Introduction
Breast cancer is a complex disease and the major
cause of cancer-related mortality among women. Approximately 70% of
breast cancers express the estrogen receptor (ER) and are thus
termed ER-positive (ER+) breast cancers, which represent
the primary luminal molecular subtype of breast cancer (1–3).
Generally, patients with ER+ breast cancer have a better
efficacy and prognosis than those with ER-negative (ER−)
breast cancer (4). ER can be
categorized into two structurally related genes, ERα and ERβ, in
which ERα is a major regulator of breast cancer development and
progression (5,6). Therefore, therapies targeting ER,
known as endocrine therapies, have become the mainstay of
prevention and treatment of all stages of ER+ breast
cancers (7,8). The common drugs used in endocrine
therapy in breast cancer include the following: i) Selective ER
modulators (SERM), such as tamoxifen, which directly inhibit ERs by
selecting estrogen modulators with mixed agonistic/antagonistic
activities; ii) selective ER downregulators (SERD), such as
fulvestrant, which inhibit ER signaling through the degradation of
ER expression; iii) aromatase inhibitors, such as letrozole, which
deprive the receptor's ligand by blocking estrogen production
(9,10).
Tamoxifen, the first-line endocrine drug used in the
treatment of ER+ breast cancer, has contributed to a
marked increase in the long-term survival rate (11,12).
Nevertheless, a considerable proportion of patients with localized
breast cancers and metastatic breast cancers become resistant to
endocrine therapies (13). In view
of this, high-dose tamoxifen (>100 mg daily) is used in place of
standard-dose tamoxifen (20 mg daily) for the treatment of the
above-mentioned breast cancers (14). However, this treatment is associated
with severe side-effects, including hyperplasia, venous
thromboembolic disease (15) and
acquired tamoxifen resistance (16,17).
To overcome this limitation, novel strategies to
reduce the dose of tamoxifen, while still maintaining its
anticancer functions are currently under investigation (18). Some clinically used drugs, which
were not developed for the treatment of cancer previously, may have
some antitumor effects which may enhance the sensitivity of
ER+ tumors to tamoxifen. For example, the anti-diabetic
drug, metformin, has been shown to enhance the tamoxifen-mediated
tumor growth inhibition of ER+ breast cancer (19). The selective cyclooxygenase (COX)-2
inhibitor, celecoxib, has also been shown to alleviate the
tamoxifen-induced angiogenic effects in metastatic ER+
breast cancer (20). Recently, it
has been reported that the activation of mammalian target of
rapamycin (mTOR) signaling leads to multiple agent therapeutic
resistance in ER+ breast cancer (21). Rapamycin, an mTOR inhibitor, which
is a macrolide immunosuppressant and was originally used for the
prevention of organ transplant rejection and approved by the US
Food and Drug Administration (FDA) in September, 1999 for its
safety, has been found to synergize with cisplatin in the treatment
for basal-like breast cancer cell (22). Moreover, a phase II neoadjuvant
endocrine therapy clinical trial demonstrated the synergistic
effects of the combination of the mTORC1 inhibitor, everolimus,
with letrozole in the treatment of breast cancer (23). However, to the best of our
knowledge, there were few studies to date which have investigated
whether rapamycin has the potential to enhance the sensitivity of
ER+ breast cancers to tamoxifen.
In the present study, we found that rapamycin indeed
enhanced the sensitivity of ER+ breast cancer cells to
tamoxifen both in vitro and in vivo. Moreover, we
found that this synergistic effect may be mediated partly through
the upregulation of ER expression following the induction of p73.
Taken together, combination therapy with rapamycin and tamoxifen
may provide new insight and may aid in the development of novel
therapeutic strategies for the treatment of ER+ breast
cancer.
Materials and methods
Cell culture and treatment
The human breast cancer cell lines, MCF-7 and
ZR-75-1, were obtained from the American Type Culture Collection
(ATCC; Manassas, VA, USA). The cells were cultured in complete
medium consisting of high glucose Dulbecco's modified Eagle's
medium (DMEM; Wisent Biotechnology, Nanjing, China) supplemented
with 10% fetal bovine serum (FBS), 100 µg/ml
penicillin-streptomycin (HyClone, Logan, UT, USA) at 37°C in a
humidified atmosphere containing 5% CO2.
For drug treatment, the MCF-7 and ZR-75-1 cells were
first treated with tamoxifen (0–25 µM; Sigma-Aldrich, Dorset, UK)
or rapamycin (0–6 µM; Sigma-Aldrich) individually for 48 h.
Following rapamycin treatment, we found the effective concentration
of rapamycin started began from 40 nM with an ~20% inhibitory rate.
We selected 40 nM rapamycin to further investigate the effects of
the combination of the two drugs on breast cancer. The changes in
p73 and ERα expression following rapamycin treatment are shown in
Fig. 1A and B.
 | Figure 1.Rapamycin functions synergistically
with tamoxifen in the MCF-7 and ZR-75-1 cells. (A and B) Changes in
p73 and ERα expression following rapamycin treatment (C and D)
MCF-7 and ZR-75-1 cells were treated with tamoxifen at various
concentrations (0, 1, 2, 4, 6, 8, 16, 20 and 24 µM), or a
combination of tamoxifen and rapamycin (40 nM). After 48 h, cell
viability was measured by CCK-8 assays. (E and F) MCF-7 and ZR-75-1
cells were treated with 9 µM tamoxifen, 40 nM rapamycin, or a
combination of the two agents for 48 h, and cell viability was
measured. Histograms represent the quantification of cell
viability. These data were calculated from 3 separate experiments
and presented as the means ± SEM, *P<0.05 for the TAM (9 µM)
group vs. the TAM (9 µM) + Rapa (40 nM) group. TAM, tamoxifen;
Rapa, rapamycin. |
Cell viability assay
Cell viability was measured using CCK-8 kits
(Dojindo, Kumamoto, Japan) following the manufacturer's
instruction. Briefly, the MCF7 and ZR-75-1 cells grown in
monolayers were harvested and dispensed in 96-well culture plates
in 200 µl of DMEM at a concentration of 5×103 cells per
well. After 12 h, the differential drug concentrations of tamoxifen
(0–24 µM), rapamycin 40 nM, or both 0–24 µM tamoxifen plus 40 nM
rapamycin were added to the cells. After 48 h, the medium in each
well was replaced with 100 µl DMEM containing 10% CCK-8 to measure
the growth rate of cells. The plates were incubated at 37°C for 2.5
h and the optical density (OD) values at 450 nm were measured using
a microplate reader (Tecan Austria GmbH, Grödig, Austria). Each
test was performed in triplicate.
Apoptosis assays
Apoptosis measurements were conducted using the
Annexin V apoptosis detection kit (BD Biosciences, Heidelberg,
Germany) according to the manufacturer's instructions. Early and
late apoptotic, as well as viable cell populations were identified
by plotting phycoerythrin (PE), Annexin V vs. 7-AAD
(7-amino-actinamycin D). For each measurement, 3 independent
samples were pooled.
Drug combination analysis
The combination analysis was conducted using the
method previously described by Chou and Talalay (24). Cell viability was measured by CCK-8
assay. Drug dose-effect calculations and the combination indices
(CI) for 50% growth inhibition were obtained using GraphPad Prism
7.0 software (GraphPad, La Jolla, CA, USA), and the Student's t
test was applied to verify whether the CI values at 50% growth
inhibition were significantly different from CI = 1. As regards the
CI combination indices, CI <1 indicates synergism, CI = 1
indicates addivity, and CI >1 indicates antagonism.
Animal model in vivo
Nude mice were used in this study, 30 female nude
mice, aged 4 weeks and weighing 12–15 g, were purchased from the
Animal Core Facility of Nanjing Medical University (Nanjing,
China). The study was approved by the Institutional Animal Care and
Use Committee for Animal Use (the Animal Ethics Committee of
Nanjing Medical University). The mice were kept under the following
conditions: Relative humidity, 40–70%; room temperature, 20–26°C;
food and water, 5 g food and 100 ml water per 100 g body weight per
day. Estrogen (E2; 0.9 mg/kg) was injected into the abdomen of the
4-week-old female nude mice every 3 days. Subsequently,
5×106 ZR-75-1 cells were injected into the abdominal
mammary fat pad of the mice. When the tumor volume reached ~200
mm3, the mice were randomly divided into 4 groups as
follows: i) The control group, in which the mice received
phosphate-bufferred saline (PBS); ii) the rapamycin group, in which
the mice received rapamycin (0.25 mg/kg body weight, p.o.); iii)
the tamoxifen group, in which mice received tamoxifen (60 mg/kg
body weight, p.o.); and iv) the combination group, in which mice
received a combination of the two drugs in their drinking water.
Tumor growth was measured using a caliper each week. After 4 weeks,
the mice were sacrificed and the tumors removed. The excised tumor
portions were fixed in 4% paraformaldehyde for further
analysis.
Immunohistochemical (IHC) staining and
analysis
Breast tissue samples (n=82) were obtained from the
First Affiliated Hospital of Nanjing Medical University, China,
between 2004 and 2007. The collection and use of the samples was
reviewed and approved by the Institutional Ethics Committee of the
First Affiliated Hospital of Nanjing Medical University and
informed consent was obtained from all patients prior to sample
collection. The TNM staging was defined according to the American
Joint Committee on Cancer (AJCC) (6th version, 2002). IHC staining
of the same tissue samples with p73 (diluted 1:50; cat. no.
PA5-35368; Thermo Fisher Scientific, Waltham, MA, USA) and ERα
antibodies (diluted 1:1,00; cat. no. 13258; Cell Signaling
Technology, Danvers, MA, USA) was conducted and analyzed as
previously described (25).
Plasmids and siRNA transfection
Plasmids and siRNA constructs of p73 siRNA targeting
p73 (siRNA1, siRNA2) and a negative control (CTRi), plasmid
targeting p73 and the control (Vector) were obtained from
GenePharma (Shanghai, China). Briefly, the MCF-7 and ZR-75-1 cells
were transfected with the plasmid or siRNAs using Lipofectamine
3000 (Invitrogen/Thermo Fisher Scientific) according to the
manufacturer's instructions. The cells were cultured in a 6-well
plate for 24 to 48 h and the expression level was detected by
western blot analysis and reverse transcription-quantitative PCR
(RT-qPCR) to determine the transfection efficiency.
Western blot analysis
Western blot analysis was carried out as previously
described (26). The
radioimmunoprecipitation assay (RIPA) kit (Beyotime Institute of
Biotechnology, Shanghai, China) was used to extract protein from
the breast cancer cells according to the manufacturer's
instructions. The icinchoninic Acid Protein Assay kit (BCA) was
used to determine the protein concentration. A total of 20 µg of
proteins with different molecular weights were separated on 10%
SDS-PAGE gels, and transferred onto polyvinylidene fluoride (PVDF)
membranes (EMD Millipore, Bedford, MA, USA) in transfer buffer. The
membranes were then blocked in 5% non-fat milk at room temperature
for ~2 h and incubated in the specific primary antibodies at 4°C
overnight. After washing in TBST, the membranes were incubated with
secondary antibodies at room temperature for ~2 h. ECL Plus (EMD
Millipore) was used to detect the protein bands with the
Bio-Imaging System. The following detection antibodies were used:
p73 (diluted 1:1,000; cat. no. 14620), ERα (diluted 1:1,000; cat.
no. 13258), anti-rabbit secondary antibodies (diluted 1:1,000; cat.
no. 7074), anti-mouse secondary antibodies (diluted 1:1,000; cat.
no. 7076) (all from Cell Signaling Technology) and β-actin (diluted
1:1,000; cat. no. AA128; Beyotime).
RNA extraction and RT-qPCR
Total RNA was extracted using TRIzol reagent
(Takara, Kusatsu, Japan). Reverse transcription and qPCR were
performed as previously described (27). The following PCR primers were used
to amplify the relevant genes: β-actin forward,
5′-GCTGTGCTATCCCTGTACGC-3′ and reverse, 5′-TGCCTCAGCGCAGCGGAACC-3′;
p73 forward, 5′-CGGGCCATGCCTGTTTACA-3′ and reverse,
5′-TGTCCTTCGTTGAAGTCCCTC-3′; ERα forward,
5′-CCCACTCAACAGCGTGTCTC-3′ and reverse,
5′-CGTCGATTATCTGAATTTGGCCT-3′. the method of quantification was
2−ΔΔCt (28).
Dual-luciferase reporter assay
Dual-luciferase reporter assays were conducted in
triplicate using respective kits (Promega, Madison, WI, USA)
according to the manufacturer's instructions. Briefly, 200 ng of a
pGL3 reporter containing target regions, an internal control, and 5
ng of Renilla luciferase vector (pRL-TK; Promega) were
co-transfected into the breast cancer cells. After 48 h, the cells
were harvested to measure the luciferase activity. All the
experiments were conducted at least 3 times.
Chromatin immunoprecipitation
(ChIP)
ChIP assays were performed using chromatin
immunoprecipitation kits (17–371, EZ-ChIP; EMD Millipore,
Billerica, MA, USA) according to the manufacturer's instructions as
described previously (29). The
primary antibody used was anti-rabbit p73. An aliquot (2 µl) of
each sample was analyzed by PCR using specific primers listed as
follows: Sense, 5′-GCACTTAGAAATGGTCCTGGTAA-3′ and antisense,
5′-CCTGCTCAATGACAATCACACT-3′.
Statistical analysis
Each experiment in this study was repeated in
triplicate, unless otherwise specified. The data were analyzed
using SPSS software (version, 22.0). The association between p73
and the patient clinicopathological parameters was analyzed using
χ2 tests. The correlation between the expression levels
of p73 and ERα in the breast cancer specimens was analyzed using a
2-tailed Spearman's correction analysis. The other data are
presented as the means ± standard error of the mean (SEM) and
differences between groups were analyzed using a Student's t-test
or ANOVA (with Dunnett's post hoc test). A value of P<0.05 was
considered to indicate a statistically significant difference.
Results
Rapamycin sensitizes the ZR-75-1 and
MCF-7 cells to tamoxifen in vitro
To examine the effects of the combination of
tamoxifen and rapamycin on cell viability, the MCF-7 and ZR-75-1
cells were treated with tamoxifen (0–24 µM) and/or rapamycin (40
nM) and examined by CCK-8 assay (Fig.
1C and D). As shown in Fig. 1E and
F, the inhibitory rates observed with the combination treatment
with rapamycin plus tamoxifen reached 83% compared to those
observed with treatment with tamoxifen alone (50%) in the ZR-75-1
cells. Likewise, treatment with rapamycin in combination with
tamoxifen suppressed cell growth by 79% compared to treatment with
tamoxifen alone (55%) in the MCF-7 cells. When the growth
inhibition rates reached 50%, the combination indices (CI) achieved
were 0.699 and 0.745 in ZR-75-1 and MCF-7 cells, respectively,
suggesting that rapamycin functions synergistically with tamoxifen
to inhibit the growth of ER+ breast cancer cells
(Table I).
 | Table I.Multiple drug dose-effect
calculations and the combination index generated using GraphPad
Prism software. |
Table I.
Multiple drug dose-effect
calculations and the combination index generated using GraphPad
Prism software.
| Cell line | RAPA (µM) | TAM (µM) | Growth inhibition
(%) | CI | Effect | P-value |
|---|
| ZR75-1 | 3 | 10.2 | 50 | 0.699 | Synergy | <0.05 |
| MCF-7 | 2.5 | 7.4 | 50 | 0.745 | Synergy | <0.05 |
Rapamycin enhances the
tamoxifen-induced apoptosis of ZR-75-1 and MCF-7 cells
Treatment with rapamycin plus tamoxifen led to a 40%
induction of cell apoptosis compared to treatment with tamoxifen
alone (19%) in the ZR-75-1 cells (Fig.
2A and C). The apoptotic rate observed with treatment with
tamoxifen in combination with rapamycin was 45% compared to
treatment with tamoxifen alone (22%) in the MCF-7 cells (Fig. 2B and D). These results demonstrate
that rapamycin is capable of functioning synergistically with
tamoxifen to enhance the tamoxifen-induced apoptosis of the ZR-75-1
and MCF-7 cells.
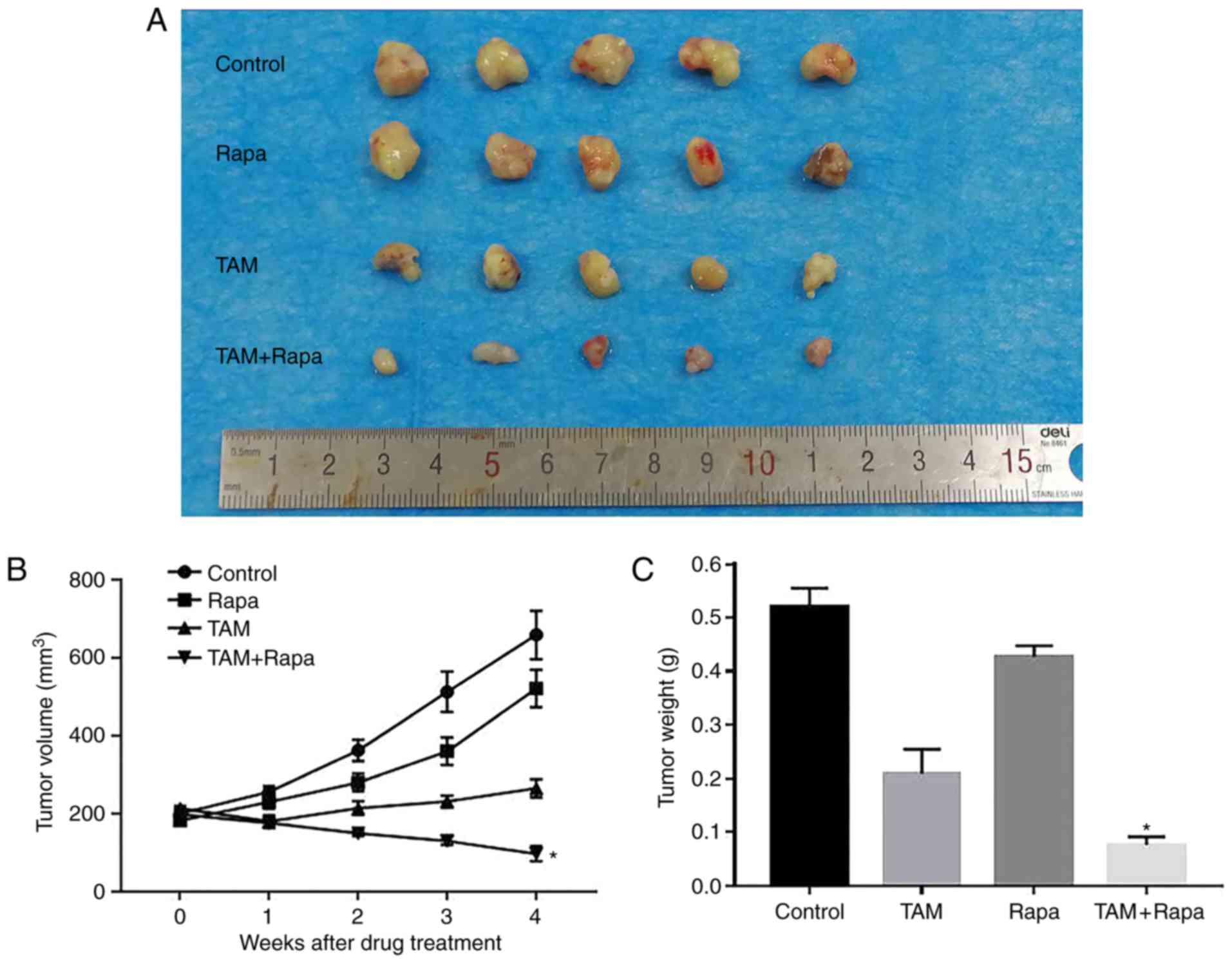
Effects of rapamycin and tamoxifen on
the growth of ER+ breast cancer in vivo
As shown in Fig. 3A and
B, compared to the control group, the combination treatment
group (rapamycin + tamoxifen) exhibited a significant inhibition of
tumor growth by 79.1% at 4 weeks. However, the groups which
received rapamycin or tamoxifen alone exhibited a suppression of
tumor growth of only 29.9 and 58.8%, respectively, compared with
the control group. Furthermore, the tumor weight of the combination
treatment group was the lightest of the 4 groups (Fig. 3C). These results indicated that the
combination of rapamycin or tamoxifen significantly and
synergistically inhibited tumor growth in vivo.
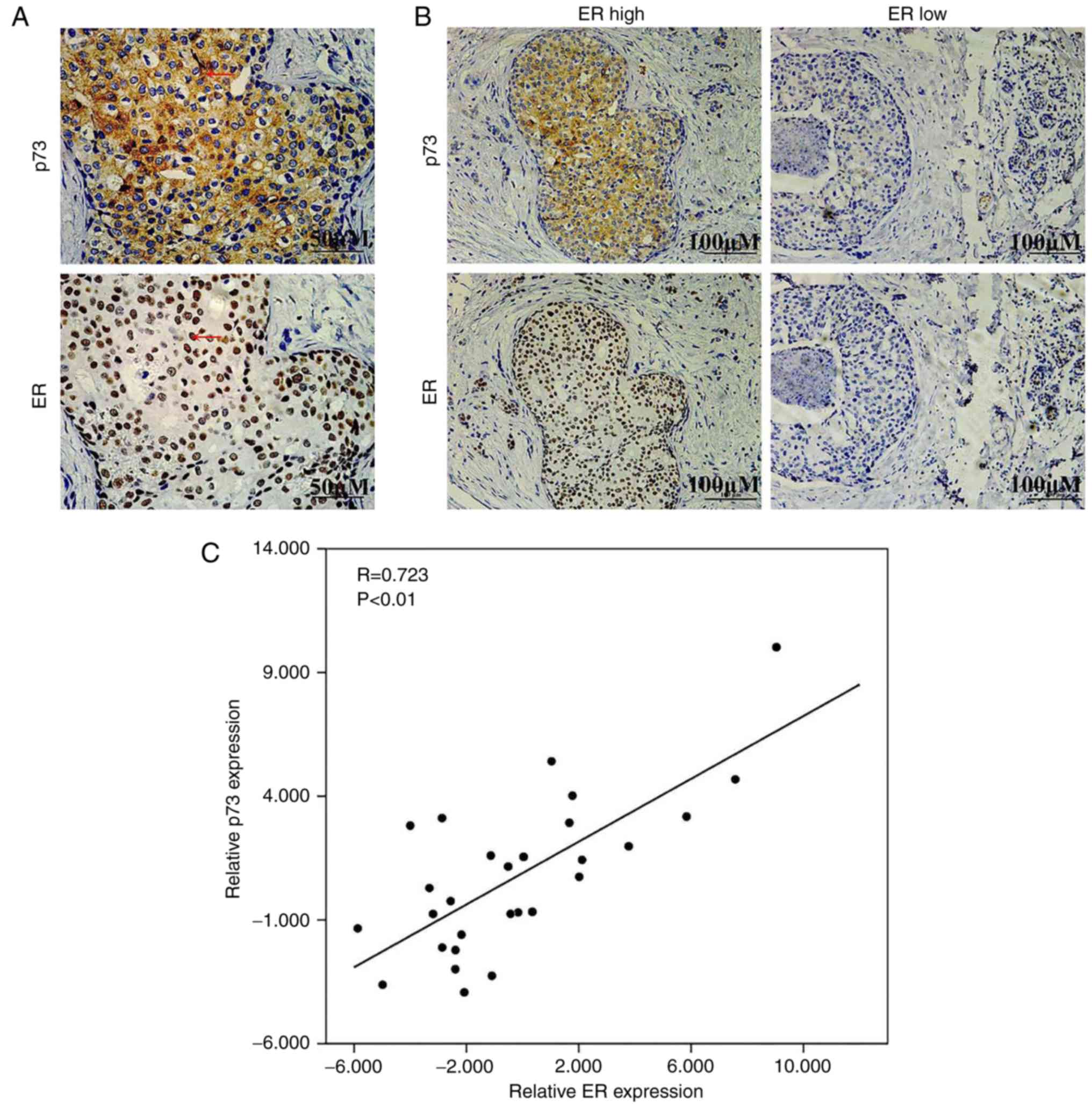
IHC staining of p73 and ERα in human
breast cancer tissues
As rapamycin is an inducer of p73, we hypothesized
that this synergistic effect may be mediated partly through the
upregulation of ER expression following the induction of p73. To
examine the association between p73 and ERα, IHC staining was
performed in 82 breast cancer tissues. As shown in Fig. 4A, p73 was mainly expressed in the
cytoplasm and ERα was mainly expressed in the nucleus.
Representative images of p73 expression in breast cancer tissues
expressing high and low levels of ERα are presented in Fig. 4B. Table
II shows the analysis of the association of p73 expression and
the clinicopathological characteristics of the breast cancer
patients. Additionally, we also found that there was a positive
correlation between the expression levels of p73 and ERα in the
breast cancer specimens (two-tailed Spearman's correlation
analysis, r=0.723, P<0.05) (Fig.
4C). On the whole, these data suggested that ERα expression
positively correlated with p73 expression in breast cancer
tissues.
 | Table II.Association of p73 with ERα and
clinicopathological characteristics of breast cancer patients. |
Table II.
Association of p73 with ERα and
clinicopathological characteristics of breast cancer patients.
|
| p73 expression |
|---|
|
|
|
|---|
| Clinicopathological
characteristics | No. of cases | Low (%) | High (%) | P-value |
|---|
| Age, years |
|
|
| 0.247 |
|
<50 | 35 | 26 (74.3) | 9 (25.7) |
|
|
≥50 | 47 | 29 (61.7) | 18 (37.9) |
|
| Pathological
grade |
|
|
| 0.482 |
|
I–II | 43 | 27 (62.8) | 16 (37.2) |
|
|
III | 39 | 28 (71.8) | 11 (28.2) |
|
| TNM stage |
|
|
| 0.983 |
|
I–II | 76 | 51 (67.1) | 25 (32.9) |
|
|
III | 6 | 4 (66.7) | 2 (33.3) |
|
| Tumor size
(cm) |
|
|
| 0.459 |
| ≤2 | 28 | 17 (60.7) | 11 (39.3) |
|
|
>2 | 54 | 38 (70.4) | 16 (29.6) |
|
| Lymph node
metastasis |
|
|
| 0.920 |
| N0 | 51 | 34 (66.7) | 17 (33.3) |
|
|
N1-N3 | 31 | 21 (67.7) | 10 (32.3) |
|
| ER |
|
|
| 0.019 |
|
Negative | 46 | 36 (78.3) | 10 (21.7) |
|
|
Positive | 36 | 19 (52.8) | 17 (47.2) |
|
| PR |
|
|
| 0.945 |
|
Negative | 68 | 46 (67.6) | 22 (32.4) |
|
|
Positive | 14 | 9 (64.3) | 5 (35.7) |
|
| Her2 |
|
|
| 0.232 |
|
Negative | 33 | 25 (75.8) | 8 (24.2) |
|
|
Positive | 49 | 30 (61.2) | 19 (38.8) |
|
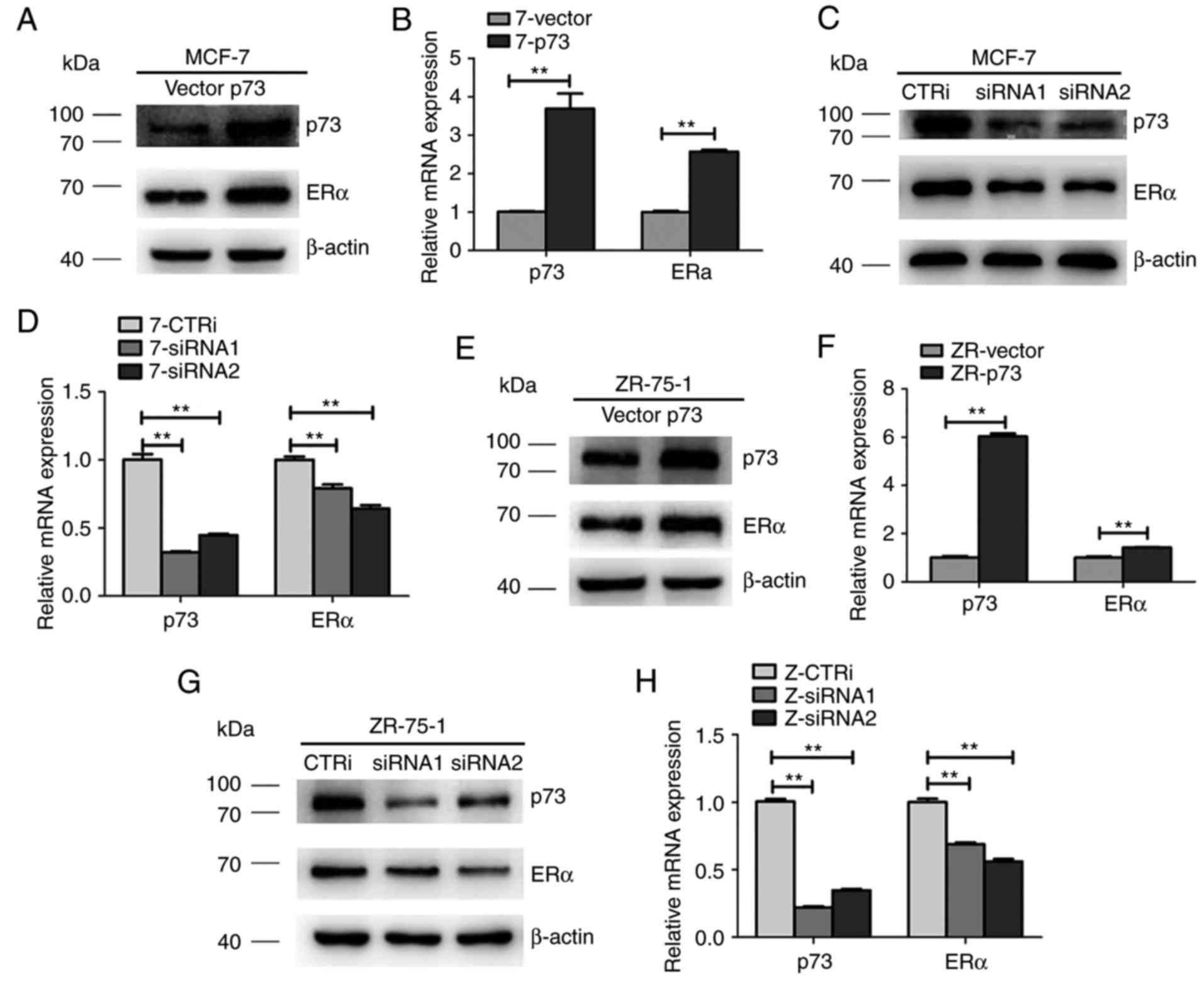
p73 regulates ERα expression in MCF-7
and ZR-75-1 cells
To examine whether p73 regulates ERα expression in
breast cancer, the MCF-7 and ZR-75-1 cells were transiently
transfected with siRRNA and scramble siRNA. As shown in Fig. 5C and D, the expression of ERα in the
MCF-7 cells was effectively downregulated by siRNA against p73
compared with the cells transfected with the scramble siRNA (CTRi)
at both the protein and mRNA level. Moreover, a p73 overexpression
plasmid was transfected into the MCF-7 cell lines and the effects
of p73 on ERα expression were investigated by RT-qPCR and western
blot analysis. As shown in Fig. 5A and
B, the expression of ERα was upregulated in the MCF-7 cells
transfected with the p73 overexpression plasmid compared with that
of the empty vector-transfected cells. Similar results were also
observed in the ZR-75-1 cells (Fig.
5E-H). These data suggest that p73 positively regulates ERα
expression in ER+ breast cancer cells.
p73 directly binds to the ERα promoter
region
To further investigate whether p73 regulates ERα
transcription, we hypothesized that p73 regulates the expression of
ERα by directly binding to ERα DNA. A shown by the schematic
diagram in Fig. 6A, the luciferase
reporter constructs contain the E-box in the promoter region of the
ERα gene. Thus, the MCF-7 and ZR-75-1 cells were transfected with a
luciferase reporter containing promoter of the ERα gene in order to
determine whether p53 directly controls the transcription of the ER
gene. The results revealed a 1.6- and 1.3-fold increase in
luciferase activity in the MCF-7 and ZR-75-1 cells compared to the
control vector-transfected cells, respectively (Fig. 6B and C). Furthermore, ChIP assays
revealed that p73 binds to this E-box in the promoter region of ERα
in the MCF-7 cells (Fig. 6D).
GAPDH, which represented a negative control, was not bound by p73.
These data indicate that p73 directly regulates ERα expression by
binding to the E-box elements in the promoter region of the ERα
gene.
Discussion
In the present study, we found that the mTOR
inhibitor, rapamycin, which has been approved by the FDA for the
prevention of organ transplant rejection, enhances the sensitivity
of ER+ breast cancer cells to tamoxifen partly through
the upregulation of ER expression following the induction of
p73.
In patients with metastatic breast cancers,
tamoxifen treatment would leads to disease regression in ~30% of
cases (30). Patients who receive
tamoxifen treatment for 0 to 4 years obtain maximal benefits, with
a reduced recurrence rate 51% and a reduced death rate by 28%. The
reduction in recurrence and mortality is sustained in year 5 and
beyond (30,31). In a recent study, the worldwide
Adjuvant Tamoxifen: Longer Against Shorter (ATLAS) trial, indicated
that 10 years of tamoxifen treatment reduced breast cancer
recurrence and mortality more effectively than treatment for 5
years (32). This fully affirmed
the efficacy of tamoxifen in the treatment for ER+
breast cancer, while its severe side-effects following long-term
and high-dose treatment must still be seriously considered. In this
study, we observed the synergistic effects of tamoxifen and
rapamycin used in combination in ER+ breast cancer cell
lines. In addition, this synergistic effect of rapamycin and
tamoxifen was confirmed using a nude mouse model in vivo.
The dose of rapamycin (40 nM) used in combination with tamoxifen
was lower than that of metformin (5 mM) and celecoxib (30 µM) used
in previous studies (19,20). This indicates that rapamycin may be
a more desirable drug for the prevention of breast cancers and the
growth inhibition of existing tumors in women. To date, apart from
breast cancer, the combination of rapamycin and CC-5013 (Revlimid)
has been shown to improve patient outcome in multiple myeloma
(33). Of note, in this study, the
concentration of tamoxifen required for growth inhibition was
substantially reduced when rapamycin was combined with
tamoxifen.
The mTOR pathway plays a crucial role in multiple
cellular processes and is the most frequently activated signaling
pathway, which promotes tumor growth and progression (34). Gene alterations in the mTOR pathway
are frequently observed in ER+ breast cancer. These
include insulin-like growth factor 1 receptor (IGF-1R),
phosphatidylinositide 3-kinase (PI3K) and human epidermal growth
factor receptor 2 (HER2) gene amplifications (35–37) or
phosphatase and tensin homolog (PTEN) gene function loss (38). Correspondingly, the loss of PTEN
expression is associated with low ERα levels and high PI3K
activity, which may result in a poor response to tamoxifen
treatment (39,40). Hence, the hyperactivation of the
mTOR pathway may lead to the downregulation of ERα expression and
may promote hormone-independent cell growth. mTOR inhibitors (e.g.,
rapamycin) may reverse this process by increasing ERα levels,
thereby restoring hormone dependence and sensitivity to endocrine
therapy (41).
A crosstalk exists between the mTOR pathway, and ERα
and p53-family members. p53 has been reported to regulate ERα
expression through transcriptional control by binding to the ERα
promoter (42). p73 is structurally
and functionally related to p53; however, to date, at least to the
best of our knowledge, there are no studies available on the
interaction between p73 and ERα. Additionally, rapamycin can
selectively increase p73 occupancy at its binding sites and
modulate its activity and function (43). As both ER and p73 are involved in
the mTOR pathway, we hypothesized that the activation of p73 may
regulate ERα expression. In this stuyd, IHC and tissue microarray
analysis were applied to confirm the association between p73 and
ERα. Moreover, the upregulation of p73 resulted in an increased ERα
mRNA and protein expression, whereas the knockdown of p73 decreased
the levels of ERα protein and transcript in the ER+
breast cancer cells. These data suggest that the expression of p73
and ERα is linked in ER+ breast cancer, which would be
expected to account for the synergistic effects of rapamycin plus
tamoxifen.
The combination of tamoxifen and rapamycin, though,
has not been previously investigated in clinical trials, at least
to the best of our knowledge. However, was previously reported that
the mTORC1 inhibitor, everolimus, plus tamoxifen increased the
6-month clinical benefit rate by 61% compared to 42% with tamoxifen
alone and reduced the risk of death by 55% in women with metastatic
breast cancer. Moreover, the progression time appeared to be more
prolonged with the combination vs. tamoxifen alone (8.6 months vs.
4.5 months, hazard ratio 0.54) (44).
In conclusion, in the present study, we revealed a
novel mechanism in that p73 transcriptionally regulates ERα
expression by directly binding in its promoter region. In addition,
combination therapy with an mTOR inhibitor and tamoxifen, leading
to the activation of p73 may provide new insight and may aid in the
development of novel strategies for the treatment of patients with
ER+ breast cancers.
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science
Foundation of China (grant nos. 81572595, 81602336 and 81202077)
and the Key Medical Subjects of Jiangsu Province (grant no. H201110
to QD).
Availability of data and materials
The datasets analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
QD, JW, JFW and LZ contributed to the design of this
study. XXL, LS and JW contributed to the experimental work. JYQ,
TSX and WBZ contributed to the data collection and analysis. XS,
XJZ contributed to the interpretation of the data and the drafting
of the manuscript. All authors read and approved the manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
The collection and use of the samples was reviewed
and approved by the Institutional Ethics Committee of the First
Affiliated Hospital of Nanjing Medical University and informed
consent was obtained from all patients prior to sample collection.
The use of animals in this study was approved by the Institutional
Animal Care and Use Committee for Animal Use (the Animal Ethics
Committee of Nanjing Medical University).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumors. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Creighton CJ: The molecular profile of
luminal B breast cancer. Biologics. 6:289–297. 2012.PubMed/NCBI
|
|
4
|
Katzenellenbogen BS and Katzenellenbogen
JA: Estrogen receptor transcription and transactivation: Estrogen
receptor alpha and estrogen receptor beta: Regulation by selective
estrogen receptor modulators and importance in breast cancer.
Breast Cancer Res. 2:335–344. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burns KA and Korach KS: Estrogen receptors
and human disease: An update. Arch Toxicol. 86:1491–1504. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Higa GM and Fell RG: Sex hormone receptor
repertoire in breast cancer. Int J Breast Cancer. 2013:2840362013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yager JD and Davidson NE: Estrogen
carcinogenesis in breast cancer. N Engl J Med. 354:270–282. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shanle EK and Xu W: Selectively targeting
estrogen receptors for cancer treatment. Adv Drug Deliver Rev.
62:1265–1276. 2010. View Article : Google Scholar
|
|
9
|
Britton DJ, Hutcheson IR, Knowlden JM,
Barrow D, Giles M, McClelland RA, Gee JMW and Nicholson RI:
Bidirectional cross talk between ERalpha and EGFR signalling
pathways regulates tamoxifen-resistant growth. Breast Cancer Res
Treat. 96:131–146. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jeselsohn R, Buchwalter G, De Angelis C,
Brown M and Schiff R: ESR1 mutations-a mechanism for acquired
endocrine resistance in breast cancer. Nat Rev Clin Oncol.
12:573–583. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morandi A, Martin L, Gao Q, Pancholi S,
Mackay A, Robertson D, Zvelebil M, Dowsett M, Plaza-Menacho I and
Isacke CM: GDNF-RET signaling in ER-positive breast cancers is a
key determinant of response and resistance to aromatase inhibitors.
Cancer Res. 73:3783–3795. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang P, Bahreini A, Gyanchandani R, Lucas
PC, Hartmaier RJ, Watters RJ, Jonnalagadda AR, Trejo Bittar HE,
Berg A, Hamilton RL, et al: Sensitive detection of mono- and
polyclonal ESR1 mutations in primary tumors, metastatic lesions and
cell free DNA of breast cancer patients. Clin Cancer Res.
22:1130–1137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ali S and Coombes RC: Endocrine-responsive
breast cancer and strategies for combating resistance. Nat Rev
Cancer. 2:101–112. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stathopoulos GP and Trafalis D: High-dose
tamoxifen in breast cancer bone metastasis. J BUON. 18:532–534.
2013.PubMed/NCBI
|
|
15
|
Hendrick A and Subramanian VP: Tamoxifen
and thromboembolism. JAMA. 243:514–515. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Musgrove EA and Sutherland L: Biological
determinants of endocrine resistance in breast cancer. Nat Rev
Cancer. 9:631–643. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lorizio W, Wu AH, Beattie MS, Rugo H, Tchu
S, Kerlikowske K and Ziv E: Clinical and biomarker predictors of
side effects from tamoxifen. Breast Cancer Res Treat.
132:1107–1118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Castrellon AB: Novel strategies to improve
the endocrine therapy of breast cancer. Oncol Rev. 11:3232017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma J, Guo Y, Chen S, Zhong C, Xue Y, Zhang
Y, Lai X, Wei Y, Yu S, Zhang J, et al: Metformin enhances
tamoxifen-mediated tumor growth inhibition in ER-positive breast
carcinoma. BMC Cancer. 14:1722014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kumar BN, Rajput S, Dey KK, Parekh A, Das
S, Mazumdar A and Mandal M: Celecoxib alleviates
tamoxifen-instigated angiogenic effects by ROS-dependent
VEGF/VEGFR2 autocrine signaling. BMC Cancer. 13:2732013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hare SH and Harvey AJ: mTOR function and
therapeutic targeting in breast cancer. Am J Cancer Res. 7:383–404.
2017.PubMed/NCBI
|
|
22
|
Wong SW, Tiong KH, Kong WY, Yue YC, Chua
CH, Lim JY, Lee CY, Quah SI, Fow C, Chung C, et al: Rapamycin
synergizes cisplatin sensitivity in basal-like breast cancer cells
through up-regulation of p73. Breast Cancer Res Treat. 128:301–313.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Martin L, Pancholi S, Farmer I, Guest S,
Ribas R, Weigel MT, Thornhill AM, Ghazoui Z, A'Hern R, Evans DB, et
al: Effectiveness and molecular interactions of the clinically
active mTORC1 inhibitor everolimus in combination with tamoxifen or
letrozole in vitro and in vivo. Breast Cancer Res. 14:R1322012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chou T and Talalay P: Quantitative
analysis of dose-effect relationships: The combined effects of
multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 22:27–55.
1984. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schindlbeck C, Jeschke U, Schulze S,
Karsten U, Janni W, Rack B, Krajewski S, Sommer H and Friese K:
Prognostic impact of Thomsen-Friedenreich tumor antigen and
disseminated tumor cells in the bone marrow of breast cancer
patients. Breast Cancer Res Treat. 101:17–25. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lou P, Li C, Shi L, Xia TS, Zhou W, Wu J,
Zhou X, Li X, Wang Y, Wei JF, et al: RNPC1 enhances progesterone
receptor functions by regulating its mRNA stability in breast
cancer. Oncotarget. 8:16387–16400. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shi L, Xia T, Wei X, Zhou W, Xue J, Cheng
L, Lou P, Li C, Wang Y, Wei JF, et al: Estrogen receptor (ER) was
regulated by RNPC1 stabilizing mRNA in ER positive breast cancer.
Oncotarget. 6:12264–12278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li XX, Shi L, Zhou XJ, Wu J, Xia TS, Zhou
WB, Sun X, Zhu L, Wei JF and Ding Q: The role of c-Myc-RBM38 loop
in the growth suppression in breast cancer. J Exp Clin Cancer Res.
36:492017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tamoxifen for early breast cancer: An
overview of the randomised trials. Early breast cancer trialists'
collaborative group. Lancet. 351:1451–1467. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Osborne CK: Tamoxifen in the treatment of
breast cancer. N Engl J Med. 339:1609–1618. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Davies C, Pan H, Godwin J, Gray R,
Arriagada R, Raina V, Abraham M, Medeiros Alencar VH, Badran A,
Bonfill X, et al: Long-term effects of continuing adjuvant
tamoxifen to 10 years versus stopping at 5 years after diagnosis of
oestrogen receptor-positive breast cancer: ATLAS, a randomised
trial. Lancet. 381:805–816. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Raje N, Kumar S, Hideshima T, Ishitsuka K,
Chauhan D, Mitsiades C, Podar K, Le Gouill S, Richardson P, Munshi
NC, et al: Combination of the mTOR inhibitor rapamycin and CC-5013
has synergistic activity in multiple myeloma. Blood. 104:4188–4193.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Markman B, Dienstmann R and Tabernero J:
Targeting the PI3K/Akt/mTOR pathway-beyond rapalogs. Oncotarget.
1:530–543. 2010.PubMed/NCBI
|
|
35
|
Ellis MJ, Tao Y, Young O, White S, Proia
AD, Murray J, Renshaw L, Faratian D, Thomas J, Dowsett M, et al:
Estrogen-independent proliferation is present in estrogen-receptor
HER2-positive primary breast cancer after neoadjuvant letrozole. J
Clin Oncol. 24:3019–3025. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Law JH, Habibi G, Hu K, Masoudi H, Wang
MY, Stratford AL, Park E, Gee JM, Finlay P, Jones HE, et al:
Phosphorylated insulin-like growth factor-i/insulin receptor is
present in all breast cancer subtypes and is related to poor
survival. Cancer Res. 68:10238–10246. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Creighton CJ, Fu X, Hennessy BT, Casa AJ,
Zhang Y, Gonzalez-Angulo AM, Lluch A, Gray JW, Brown PH, Hilsenbeck
SG, et al: Proteomic and transcriptomic profiling reveals a link
between the PI3K pathway and lower estrogen-receptor (ER) levels
and activity in ER+ breast cancer. Breast Cancer Res. 12:R402010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Saal LH, Johansson P, Holm K,
Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA,
Malmström P, Memeo L, et al: Poor prognosis in carcinoma is
associated with a gene expression signature of aberrant PTEN tumor
suppressor pathway activity. Proc Natl Acad Sci USA. 104:7564–7569.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gonzalez-Angulo AM, Ferrer-Lozano J,
Stemke-Hale K, Sahin A, Liu S, Barrera JA, Burgues O, Lluch AM,
Chen H, Hortobagyi GN, et al: PI3K pathway mutations and PTEN
levels in primary and metastatic breast cancer. Mol Cancer Ther.
10:1093–1101. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pérez-Tenorio G, Alkhori L, Olsson B,
Waltersson MA, Nordenskjöld B, Rutqvist LE, Skoog L and Stål O:
PIK3CA mutations and PTEN loss correlate with similar prognostic
factors and are not mutually exclusive in breast cancer. Clin
Cancer Res. 13:3577–3584. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ciruelos Gil EM: Targeting the
PI3K/AKT/mTOR pathway in estrogen receptor-positive breast cancer.
Cancer Treat Rev. 40:862–871. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shirley SH, Rundhaug JE, Tian J,
Cullinan-Ammann N, Lambertz I, Conti CJ and Fuchs-Young R:
Transcriptional regulation of estrogen receptor-alpha by p53 in
human breast cancer cells. Cancer Res. 69:3405–3414. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rosenbluth JM, Mays DJ, Jiang A, Shyr Y
and Pietenpol JA: Differential regulation of the p73 cistrome by
mammalian target of rapamycin reveals transcriptional programs of
mesenchymal differentiation and tumorigenesis. Proc Natl Acad Sci
USA. 108:2076–2081. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bachelot T, Bourgier C, Cropet C,
Ray-Coquard I, Ferrero J, Freyer G, Abadie-Lacourtoisie S, Eymard
J, Debled M, Spaëth D, et al: Randomized phase II trial of
everolimus in combination with tamoxifen in patients with hormone
receptor-positive, human epidermal growth factor receptor
2-negative metastatic breast cancer with prior exposure to
aromatase inhibitors: A GINECO study. J Clin Oncol. 30:2718–2724.
2012. View Article : Google Scholar : PubMed/NCBI
|