Introduction
The incidence of cancer is on the increase and one
of the main causes of global mortality (1). Hepatocellular carcinoma (HCC) is a
primary malignant tumor and the leading cause of cancer among
cirrhotic patients, making it a global health issue (2,3). HCC
usually develops in the context of inflammation and organ injury
(4). The pathogenic factors are
varied. Hepatitis B and C viruses, and autoimmune hepatitis cause
progressive liver disease and are major risk factors for the
development of HCC (5–9). Due to the lack of effective therapies,
such as standard chemotherapeutic agents, and the challenges
experienced in early diagnosis, HCC patients have a poor prognosis
(10). Therefore, new therapeutic
targets may be identified from investigations into the molecular
mechanism involved in liver cancer (11).
The membrane protein is a unique structure of
protein that plays an important role in cell contact, signal
transduction and enzyme activity. It has various functions and
becomes the ideal drug target. CD151, as a 4 transmembrane protein
gene, is associated with the invasion and metastasis of HCC
(12). The transmembrane protein
(TMEM) family sequence functions remain unknown only individual
protein function has been reported. TMEM9 was characterized as a
novel human transmembrane protein, belonging to a new protein
family (13). The gene is localized
to chromosome 1q41. To the best of our knowledge, the role of TMEM9
in HCC studies remains to be investigated.
In the present study, we investigated the function
of the TMEM9 gene in HCC. The results suggest that this gene
is closely associated with liver cancer. Thus, this may be a
candidate gene for further study of molecular or therapeutic
targets.
Materials and methods
Patients and tissue samples
Between 2008 and 2013, 70 HCC patients presenting to
the Zhongnan Hospital of Wuhan University (Hubei, China) were
enrolled in the present study. All the patients had complete
clinical and pathological follow-up data. Adjacent normal
hepatocellular tissues were also collected as negative controls.
These normal hepatocellular tissues were resected within at least 5
cm of the tumor margin when the patients underwent definitive
surgery. Clinical fresh tissue samples were detected by qPCR.
Approval for the study was provided by the independent Ethics
Committee of the Zhongnan Hospital of Wuhan University. Informed
and written consent was obtained from all the patients or their
advisers according to the ethics committee guidelines.
Cell culture and transfection
conditions
Human 97H, 97L, HepG2, 7721, 7404 and HuH7 HCC cell
lines were obtained from the American Type Culture Collection
(ATCC; Manassas, VA, USA). The cells were grown in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% fetal bovine
serum (FBS) and 1% double antibiotics (penicillin/streptomycin) and
maintained in a 37°C incubator with a 5% CO2 humidified
atmosphere. Transfections were performed using the Lipofectamine™
2000 reagent according to the manufacturer's instructions
(Invitrogen Life Technologies, Carlsbad, CA, USA). After 48 h of
transfection, the cells were used for cell proliferation assays,
cell cycle analysis, and apoptosis, Matrigel invasion, migration
and adhesion assays. Silencer negative control siRNA was used as a
negative control.
RT-qPCR analysis
Cellular RNA was isolated using the TRIzol kit
(Invitrogen Life Technologies). SYBR-Green RT-qPCR was performed to
detect the mRNA expression. GADPH was used to normalize the RNA
inputs. The primers used were: TMEM9 sense,
5′-GGGCACATTTACAACCAG-3′ and antisense, 5′-ATCAGGAAGGCCATGTAG-3′;
GADPH sense, 5′-CACCCACTCCTCCACCTTTG-3′ and antisense,
5′-CCACCACCCTGTTGCTGTAG-3′.
Cell proliferation assay
Viability of cells 72 h after transfection was
assessed using the Cell Counting Kit-8 (CCK-8) (Qihai, Shanghai,
China). Briefly, cells were seeded at a density of 3×104
in each 96-well plate and cultured for 0, 24, 48 and 72 h,
respectively. CCK-8 reagent (100 µl/well) was added to each
well and incubated for 1 h at 37°C. The optical density (OD) values
were determined at 450 nm using a microplate reader. Three
different experiments were performed for each experimental
condition.
Flow cytomery
The cell cycle was assessed by flow cytometric
analysis at different time points using a propidium iodide (PI)
cell cycle detection kit (Beyotime, Shanghai, China). The cells
were collected, treated and stained with PI according to the
manufacturer's instructions. The cell cycle was detected using a
flow cytometer (BD Biosciences, Heidelberg, Germany).
Apoptotic cells were visualized using an Annexin
V-FITC/PI kit (BD Biosciences, San Jose, CA, USA). The apoptosis of
HCC-transfected cells were determined by flow cytometric (FCM)
analysis using a FACSCalibur.
Cell invasion and migration
After transfection, the cells were detached and
washed twice in PBS. Then, 1×105 cells/ml were seeded in
the upper chamber of a Transwell insert (8-µm pore size)
coated (invasion) or not coated (migration) with 80 µl
Matrigel (BD Biosciences). The lower chamber was filled with 0.75
ml of DMEM. After a 48-h (invasion) or 24-h (migration) incubation
period, the non-migrated cells in the upper chamber were scraped
away, and adherent cells were stained with formaldehyde solution.
Any cells on the underside were counted and photographed under ×200
microscope fields.
Cell adhesion
To determine the adhesion cells, 12-well plates were
used. Cell suspension (1×105 cells/ml) was added to the
well and incubated for 1 h at 37°C. Adherent cells were fixed with
4% methanol and stained with crystal violet for 20 min. The number
of adherent cells were photographed and counted from three random
selected ×200 fields of microscope.
Channel protein expression detection
To detect the role of TMEM9 in liver cancer cells,
we selected the proteins CCNB1, CCNB2, CDK1, PRL10A, S100A10 and
EIF3H to detect the protein expression using western blotting.
Protein lysates were prepared. Equal amounts of samples were
resolved by SDS-PAGE and transferred to nitrocellulose membranes.
The membranes were then blocked with 5% low-fat milk for 1 h or
overnight at 4°C; incubated with CCNB1, CCNB2, CDK1, PRL10A,
S100A10 and EIF3H with primary antibodies for 2 h, followed by
secondary antibodies for 1 h at room temperature; and analyzed.
GADPH protein levels were determined as a loading control.
Statistical analysis
Statistical significances were determined using the
GraphPad Prism v5.0 software (GraphPad Software, La Jolla, CA,
USA). Kaplan-Meier analysis was used to determine that the overall
survival time between low and high expression of HCC. Data are
presented as the mean ± SD of at least three independent
replicates. Differences were considered significant when P<0.05
or P<0.01.
Results
TMEM9 is highly expressed in HCC with
poor patient survival
To investigate the expression of TMEM9, we used
RT-qPCR to investigate in the HCC tissues of 30 patients. The
results showed a higher level of TMEM9 expression (Fig. 1A). Then we investigated the
correlation between TMEM9 expression and prognosis of the patients
with HCC. As shown in Fig. 1B,
Kaplan-Meier analysis showed that the overall survival time of
lower-TMEM9-expressing patients was notably higher than that of
higher-TMEM9-expressing patients. The expression levels of TMEM9 in
the six HCC cells were also evaluated by qPCR and western blotting
(Fig. 1C and D). The HepG2 and 7721
cell lines had a higher TMEM9 mRMA and protein expression.
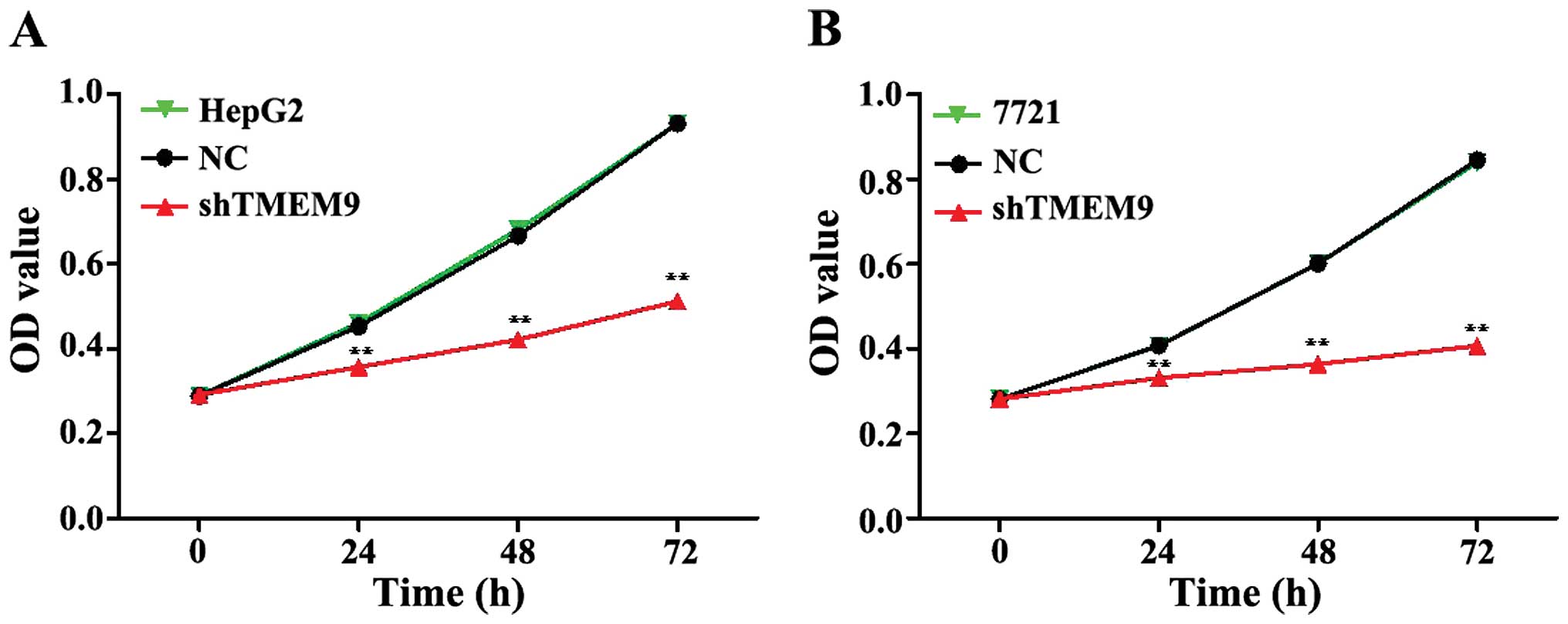
Knockdown of TMEM9 inhibits cell
proliferation and induces apoptosis
To assess the potential effects of RNAi silencing
TMEM9 on proliferation, CCK-8 analysis was performed 72 h after
transfection. The proliferative ability of HepG2 and 7721 cells was
significantly inhibited at 24, 48 and 72 h (Fig. 2).
Moreover, we investigated the effects of TMEM9 on
cell cycle and apoptosis in HCC. The flow cytometric analysis
revealed that the population of G0/G1 phase was significantly
increased but that of S and G2/M phase was decreased in HepG2 and
7721 cells, when compared with the negative control (NC)
(P<0.01) (Fig. 3).
In addition, we assessed the apoptotic function of
TMEM9 in HepG2 and 7721 cells using the Annexin V-FITC/PI staining
assay. As shown in Fig. 4, the
results showed that knockdown of TMEM9 in HCC cells markedly
induced the cell apoptotic rate compared with NC (P<0.01).
Knockdown of TMEM9 decreases metastasis
of HCC cells
Cell metastasis plays an important role in cancer
progression. We determined whether TMEM9 regulated metastasis of
HCC cells. Cell invasion, migration and adhesion assays were then
used to detect the metastatic capacity. As shown in Fig. 5, the cell invasion ability was
reduced when compared with NC (P<0.01). The migration and
adhesion cells were also decreased (P<0.01) (Figs. 6 and 7).
Knockdown of TMEM9 decreases protein
expression in HCC
Signaling pathways are often activated in tumor
cells. We assessed the protein expression using western blotting.
The results showed that the protein expression of CDK1, EIF3H,
RPL10L, S100-A10, CCNB1 and CCNB2 was decreased compared to the
control group (P<0.01) (Fig.
8).
Discussion
HCC is one of the most common types of cancer
worldwide. Of an estimated 700,000 cancer-associated mortalities
that arising in 2008, 50% occurred in China (2,14). The
5-year survival rate remains at <40% following surgery (15). HCC cause serious damage to human
health; thus, investigation of its development mechanism and
identification of effective measures of prevention, diagnosis and
treatment of HCC is crucial. In the present study, we investigated
the biological function of TMEM9 in HCC cells (16). The clinical data show that TMEM9 was
highly expressed in HCC patients. Moreover, TMEM9 expression was
associated with the patient survival rate. The in vitro
experiments showed that knockdown of TMEM9 in HCC HepG2 and 7721
cancer cells inhibited cell growth and metastasis, and promoted
cell apoptosis. Thus, TMEM9 serves as a potential target for the
treatment of HCC.
In order to elucidate the possible mechanism
involved, we identified the related protein expression. CDK1 is a
highly conserved protein and a key player in the cell cycle
regulation (17). Eukaryotic
translation initiation factors (EIFs) are involved in the protein
translation initiation process, and EIF2, EIF3, EIF4 and EIF5 have
been previously investigated (18).
EIF3H is an important subunit of the EIF3 family. The EIF3H
expression level is closely associated with a variety of tumors,
and is overexpressed in numerous malignant tumors (19,20).
Ribosomal protein L 10-like (RPL10L), a protein-coding gene, has a
structural constituent of ribosome. Furthermore, S100A10, CCNB1 and
CCNB2 are involved in mitosis and the regulation of cell cycle
progression (21–23). Our results show that the cell
cycle-related protein expressions were significantly decreased when
compared with NC.
In summary, to the best of our knowledge, the
present study provides evidence for the first time that TMEM9 is
crucial in the cell proliferation, apoptosis and metastasis of HCC
cells. Additionally, TMEM9 regulates these biological processes by
regulating cell cycle-related proteins. As TMEM9 expression level
is associated with the patient survival rate, inhibition of TMEM9
in tumor tissues provides a therapeutic strategy. However,
additional investigations should be conducted to validate its
therapeutic function in the future.
Acknowledgments
The present study was supported by the Natural
Science Fund of Hubei Province (no. 2012FFA044), the Health
Department Found of Hubei Province (no. JX6B18), and the Public
Service Platform Construction Projects of Wuhan Technology Bureau
(no. 2013060705010326).
References
|
1
|
Mathers C, Fat DM and Boerma JT: The
Global Burden of Disease: 2004 Update. World Health Organization;
2008
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bosch FX, Ribes J, Díaz M and Cléries R:
Primary liver cancer: Worldwide incidence and trends.
Gastroenterology. 127(Suppl 1): S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Villanueva A, Newell P, Chiang DY,
Friedman SL and Llovet JM: Genomics and signaling pathways in
hepatocellular carcinoma. Semin Liver Dis. 27:55–76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yuen MF, Tanaka Y, Fong DY-T, Fung J, Wong
DK, Yuen JC, But DY, Chan AO, Wong BC, Mizokami M, et al:
Independent risk factors and predictive score for the development
of hepatocellular carcinoma in chronic hepatitis B. J Hepatol.
50:80–88. 2009. View Article : Google Scholar
|
|
6
|
Yang HI, Sherman M, Su J, Chen PJ, Liaw
YF, Iloeje UH and Chen CJ: Nomograms for risk of hepatocellular
carcinoma in patients with chronic hepatitis B virus infection. J
Clin Oncol. 28:2437–2444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lok AS, Seeff LB, Morgan TR, di Bisceglie
AM, Sterling RK, Curto TM, Everson GT, Lindsay KL, Lee WM,
Bonkovsky HL, et al HALT-C Trial Group: Incidence of hepatocellular
carcinoma and associated risk factors in hepatitis C-related
advanced liver disease. Gastroenterology. 136:138–148. 2009.
View Article : Google Scholar
|
|
8
|
Yeoman AD, Al-Chalabi T, Karani JB,
Quaglia A, Devlin J, Mieli-Vergani G, Bomford A, O'Grady JG,
Harrison PM and Heneghan MA: Evaluation of risk factors in the
development of hepatocellular carcinoma in autoimmune hepatitis:
Implications for follow-up and screening. Hepatology. 48:863–870.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang X, Wang N, Cheung F, Lao L, Li C and
Feng Y: Chinese medicines for prevention and treatment of human
hepatocellular carcinoma: Current progress on pharmacological
actions and mechanisms. J Integr Med. 13:142–164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lopez PM, Villanueva A and Llovet JM:
Systematic review: Evidence-based management of hepatocellular
carcinoma - an updated analysis of randomized controlled trials.
Aliment Pharmacol Ther. 23:1535–1547. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Villanueva A, Toffanin S and Llovet JM:
Linking molecular classification of hepatocellular carcinoma and
personalized medicine: Preliminary steps. Curr Opin Oncol.
20:444–453. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ke AW, Shi GM, Zhou J, Huang XY, Shi YH,
Ding ZB, Wang XY, Devbhandari RP and Fan J: CD151 amplifies
signaling by integrin α6β1 to PI3K and induces the
epithelial-mesenchymal transition in HCC Cells. Gastroenterology.
140:1629–1641.e15. 2011. View Article : Google Scholar
|
|
13
|
Kveine M, Tenstad E, Døsen G, Funderud S
and Rian E: Characterization of the novel human transmembrane
protein 9 (TMEM9) that localizes to lysosomes and late endosomes.
Biochem Biophys Res Commun. 297:912–917. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
15
|
Fang F, Yang L, Tao Y and Qin W: FBI-1
promotes cell proliferation and enhances resistance to chemotherapy
of hepatocellular carcinoma in vitro and in vivo. Cancer.
118:134–146. 2012. View Article : Google Scholar
|
|
16
|
Zhai XF, Chen Z, Li B, Shen F, Fan J, Zhou
WP, Yang YK, Xu J, Qin X, Li LQ and Ling CQ: Traditional herbal
medicine in preventing recurrence after resection of small
hepatocellular carcinoma: a multicenter randomized controlled
trial. J Integr Med. 11:90–100. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Morgan DO: The Cell Cycle: Principles of
Control. New Science Press. 2007.
|
|
18
|
Spilka R, Ernst C, Mehta AK and Haybaeck
J: Eukaryotic translation initiation factors in cancer development
and progression. Cancer Lett. 340:9–21. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okamoto H, Yasui K, Zhao C, Arii S and
Inazawa J: PTK2 and EIF3S3 genes may be amplification targets at
8q23–q24 and are associated with large hepatocellular carcinomas.
Hepatology. 38:1242–1249. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cappuzzo F, Varella-Garcia M, Rossi E,
Gajapathy S, Valente M, Drabkin H and Gemmill R: MYC and EIF3H
coamplification significantly improve response and survival of
non-small cell lung cancer patients (NSCLC) treated with gefitinib.
J Thorac Oncol. 4:472–478. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jackman M, Firth M and Pines J: Human
cyclins B1 and B2 are localized to strikingly different structures:
B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J.
14:1646–1654. 1995.PubMed/NCBI
|
|
22
|
Porter LA, Singh G and Lee JM: Abundance
of cyclin B1 regulates γ-radiation-induced apoptosis. Blood.
95:2645–2650. 2000.
|
|
23
|
Rescher U and Gerke V: S100A10/p11:
Family, friends and functions. Pflugers Arch. 455:575–582. 2008.
View Article : Google Scholar
|