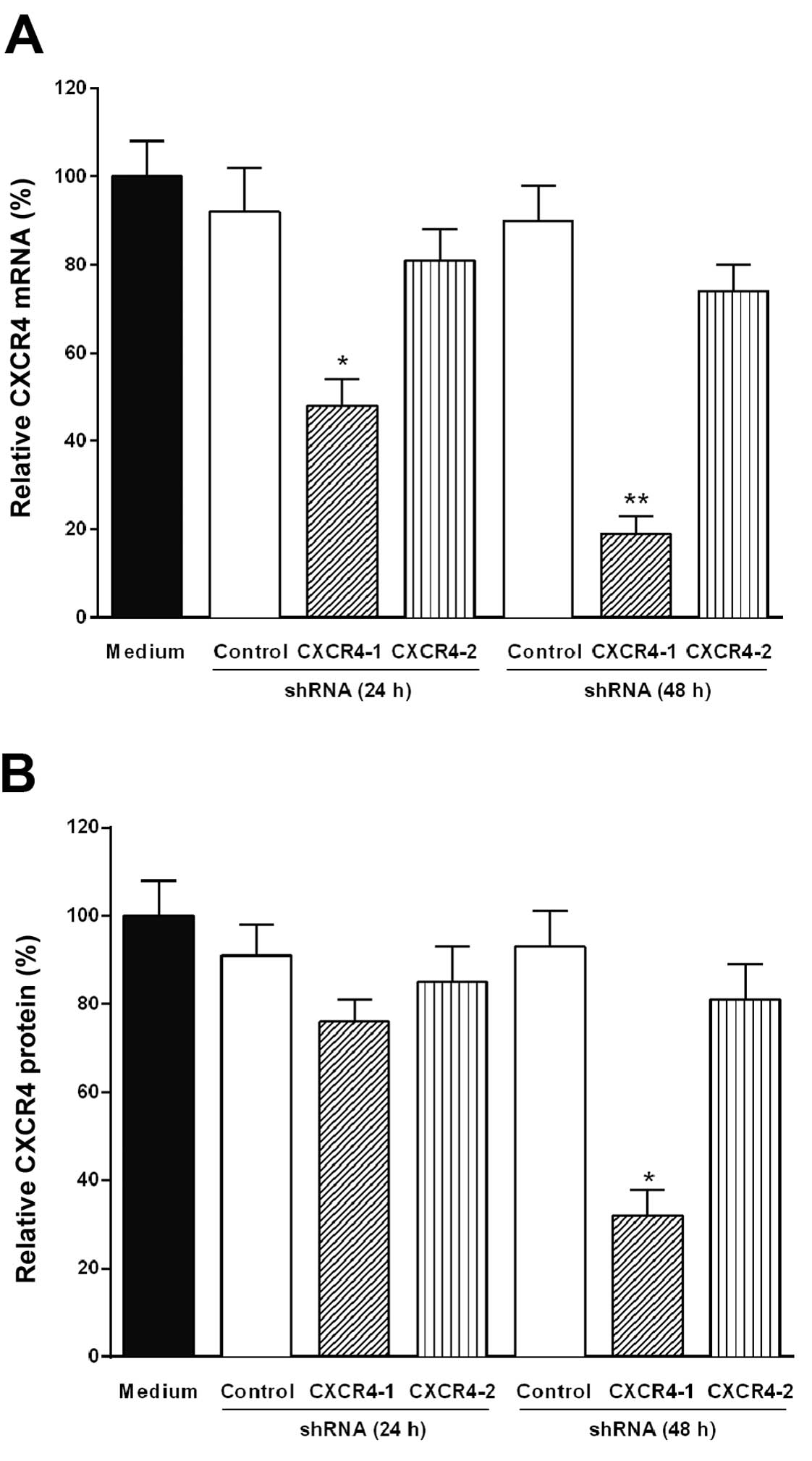
Fig. 2A shows that only the
plasmid-based CXCR4-1 shRNA significantly reduced the level of
CXCR4 mRNA (85%, P<0.001) after 48 h of transfection and this
effect remains for up to 4 days (data not shown).
Reduction of CXCR4 protein level by
transfection of B16 melanoma cells with CXCR4 shRNA
The level of CXCR4 protein in B16-F10 melanoma cells
transfected with CXCR4-1 shRNA, CXCR4-2 shRNA or control shRNA was
evaluated by western blot analysis. The downregulation of CXCR4
protein expression was also significantly (70%, P<0.001)
observed after 48 h of transfection with CXCR4-1 shRNA as shown in
Fig. 2B.
Transfection of B16-10 melanoma cells
with CXCR4 shRNA inhibits tumor growth in mice
The B16-F10 melanoma cells transfected with CXCR4-1
shRNA or control shRNA were injected subcutaneously into C57BL/6
mice. The animals were sacrificed 14 days after the inoculation of
B16-F10 cells and tumors were excised and weighted. Fig. 3 shows that tumor growth was
significantly inhibited (66%, P<0.001) when B16-F10 melanoma
cells were transfected with CXCR4-1 shRNA when compared to control
shRNA.
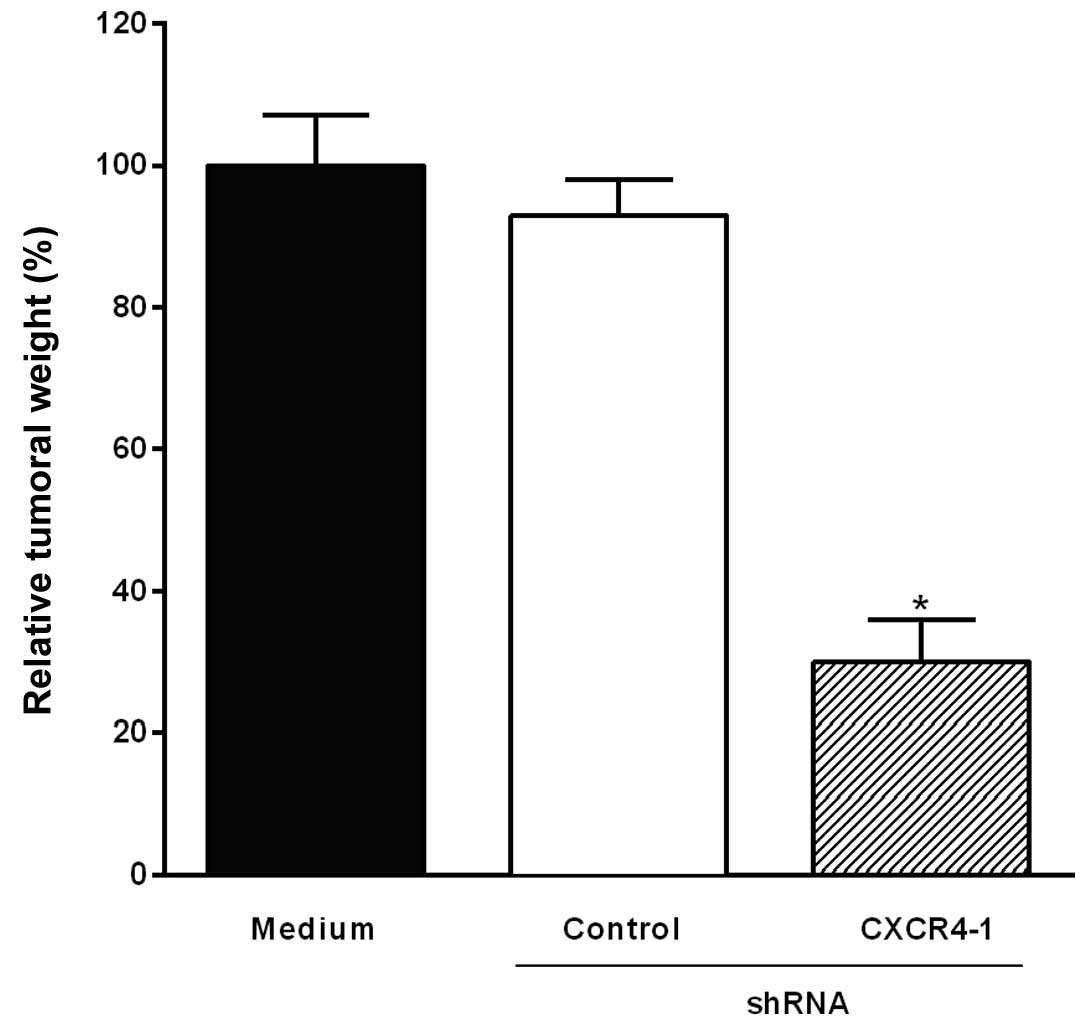
Intratumoral injection of the CXCR4-1
shRNA-expressing plasmid inhibits B16-F10 melanoma growth
To investigate the effect of the intratumoral
injection of the CXCR4-1 shRNA-expressing plasmids on melanoma
growth, mice were inoculated subcutaneously with B16-F10 melanoma
cells. Fig. 4 shows a significant
reduction of tumor weight (70%, P<0.001) in mice that had
received a single intratumoral injection of the CXCR4-1 shRNA
compared to animals injected with the plasmid-based control
shRNA.
Discussion
Current strategies for studying the involvement of
CXCR4 in cancer are based on receptor blockade in the surface of
tumor cells with specific antagonists or antibodies. In the present
study, we applied the RNAi technology to elucidate the role played
by CXCR4 in B16-F10 melanoma growth. The first step was to examine
the ability of two CXCR4-specific shRNAs expressing plasmids to
downregulate CXCR4 mRNA and CXCR4 protein in vitro. Our
results indicate that B16-F10 melanoma cells constitutively express
CXCR4 and that only CXCR4-1 shRNA significantly reduced the level
of CXCR4 mRNA and CXCR4 protein after in vitro transfection
of tumor cells. Thus, CXCR4-1 shRNA was used in all further
experiments. The next step was to investigate the effect of
silencing CXCR4 expression on B16-F10 melanoma growth in
vivo. To address this question, the transfected tumor cells
with CXCR4-1 shRNA or control shRNA in vitro were injected
subcutaneously into mice. Our findings showed that the tumor growth
was significantly reduced only in mice inoculated with B16-F10
melanoma cells transfected with CXCR4-1 shRNA, suggesting that
CXCR4 plays a critical role in B16-F10 melanoma growth.
It is known that RNAi-based therapy is effective and
elicit gene silencing response, the double-stranded RNA molecules
must be delivered to the target cells. Unfortunately, the specific
delivery of RNAi has been challenging despite many efforts made in
the last few years and the intratumoral injection of RNAi has
emerged as a promising alternative to overcome this obstacle
(19,20). We also decided to investigate the
effect of the intratumoral injection of CXCR4 shRNA expressing
plasmid in the pre-established subcutaneous B16-F10 melanoma. Our
results indicate that the intratumoral injection of CXCR4-1 shRNA
significantly inhibited tumor growth when compared to animals
injected with the plasmid-based control shRNA. It is worthwhile
that this effect was obtained with only a single injection of the
CXCR4-1 shRNA expressing plasmid and this approach for RNAi
delivery was effective for at least a week.
The role of the CXCR4/CXCL12 axis in cancer has been
extensively investigated in the last decade. Thus, it was found
that CXCR4 is overexpressed in >23 human cancers, including
melanoma and contributes to cell proliferation, cell survival,
invasion ad angiogenesis (21).
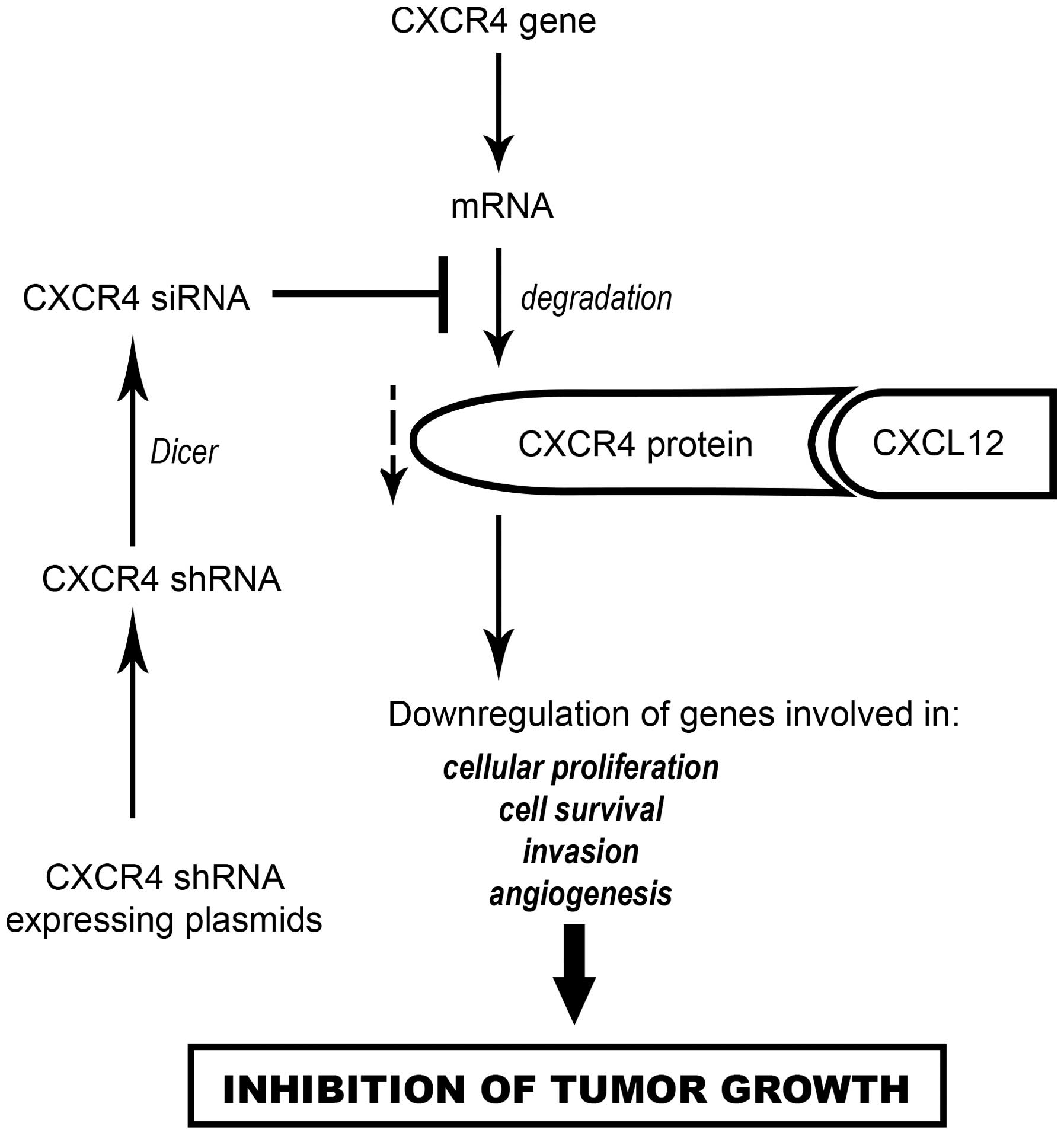
Based on these findings, we proposed that the effect
of CXCR4 shRNA expressing plasmids on B16-F10 melanoma growth could
be explained as illustrated in Fig.
5. Briefly, CXCR4 shRNA expressing plasmids were used to
transfect the B16-F10 melanoma cells in vitro or in
vivo by intratumoral injection. After the uptake of CXCR4 shRNA
expressing plasmids by tumor cells, CXCR4 shRNA are transcribed in
the nucleus, exported to the cytoplasm and processed by Dicer to
generate CXCR4 siRNA which induces the specific degradation of
CXCR4 mRNA. Therefore, the level of CXCR4 protein is decreased with
subsequent downregulation of genes involved in cell survival, cell
adhesion, invasion and angiogenesis, resulting in the inhibition of
B16-F10 melanoma growth.
The present study gives support to the concept that
a direct administration of RNAi-based therapeutics into the target
tumor is a promising approach for overcoming the hurdles of
systemic delivery. Our findings also suggest that the intratumoral
injection of CXCR4-1 shRNA expressing vector may be a novel
therapeutic approach for human solid tumors such as cutaneous
melanoma and breast cancer since CXCR4 is overexpressed in these
tumors.
It should be emphasized that the present study is
the first demonstration that CXCR4 plays a critical role in the
growth of B16-F10 melanoma. However, further work is required to
elucidate the molecular mechanisms involved in this phenomenon.
Currently there is great interest in the discovery of antagonists
for therapeutic targeting CXCR4 expression. Considering that our
results indicate that CXCR4 is implicated in the early stages of
B16-F10 melanoma growth, its role well established in metastasis
and the fact that this chemokine receptor is highly conserved
between human and mouse (22), this
experimental model of cancer may contribute for the discovery of
CXCR4 antagonists with clinical implications.
Acknowledgments
The present study was supported by FAPESP
(06/57963-1). We thank Cacilda D. Pereira and Zuleica A. S. Moraes
for the technical assistance.
References
|
1
|
Murdoch C: CXCR4: Chemokine receptor
extraordinaire. Immunol Rev. 177:175–184. 2000. View Article : Google Scholar
|
|
2
|
Zlotnik A: Chemokines in neoplastic
progression. Semin Cancer Biol. 14:181–185. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Toyozawa S, Kaminaka C, Furukawa F,
Nakamura Y, Matsunaka H and Yamamoto Y: Chemokine receptor CXCR4 is
a novel marker for the progression of cutaneous malignant
melanomas. Acta Histochem Cytochem. 45:293–299. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mitchell B, Leone D, Feller JK, Bondzie P,
Yang S, Park HY and Mahalingam M: Correlation of chemokine receptor
CXCR4 mRNA in primary cutaneous melanoma with established
histopathologic prognosticators and the BRAF status. Melanoma Res.
24:621–625. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Payne AS and Cornelius LA: The role of
chemokines in melanoma tumor growth and metastasis. J Invest
Dermatol. 118:915–922. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Longo-Imedio MI, Longo N, Treviño I,
Lázaro P and Sánchez-Mateos P: Clinical significance of CXCR3 and
CXCR4 expression in primary melanoma. Int J Cancer. 117:861–865.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Scala S, Ottaiano A, Ascierto PA, Cavalli
M, Simeone E, Giuliano P, Napolitano M, Franco R, Botti G and
Castello G: Expression of CXCR4 predicts poor prognosis in patients
with malignant melanoma. Clin Cancer Res. 11:1835–1841. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Scala S, Giuliano P, Ascierto PA, Ieranò
C, Franco R, Napolitano M, Ottaiano A, Lombardi ML, Luongo M,
Simeone E, et al: Human melanoma metastases express functional
CXCR4. Clin Cancer Res. 12:2427–2433. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Duda DG, Kozin SV, Kirkpatrick ND, Xu L,
Fukumura D and Jain RK: CXCL12 (SDF1α)-CXCR4/CXCR7 pathway
inhibition: An emerging sensitizer for anticancer therapies? Clin
Cancer Res. 17:2074–2080. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fidler IJ: Biological behavior of
malignant melanoma cells correlated to their survival in vivo.
Cancer Res. 35:218–224. 1975.PubMed/NCBI
|
|
11
|
André ND, Silva VAO, Ariza CB, Watanabe
MAE and De Lucca FL: In vivo silencing of CXCR4 with jetPEI/CXCR4
shRNA nanoparticles inhibits pulmonary metastasis of B16-F10
melanoma cells. Mol Med Rep. 12:8320–8326. 2015.
|
|
12
|
Li Z, Li N, Wu M, Li X, Luo Z and Wang X:
Expression of miR-126 suppresses migration and invasion of colon
cancer cells by targeting CXCR4. Mol Cell Biochem. 381:233–242.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Díaz MR and Vivas-Mejia PE: Nanoparticles
as drug delivery systems in cancer medicine: Emphasis on
RNAi-containing nanoliposomes. Pharmaceuticals (Basel).
6:1361–1380. 2013. View Article : Google Scholar
|
|
14
|
Li CX, Parker A, Menocal E, Xiang S,
Borodyansky L and Fruehauf JH: Delivery of RNA interference. Cell
Cycle. 5:2103–2109. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sakurai Y, Hatakeyama H, Sato Y, Hyodo M,
Akita H and Harashima H: Gene silencing via RNAi and siRNA
quantification in tumor tissue using MEND, a liposomal siRNA
delivery system. Mol Ther. 21:1195–1203. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deharvengt SJ, Gunn JR, Pickett SB and
Korc M: Intratumoral delivery of shRNA targeting cyclin D1
attenuates pancreatic cancer growth. Cancer Gene Ther. 17:325–333.
2010. View Article : Google Scholar :
|
|
17
|
André ND, Silva VAO, Watanabe MAE and De
Lucca FL: Intratumoral injection of PKR shRNA expressing plasmid
inhibits B16-F10 melanoma growth. Oncol Rep. 32:2267–2273.
2014.PubMed/NCBI
|
|
18
|
Kumar R, Yoneda J, Fidler IJ and Dong Z:
GM-CSF-transduced B16 melanoma cells are highly susceptible to
lysis by normal murine macrophages and poorly tumorigenic in
immune-compromised mice. J Leukoc Biol. 65:102–108. 1999.PubMed/NCBI
|
|
19
|
Deng Y, Wang CC, Choy KW, Du Q, Chen J,
Wang Q, Li L, Chung TK and Tang T: Therapeutic potentials of gene
silencing by RNA interference: Principles, challenges, and new
strategies. Gene. 538:217–227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li T, Wu M, Zhu YY, Chen J and Chen L:
Development of RNA interference-based therapeutics and application
of multi-target small interfering RNAs. Nucleic Acid Ther.
24:302–312. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chatterjee S, Behnam Azad B and Nimmagadda
S: The intricate role of CXCR4 in cancer. Adv Cancer Res.
124:31–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Heesen M, Berman MA, Benson JD, Gerard C
and Dorf ME: Cloning of the mouse fusin gene, homologue to a human
HIV-1 co-factor. J Immunol. 157:5455–5460. 1996.PubMed/NCBI
|