Introduction
Gastric cancer (GC) ranks as the fifth most common
cancer and is the third leading cause of cancer-related death
worldwide (1). Despite a steadily
declining incidence, GC remains a highly fatal disease that causes
more than 723,000 deaths per year (2). In addition, due to its silent nature
and underlying genetic and biological heterogeneity, the prognosis
of GS has improved little over time (1). Therefore, there is an urgent need to
improve GC patient outcomes by achieving a detailed understanding
of the molecular mechanism underlying GC.
MicroRNAs (miRNAs) are a class of ~22 nucleotides
long, endogenous, small, non-coding RNAs that negatively regulate
genes by triggering either mRNA degradation or translational
repression through base pairing to the 3′-untranslated region
(3′-UTR) of specific target mRNAs (3,4).
Compelling evidence indicates that more than 200 miRNAs are
involved and functional in GC pathogenesis and treatment response
(5,6). For example, oncogenic miR-21 and
miR-130b were reported to be upregulated in GC and to negatively
target the tumor suppressor genes PTEN and RUNX3 (7,8).
Reciprocally, tumor-suppressor miRNAs, such as miR-101 and miR-375,
were found to be downregulated in GC and led to overexpression of
the oncogenic genes EZH2 and PDK1 (9,10).
Apart from these miRNAs, miR-134 has been revealed to be
downregulated in many solid cancers (11–13).
In gastrointestinal stromal tumors, miR-134 was confirmed to be
downregulated (14), suggesting a
conceivable role of miR-134 in the pathogenesis of GC. However, the
detail molecular mechanism needs to be elucidated.
Golgi phosphoprotein 3 (GOLPH3), also known as GPP34
and GMx33, is a 34-kDa Golgi-localizing protein that was originally
identified in the mouse Golgi apparatus by Snyder et al
(15). GOLPH3 was identified as a
novel oncogene in several solid tumors, including lung, prostate,
breast, ovarian and pancreatic cancers, and melanomas (16). In patients with GC, GOLPH3 has been
reported to be upregulated and is associated with poor clinical
outcomes (17). These studies imply
that GOLPH3 is a promising molecular target for the prevention of
GC.
In the present study, using both in vitro and
in vivo models, we investigated the role of miR-134 in
regulating GC cell lines and tissues, and the underlying mechanism.
miR-134 was found to markedly inhibit GC cell proliferation and
target GOLPH3 via its 3′-UTR region. Moreover, miR-134 expression
was inversely correlated with GOLPH3 protein level in GC.
Overexpression of GOLPH3 was able to partially reverse the
inhibition of GC cell proliferation caused by miR-134. Thereby,
these data suggest that miR-134 functions as a tumor suppressor by
negatively regulating the GOLPH3 oncogene, which promotes GC
proliferation.
Materials and methods
Patient samples
Twenty-six human GC specimens and pair-matched
gastric adjacent non-tumor tissues were obtained from patients
undergoing surgical resection at the Department of
Gastroenterology, The Affiliated Hospital of Inner Mongolia Medical
University (Hohhot, China). GC diagnosis was confirmed by
pathologic examination. Informed consent was obtained from all
patients before collection. The use of human tissue samples was
approved by the Ethics Committee of the Inner Mongolia Medical
University.
Cell culture
Human GC cell lines AGS, SNU-1, BGC-823, MGC-803 and
SGC-7901 were purchased from the American Type Culture Collection
(ATCC; Manassas, VA, USA). Immortalized normal human gastric
epithelial cell line GES-1 was obtained from the Institute of
Biochemistry and Cell Biology at the Chinese Academy of Sciences
(Shanghai, China). The entire cell lines were maintained in
RPMI-1640 medium (Gibco, Rockville, MD, USA) supplemented with 10%
fetal bovine serum (FBS) containing 1% penicillin/streptomycin in a
humidified atmosphere containing 5% CO2 at 37̊C.
Xenograft tumor assay
Four-week-old female BALB/c athymic nude mice
weighing 15–20 g were purchased from the Experimental Animal Centre
of the Inner Mongolia Medical University, and raised in a specific
pathogen-free environment. Mice were inoculated subcutaneously with
a 100-µl injection of 1×106 BGC-823 cells transfected
either with the miR-134-overexpressing or the control vector into
the right flanks of the mice. Tumor dimensions were measured every
2 days and tumor volume was calculated according to the following
formula: Volume = tumor length × tumor width2/2. At day
50, all mice were sacrificed and tumors were weighed. All animal
care and treatments were performed in strict accordance with the
National Institutes of Health Guide for the Care and Use of
Laboratory Animals and were approved by the Institutional Animal
Care Committee of the Inner Mongolia Medical University.
Real-time PCR analysis
Total RNA from sample tissues and cell lines were
extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) according to
the manufacturer's instructions. Then, reverse transcription
reactions were carried out using the One Step PrimeScript miRNA
cDNA Synthesis kit and PrimeScript RT-PCR kit (both from Takara,
Dalian, China) for miRNA and mRNA analysis, respectively. RT-qPCR
analysis was conducted utilizing SYBR-Green qPCR Master Mix (Thermo
Fisher, Shanghai, China) and 7900HT Fast Real-Time PCR System
(Applied Biosystems, Darmstadt, Germany). The primers used were as
follows: 5′-GGTGTGACTGGTTGACCA-3′ (forward) and
5′-TGCGTGTCGTGGAGTC-3′ (reverse) for miR-134; and
5′-TGTAAGTCAGATGCTCCAACAGG-3′ (forward) and
5′-TCACCCATTTGTCAGAACGG-3′ (reverse) for GOLPH3. Relative
expression of the target genes was normalized to U6 snRNA (for
miRNA) or GAPDH (for mRNA) using the 2−ΔΔCt method.
Cell proliferation assay
To investigate the effect of miR-134 on the
proliferation of GC cell lines, 2×104 cells were
transfected with different concentrations (0, 20, 40 or 80 nM) of
pre-miR-134 (miR-134 precursors) or pre-miRNA negative control
(negative control) an placed into a 96-well plate. After incubation
for 24, 48, 72 and 96 h, 20 µl of MTT (5 mg/ml) was added to each
well and the mixture was cultured for another 4 h. Afterwards, 200
µl/well of dimethyl sulfoxide was added to dissolve the formazan
crystals. Finally, the absorbance at 490 nm was measured using an
ELISA reader (Bio-Tek Instruments Inc., Winooski, VT, USA).
Western blot analysis
Cells were lysed with RIPA buffer (Sigma-Aldrich,
St. Louis, MO, USA). Protein concentration was assayed using the
Bradford method and a total of 30 µg protein was separated through
12% SDS-PAGE gels. Then, proteins were electrophoretically
transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA,
USA). After blocking with 3% non-fat milk in TBS for 1 h at 37̊C,
the membranes were incubated with primary antibodies at 4̊C
overnight. HRP-conjugated secondary antibodies were then added and
incubated for 1 h at room temperature. Finally, protein bands were
visualized using an enhanced chemiluminescence kit (Amersham,
Little Chalfont, UK). Anti-human antibodies used in western blot
analysis were as follows: GOLPH3 (1:1,000; Abcam, Cambridge, UK),
AKT (1:1,500), p-AKT (1:1,500), mTOR (1:1,000), p-mTOR (Ser2448,
1:1,000), S6K (1:1,500), p-S6K (1:1,500), and HRP-conjugated
secondary antibody (1:2,000) (both from Santa Cruz Biotechnology,
Santa Cruz, CA, USA.
Luciferase reporter assay
To validate the target gene of miR-134, a luciferase
reporter assay was performed using the pGL3 luciferase promoter
vector (Promega, Madison, WI, USA). The cDNA fragments of the human
GOLPH3 3′-UTR containing the putative miR-134 binding sites were
amplified and subcloned into the luciferase reporter vector pGL3
between the XbaI and FseI restriction sites. As a
control, the pGL3-mut-GOLPH3 plasmids were constructed using the
3′-UTR with the miR-134 binding sites mutated. Subsequently,
1×105 HEK-293T cells were plated on 24-well plates, and
transfected with 80 ng of pGL3-GOLPH3 or pGL3-mut-GOLPH3 vectors in
the presence of 40 nM pre-miR-134 or pre-miR negative controls
using Lipofectamine 2000 (Invitrogen). After transfection for 24 h,
cells were harvested and the luciferase activities were measured
using the Dual-Luciferase Reporter Assay kit (Promega).
Statistical analysis
Statistical analysis was conducted using the
Student's t-test. Multiple comparisons were analyzed by one-way
analysis of variance. Results were considered statistically
significant at P<0.05.
Results
miR-134 is downregulated in GC
Previous research has identified consistently
decreased miR-134 expression in many types of cancers (11–13).
To investigate miR-134 expression in GC, we collected 26 human GC
specimens and pair-matched gastric adjacent non-neoplastic mucosa.
Compared with adjacent non-tumor tissues, the expression of mature
miR-134 was significantly downregulated in the GC tissues (Fig. 1A). Similarly, we observed frequent
downregulation of miR-134 in different GC cell lines compared with
the immortalized gastric epithelial cell line GES-1 with no
tumorigenic effect (Fig. 1B).
Moreover, the expression level of miR-134 was positively correlated
with the differentiation degree of GC cells by virtue of the lowest
expression of miR-134 being observed in the most aggressive cancer
cell lines, AGS and SNU1, and the highest expression being found in
the GES-1 line. These data suggest a negative correlation between
miR-134 expression and GC.
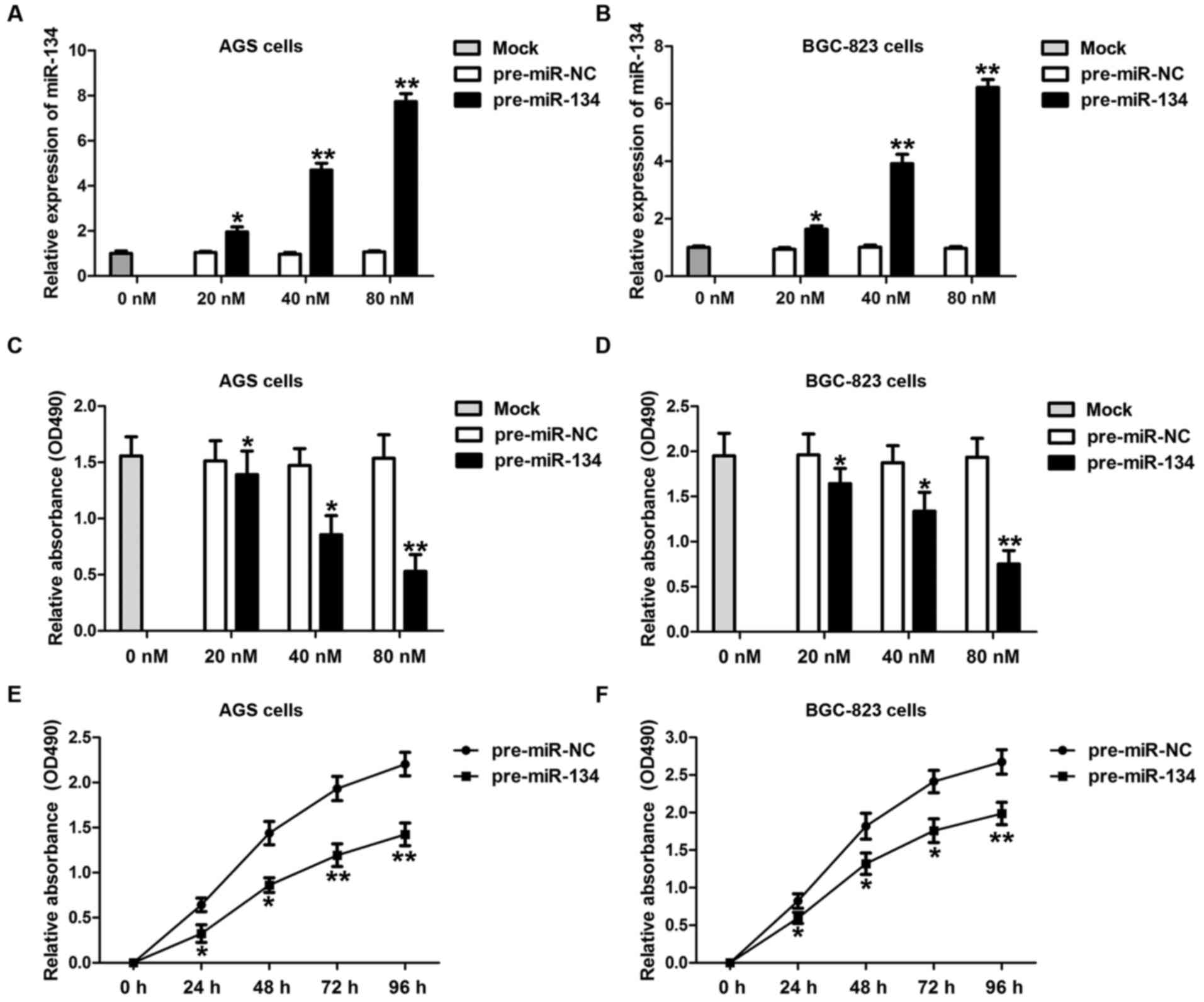
Overexpression of miR-134 suppresses
GC cell growth
To fully dissect the potential role of miR-134 in
gastric carcinogenesis, we examined the biological function of
miR-134 in AGS and BGC-823, two commonly used gastric
adenocarcinoma cell lines. GC cells were transfected with either
miR-134 precursor (pre-miR-134) or negative control precursor
(pre-miR-NC) at different concentrations (0, 20, 40 and 80 nM) and
incubated for 24, 48 and 72 h, respectively. The transfection
efficiencies of miR-134 were detected by RT-qPCR (Fig. 2A and B). As shown in Fig. 2C and D, cell proliferation was
significantly suppressed by miR-134 overexpression in both AGS and
BGC-823 cells in a dose-dependent manner. In addition, a longer
duration of miR-134 transfection markedly attenuated AGS (Fig. 2E) and BGC-823 cell (Fig. 2F) proliferation. These data suggest
that miR-134 may be important for gastric carcinogenesis.
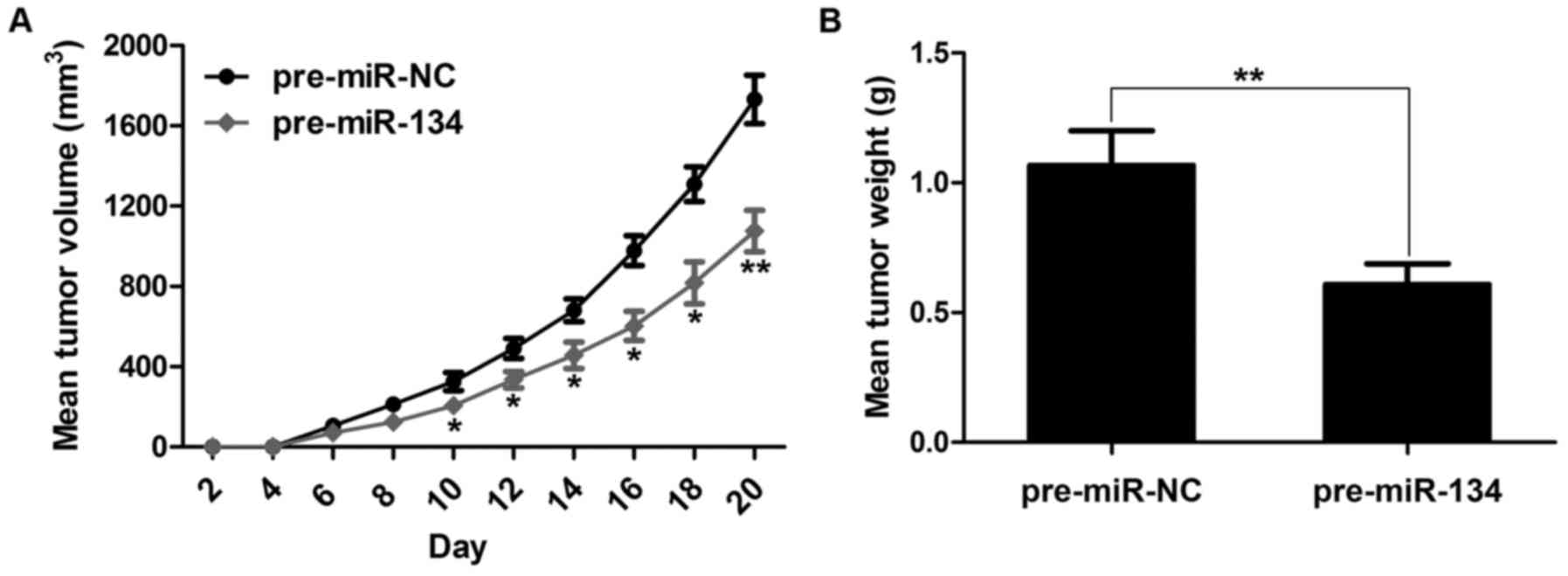
Overexpression of miR-134 suppresses
xenograft tumor growth in vivo
To verify the positive role of miR-134 in gastric
tumor progression in vivo, we engineered the miR-134
overexpression nude mouse model by subcutaneously inoculating AGS
cells with miR-134 into nude mice (AGS with miR-NC was used as a
control). Not surprisingly, overexpression of miR-134 revealed a
significant decrease in tumor volume when compared with the control
group (Fig. 3A). Additionally,
forced expression of miR-134 resulted in markedly lower tumor
weight than the control group (Fig.
3B). Altogether, these results demonstrated that miR-134 is
essential for controlling gastric tumor formation.
miR-134 targets the 3′-UTR of
GOLPH3
To delineate the underlying molecular mechanism by
which miR-134 inhibits GC, we performed bioinformatic analysis to
screen the predicted target genes of miR-134. Notably, we
discovered that in both TargetScan and PicTar databases, GOLPH3 was
a possible predicted target with conserved complementary ‘seed’
sites for miR-134 in the 3′-UTR in many species, including hsa,
rno, mmu and cfa (Fig. 4A).
Moreover, a recent study showed that the mRNA level of GOLPH3 is
upregulated in GC tissue and is positively associated with cell
proliferation (18). Given this
evidence and the observed function in miR-134-overexpressing cell
lines and mice, we speculated that GOLPH3 was a target of miR-134.
To test this hypothesis, we co-transfected pGL3-GOLPH3 or
pGL3-mut-GOLPH3 vectors in the presence of pre-miR-134 or
pre-miR-NC into HEK293T cells. The result of the luciferase
reporter assay showed that HEK293T cells transfected with the
3′-UTR of GOLPH3 and pre-miR-134 exhibited decreased luciferase
activity compared with the pre-miR-NC, while mutation at the 3′-UTR
of GOLPH3 lost this response (Fig.
4B).
Given that microRNAs regulate genes by triggering
either mRNA degradation or translational repression, we also tested
the expression of GOLPH3 mRNA and protein levels in AGS and the
BGC-823 cells. An ~63 and 74% reduction in GOLPH3 protein
expression was found in the miR-134-overexpressing AGS and BGC-823
cell lines, respectively (Fig. 4C).
However, overexpression of miR-134 had no effect on GOLPH3 mRNA
levels (Fig. 4D). Moreover,
compared with the adjacent non-tumor tissues, the expression of
GOLPH3 was markedly upregulated in GC tissues (Fig. 4E).
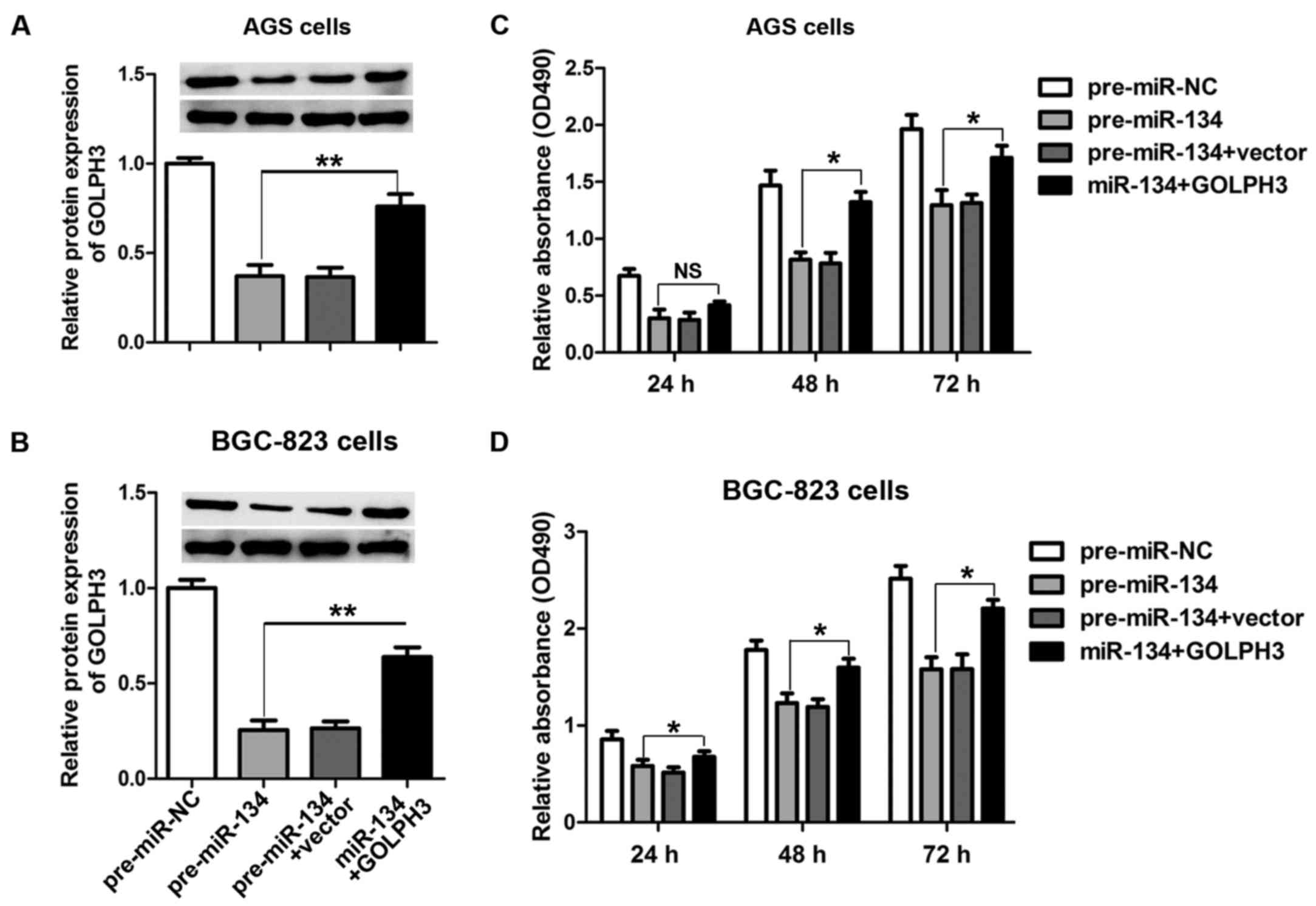
The cell growth-inhibiting activity of
miR-134 is abrogated by GOLPH3
To further validate the hypothesis that miR-134
regulates the growth of GC cells through GOLPH3, we co-transfected
pre-miR-134 and GOLPH3 overexpression vectors harboring no specific
miR-134-binding sequences in the 3′-UTR in AGS and BGC-823 cells.
Co-transfection of miR-134 and GOLPH3 induced an increased
expression of GOLPH3 when compared with pre-miR-134 transfection
alone in both AGS (Fig. 5A) and
BGC-823 cells (Fig. 5B).
Furthermore, cell proliferation was evaluated 24 h after
transfection using the MTT assay. As shown in Fig. 5C and D, transfection with
pre-miR-134 inhibited cell proliferation compared with the control,
while increasing GOLPH3 protein expression effectively reversed the
cell growth arrest induced by miR-134 overexpression in both AGS
and BGC-823 cells.
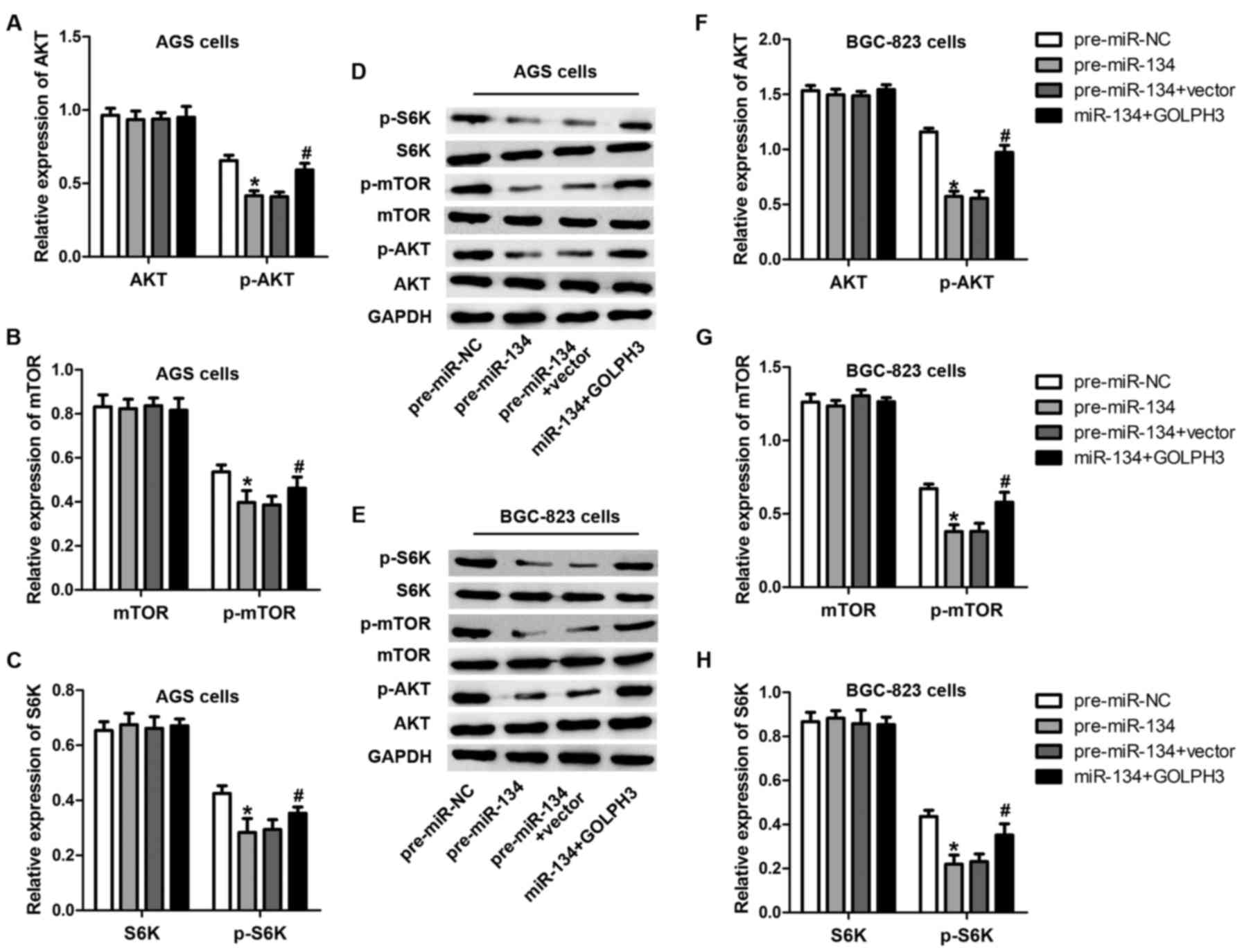
Expression of miR-134 inversely
correlates with the activation of mTOR
GOLPH3 is a Golgi-localizing protein playing an
important role in Golgi to plasma membrane trafficking (19). It has been previously reported that
GOLPH3 contributes to the activation of mTOR and p70s6k (16), which are involved in cell
proliferation and growth (20).
Considering the regulatory role of miR-134 in GOLPH3, we speculated
that miR-134 decreased mTOR and p70s6k phosphorylation and
activation through decreasing GOLPH3. To validate this hypothesis,
we examined Akt, mTOR and p70s6k phosphorylation levels after
miR-134 overexpression or miR-134-GOLPH3 overexpression using
western blot analysis. As predicted, the phosphorylation of Akt,
mTOR and p70s6k was apparently inhibited by the overexpression of
miR-134 (Fig. 6), while
overexpression of GOLPH3 restored this effect caused by miR-134.
These data imply that Akt, mTOR and p70s6k signaling were impaired
by miR-134 overexpression.
Discussion
miRNA deregulation has been correlated with the
progression and prognosis of GC since the discovery of aberrant
miRNA expression patterns in GC patients compared with normal
controls (21,22). miRNA microarrays showed that miR-1
and miR-101, whose downregulation was connected with various types
of cancers (9,23), were downregulated in gastric tissues
from GC patients (24). miR-375
targets the Janus kinase 2 (JAK2) gene in GC patient gastric
tissues (25), and may represent a
predictor of GC progression (26).
Although alterations in differentially regulated miRNAs have been
identified in GC, the present study provides insight into miR-134
and its target gene in GC, which further defines the mechanisms
underlying GC. Previous studies identified a close relationship
between miR-134 and the progression of various types of cancers. In
patients with colorectal cancer, osteosarcoma and breast cancer,
miR-134 was found to be decreased and to act as a tumor-suppressor
(11–13). In the present study, we provide
evidence that miR-134 is downregulated in GC tissues and cell
lines. Gene ontology analysis suggests that miR-134-regulated genes
are highly involved in cancer differentiation and proliferation
pathways. Potential direct targets of miR-134 that are relevant to
cell growth were identified by virtue of their upregulation in
gastric cell lines and in mice engineered to overexpress miR-134.
Indeed, we identified that overexpression of miR-134 inhibited
proliferation in GC cells and tumor formation in xenograft tumor
assays, indicating a tumor-suppressor role of miR-134 in GC.
To the best of our knowledge, miRNAs function by
regulating their target genes. Using two miRNA target prediction
algorithms (PicTar and TargetScan), we predicted >100 target
genes of miR-134. Among them, GOLPH3 was outstanding for the
following reasons. i) The 3′-UTR of GOLPH3 was predicted to be a
highly conserved binding site for miR-134 in several species: hsa,
rno, mmu and cfa. ii) GOLPH3 is an oncogene in many solid tumors
(16), and a previous clinical
study indicated that GOLPH3 is overexpressed in patients with GC
and is associated with poor clinical outcome in GC (17). iii) Apart from GOLPH3, other
proteins from the GOLPH family, such as GOLPH2, were identified to
be ectopically expressed in GCs (27). iv) Even though a full underlying
molecular mechanism was not observed, GOLPH3 was proven to play an
essential role in vesicle trafficking and Golgi structure (28). Moreover, Field et al found
that overexpression of GOLPH3 confers resistance to killing by
DNA-damaging agents and depletion of GOLPH3 could enhance the
ability of DNA-damaging agents to kill cells (29). Among potential direct targets of
miR-134, GOLPH3 was identified by the presence of a match to the
miR-134 seed sequence in the 3′-UTR of GOLPH3 by luciferase assay.
The result showed that HEK293T cells transfected with the 3′-UTR of
GOLPH3 and pre-miR-134 exhibited decreased luciferase activity
compared with the pre-miR-NC, while mutation at the 3′-UTR of
GOLPH3 lost this response.
In addition to the gene regulation discussed in the
present study, we identified the pathway involved in this process.
It has been previously reported that GOLPH3 functions in the mTOR
pathway based in cancer (16,30).
Similarly, Murayama et al reported that p-mTOR expression
was positively correlated with GC tumor invasion and tumor stage
(31). In addition, a preliminary
study revealed that GOLPH3 was positively correlated with Akt/mTOR
activation in GC (18). In line
with these studies, we showed in the present study that the
phosphorylation of AKT, mTOR and mTOR downstream substrate protein
S6K were impaired by miR-134 overexpression, while overexpression
of GOLPH3 restored this effect caused by miR-134, suggesting that
miR-134 regulated this AKT/mTOR/S6K signaling pathway via a
GOLPH3-dependent mechanism.
In conclusion, miR-134, which is downregulated in
gastric tumor tissues and cell lines, negatively regulates gastric
tumor growth and cell proliferation. Furthermore, miR-134 inhibits
GC cell proliferation by targeting GOLPH3 and subsequently, the
AKT/mTOR/S6K pathway. To the best of our knowledge, this is the
first time that miR-134 has been shown to target GOLPH3 in GC
cells. Therefore, further research exploring the anticancer role of
miR-134 may contribute to the development of new therapeutic
strategies for GC.
Acknowledgements
The present study was supported by the Scientific
Research Projects of Inner Mongolia Autonomous Region Colleges and
Universities in 2016 (grant no. NJZY16113).
References
|
1
|
Tan P and Yeoh KG: Genetics and molecular
pathogenesis of gastric adenocarcinoma. Gastroenterology.
149:1153–1162.e3. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality
Worldwide: IARC CancerBase No. 11 (Internet). International Agency
for Research on Cancer Lyon, France: 2013, http://globocan.iarc.fr
|
|
3
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song JH and Meltzer SJ: MicroRNAs in
pathogenesis, diagnosis, and treatment of gastroesophageal cancers.
Gastroenterology. 143:35–47.e2. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song S and Ajani JA: The role of microRNAs
in cancers of the upper gastrointestinal tract. Nat Rev
Gastroenterol Hepatol. 10:109–118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY and
Yan M: microRNA-21 promotes tumor proliferation and invasion in
gastric cancer by targeting PTEN. Oncol Rep. 27:1019–1026.
2012.PubMed/NCBI
|
|
8
|
Lai KW, Koh KX, Loh M, Tada K, Subramaniam
MM, Lim XY, Vaithilingam A, Salto-Tellez M, Iacopetta B, Ito Y, et
al: Singapore Gastric Cancer Consortium: MicroRNA-130b regulates
the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer.
46:1456–1463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Varambally S, Cao Q, Mani RS, Shankar S,
Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al:
Genomic loss of microRNA-101 leads to overexpression of histone
methyltransferase EZH2 in cancer. Science. 322:1695–1699. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsukamoto Y, Nakada C, Noguchi T, Tanigawa
M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M, et
al: MicroRNA-375 is downregulated in gastric carcinomas and
regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer
Res. 70:2339–2349. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie Y, Song J, Zong Q, Wang A, Yang Y, Liu
F and Meng X: Decreased expression of miR-134 and its clinical
significance in human colorectal cancer. Hepatogastroenterology.
62:615–619. 2015.PubMed/NCBI
|
|
12
|
Bao Y, Peng L, Ma J, Liu K and Li W:
Decreased miR-134 expression and its tumor-suppressive function in
human osteosarcoma. Genet Mol Res. 14:16771–16781. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Ma Y, Wang S, Chen F and Gu Y:
C/EBPα inhibits proliferation of breast cancer cells via a novel
pathway of miR-134/CREB. Int J Clin Exp Pathol. 8:14472–14478.
2015.PubMed/NCBI
|
|
14
|
Haller F, von Heydebreck A, Zhang JD,
Gunawan B, Langer C, Ramadori G, Wiemann S and Sahin O:
Localization- and mutation-dependent microRNA (miRNA) expression
signatures in gastrointestinal stromal tumours (GISTs), with a
cluster of co-expressed miRNAs located at 14q32.31. J Pathol.
220:71–86. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Snyder CM, Mardones GA, Ladinsky MS and
Howell KE: GMx33 associates with the trans-Golgi matrix in a
dynamic manner and sorts within tubules exiting the Golgi. Mol Biol
Cell. 17:511–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Scott KL, Kabbarah O, Liang MC, Ivanova E,
Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, et al:
GOLPH3 modulates mTOR signalling and rapamycin sensitivity in
cancer. Nature. 459:1085–1090. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu BS, Hu H, Zhu CY, Gu YL and Li JP:
Overexpression of GOLPH3 is associated with poor clinical outcome
in gastric cancer. Tumour Biol. 34:515–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peng J, Fang Y, Tao Y, Li K, Su T, Nong Y,
Xie F and Lai M: Mechanisms of GOLPH3 associated with the
progression of gastric cancer: A preliminary study. PLoS One.
9:e1073622014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dippold HC, Ng MM, Farber-Katz SE, Lee SK,
Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu
S, et al: GOLPH3 bridges phosphatidylinositol-4-phosphate and
actomyosin to stretch and shape the Golgi to promote budding. Cell.
139:337–351. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang Q and Guan KL: Expanding mTOR
signaling. Cell Res. 17:666–681. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu C, Ren C, Han J, Ding Y, Du J, Dai N,
Dai J, Ma H, Hu Z, Shen H, et al: A five-microRNA panel in plasma
was identified as potential biomarker for early detection of
gastric cancer. Br J Cancer. 110:2291–2299. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Han C, Zhou Y, An Q, Li F, Li D, Zhang X,
Yu Z, Zheng L, Duan Z and Kan Q: MicroRNA-1 (miR-1) inhibits
gastric cancer cell proliferation and migration by targeting MET.
Tumour Biol. 36:6715–6723. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Riquelme I, Tapia O, Leal P, Sandoval A,
Varga MG, Letelier P, Buchegger K, Bizama C, Espinoza JA, Peek RM,
et al: miR-101-2, miR-125b-2 and miR-451a act as potential tumor
suppressors in gastric cancer through regulation of the
PI3K/AKT/mTOR pathway. Cell Oncol. 39:23–33. 2016. View Article : Google Scholar
|
|
25
|
Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y,
Yao H, Liu X, Ke Y, Si J, et al: MiR-375 frequently downregulated
in gastric cancer inhibits cell proliferation by targeting JAK2.
Cell Res. 20:784–793. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang X, Yan Z, Zhang J, Gong L, Li W, Cui
J, Liu Y, Gao Z, Li J, Shen L, et al: Combination of hsa-miR-375
and hsa-miR-142-5p as a predictor for recurrence risk in gastric
cancer patients following surgical resection. Ann Oncol.
22:2257–2266. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen LG, Wang HJ, Yao HB, Guan TP, Wu F,
He XJ, Ma YY, Tao HQ and Ye ZY: GP73 is down-regulated in gastric
cancer and associated with tumor differentiation. World J Surg
Oncol. 11:1322013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sechi S, Frappaolo A, Belloni G, Colotti G
and Giansanti MG: The multiple cellular functions of the
oncoprotein Golgi phosphoprotein 3. Oncotarget. 6:3493–3506. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Farber-Katz SE, Dippold HC, Buschman MD,
Peterman MC, Xing M, Noakes CJ, Tat J, Ng MM, Rahajeng J, Cowan DM,
et al: DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3.
Cell. 156:413–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hohenberger P and Gretschel S: Gastric
cancer. Lancet. 362:305–315. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Murayama T, Inokuchi M, Takagi Y, Yamada
H, Kojima K, Kumagai J, Kawano T and Sugihara K: Relation between
outcomes and localisation of p-mTOR expression in gastric cancer.
Br J Cancer. 100:782–788. 2009. View Article : Google Scholar : PubMed/NCBI
|