Introduction
Gastric cancer has become the second highest leading
cause of cancer-related deaths worldwide (1). Since the development of new diagnostic
techniques as well as improvements in radical lymphadenectomy
surgical approaches, the prognosis of gastric cancer patients has
improved, but the incidence and mortality rates remain high. The
5-year overall survival rate is still at ~20% (2–4), and
the 5-year overall survival rate of patients with late stage is
nearly 4% (5). Therefore, it is
urgent to identify potential predictive markers and effective
molecular therapeutic targets of gastric cancer.
miRNAs (miRs), a class of small non-coding RNAs,
have been reported to suppress the expression of multiple target
genes by directly binding to a recognition sequence in the
3′-untranslated regions (3′-UTRs) of the mRNA of the target genes,
causing mRNA degradation or translational repression (6–8).
Increasing evidence has revealed that the expression of miRs is
aberrant in various cancers, and the dysregulation of miRs plays
crucial functions in the development and progression of cancers
(9–12). Recently, miRs, including miR-93,
miR-155 and miR-582 were revealed to promote or suppress gastric
cancer proliferation and metastasis (13–15).
Previous studies revealed that miR-95 was aberrantly expressed in
multiple types of cancer and regulated tumor development (16–18).
Chen et al revealed that miR-95 was downregulated in the
GSRCC type of gastric cancer (19).
However, the underlying mechanism of miR-95 has not yet been
elucidated.
In the present study, it was demonstrated that
miR-95 was downregulated in gastric cancer tissues and cell lines,
consistent with a previously study (19). Moreover, the expression of miR-95
was significantly associated with tumor size, tumor-node-metastasis
(TNM) stage and lymph node metastasis. Overexpression of miR-95
suppressed gastric cancer cell proliferation, migration and
invasion. Additionally, miR-95 also regulated EMT in gastric cancer
by directly inhibiting Slug. Collectively, our findings
demonstrated that miR-95 is a tumor suppressor in gastric
cancer.
Materials and methods
Patients and tissue samples
Patients admitted to the Affiliated Hospital of
Jining Medical University between February 2012 and October 2016
were evaluated. These patients with gastric cancer included 41
males and 22 females aged between 32–86 years, with a mean age of
60.6 years. Clinical stages were classified according to the
International Union against Cancer TNM classification system
(20). The Research Ethics
Committee of the Affiliated Hospital of Jining Medical University
approved the present study (JN2017015), and all patients provided
written informed consent. All tissue samples were stored at −80°C
before use.
Cell lines and cell culture
Human gastric cancer cell lines, such as CTC-141
(Laboratory of Stem Cell Biology of Sichuan University, Sichuan,
China) and MKN45 [American Type Culture Collection (ATCC) Manassas,
VA, USA], as well as normal human gastric epithelium cell line
GES-1 (Bogu Biotechnology, Shanghai, China) were maintained in the
Dulbecco's modified Eagle's medium (DMEM) supplemented with 100
U/ml penicillin, and 100 µg/ml streptomycin as well as 10% fetal
bovine serum (FBS) at 37°C with 5% CO2.
Cell transfection
Mimic control and miR-95 mimics (miR-95 mimic), as
well as inhibitor control and miR-95 inhibitors (miR-95 inhibitor)
were purchased from Qiagen (Duesseldorf, Germany). Cells were
transfected using Lipofectamine 3000 according to the
manufacturer's instructions (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). After transfection for 48 h, the
transfected cells were used for further experiments.
Quantitative real-time polymerase
chain reaction (qRT-PCR)
Total RNA was extracted from the gastric cancer
tissue samples and cells using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol and
was quantified using NanoDrop 2000 (Thermo Fisher Scientific,
Inc.). An RNA sample (2 µg) was used to synthesize cDNA using
RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher
Scientific, Inc.). A SYBR-Green (Roche Molecular Diagnostics,
Pleasanton, CA, USA) was used to determine relative mRNA expression
with an ABI PRISM 7500 Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.) following the instructions of the
manufacturer. Primers for miR-95 were purchased from GeneCopoeia
Co. (Guangzhou, China). The cycling parameters were as follows:
95°C for 5 min and then 40 cycles of 95°C for 15 sec and
annealing/extension at 60°C for 1 min. E-cadherin:
5′-ACCTGGTTCAGATCAAATCC-3′ (forward) and 5′-TCATTCTGATCGGTTACCGT-3′
(reverse); N-cadherin: 5′-CAGAGTTTACTGCCATGACG-3′ (forward) and
5′-AAAGTCGATTGGTTTGACCA-3′ (reverse); vimentin:
5′-ATTGAGATTGCCACCTACAG-3′ (forward) and
5′-ATCCAGATTAGTTTCCCTCAG-3′ (reverse); Slug:
5′-AGATGCATATTCGGACCCACA-3′ (forward) and
5′-CCTCATGTTTGTGCAGGAGAG-3′ (reverse); GAPDH:
5′-GAGAAGTATGACAACAGCCTC-3′ (forward) and
5′-ATGGACTGTGGTCATGAGTC-3′ (reverse); miR-95:
5′-CTGGTGGAGGGATGGATGAA-3′ (forward) and 5′-GGCCCGATCACAAACTCATC-3′
(reverse); U6: 5′-CTCGCTTCGGCAGCACA-3′ (forward) and
5′-AACGCTTCACGAATTTGCGT-3′ (reverse) GAPDH mRNA or small nuclear
RNA U6 were used as internal controls. The relative mRNA expression
was calculated via the 2−∆∆Cq method (21).
Western blot analysis
Whole protein was isolated from the transfected
cells using RIPA lysis buffer (Beyotime Institute of Biotechnology,
Jiangsu, China), and protein concentrations were assessed using
Pierce BCA Protein Assay kit (Pierce; Thermo Fisher Scientific,
Inc.) following the manufacturer's instructions. Protein ~45 µg was
separated via 10% SDS-PAGE. After being transferred to
nitrocellulose filter membranes (EMD Millipore, Bedford, MA, USA),
the membranes were blocked with 5% skimmed milk at room temperature
for 1 h. Subsequently, the membranes were incubated with indicated
primary antibodies at 4°C overnight. After being washed with
phosphate-buffered saline Tween-20 (PBST) three times at room
temperature, the membranes were incubated with HRP-conjugated
secondary antibodies at room temperature for 1 h and washed with
PBST three times at room temperature. Finally, the blots were
visualized by ECL kit (Pierce; Thermo Fisher Scientific, Inc.).
Each independent experiment was performed three times. The anbodies
as follow: EMT kit (1:1,000; cat. no. 9782; Cell Signaling
Technology, Danvers, MA, USA), β-actin (1:2,000; cat. no. ab6276;
Abcam, Cambridge, UK), goat anti-rabbit (HRP) (1:5,000; cat. no.
ab205718; Abcam), goat anti-mouse (HRP) (1:3,000; cat. no.
ab205719, Abcam). The densitometry of blots was determined using
ImageJ software (version 4.1; National Institutes of Health,
Bethesda, MD, USA).
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assay was used to detect the effect of miR-95
on cell proliferation. In brief, 3,000 transfected CTC-141 or MKN45
cells with 200 µl media were seeded in 96-well plates. After
transfection for 48 h, 20 µl CCK-8 solution (Beyotime Institute of
Biotechnology) was added to the culture medium at 0, 24, 48 and 72
h, and incubated for 30 min at 37°C. The absorbance of 450 nm was
measured using a microplate reader (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Each independent experiment was performed three
times.
Wound healing assay
A wound healing assay was used to determine the
effect of miR-95 on cell migration. Briefly, 5×105
transfected CTC-141 or MKN45 cells were seeded in 6-well plates.
When cell density was almost 90–100%, a linear wound was generated
using a 20-µl pipette tip, and the detached cells were washed with
PBS three times. The distance of wound healing was assessed at 0
and 24 h with a light microscope. Each independent experiment was
performed three times.
Transwell invasion assay
A Transwell assay was used to determine the effect
of miR-95 on cell invasion. In brief, 80 µl Matrigel was coated on
the Transwell chambers (BD Biosciences, Bedford, MA, USA) and
maintained at 37°C for 30 min. Approximately 4×104
transfected CTC-141 or MKN45 cells in 400 µl serum-free medium were
placed in the upper chamber in 24-well culture plates, and 500 µl
RPMI-1640 medium containing 10% FBS was added to the lower chamber.
Cells were maintained at 37°C with 5% CO2 for 16 h.
Subsequently, the cells were stained with 0.5% crystal violet at
room temperature for 10 min. The cells on the surface of the upper
membranes were removed by cotton swab, and the number of invading
cells was counted under a light microscope. Each independent
experiment was performed three times.
EMT induction
When cell density was almost 60%, cells were
cultured with serum-free media overnight. Recombinant human TGF-β1
(10 ng/ml) was added into media for 72 h.
Luciferase reporter assay
TargetScan Human version 7.0 (www.targetscan.org) predicted that Slug was a
potential target of miR-95. The 3′-UTR of Slug was cloned into the
pGL3 luciferase vector (Invitrogen; Thermo Fisher Scientific,
Inc.). For the luciferase assay, CTC-141 and MKN-45 cells were
co-transfected with Renilla, pGL3-Slug 3′-UTR or and miR-95
mimics or miR-95 inhibitor. After transfection for 24 h, a
luciferase reporter assay was performed using a Dual-Luciferase
Reporter Assay kit according to the manufacturer's instructions
(Promega Corp., Madison, Wisconsin, USA). The Renilla
luciferase activity was used to normalize firefly luciferase
activity. Each independent experiment was performed three
times.
Statistical analysis
All data were analyzed using SPSS 18.0 statistical
software (SPSS, Inc., Chicago, IL, USA) and presented as the mean ±
SD. A Student's t-test or one-way ANOVA followed by Tukey's were
used to analyze the differences between groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-95 is aberrantly downregulated in
gastric cancer tissues as well as cell lines, and associated with
poor tumor progression
To investigate the functions of miR-95 in gastric
cancer, we first detected the expression of miR-95 in gastric
cancer tissues. We performed qRT-PCR to analyze the miR-95
expression levels in 63 gastric tumor tissue samples and adjacent
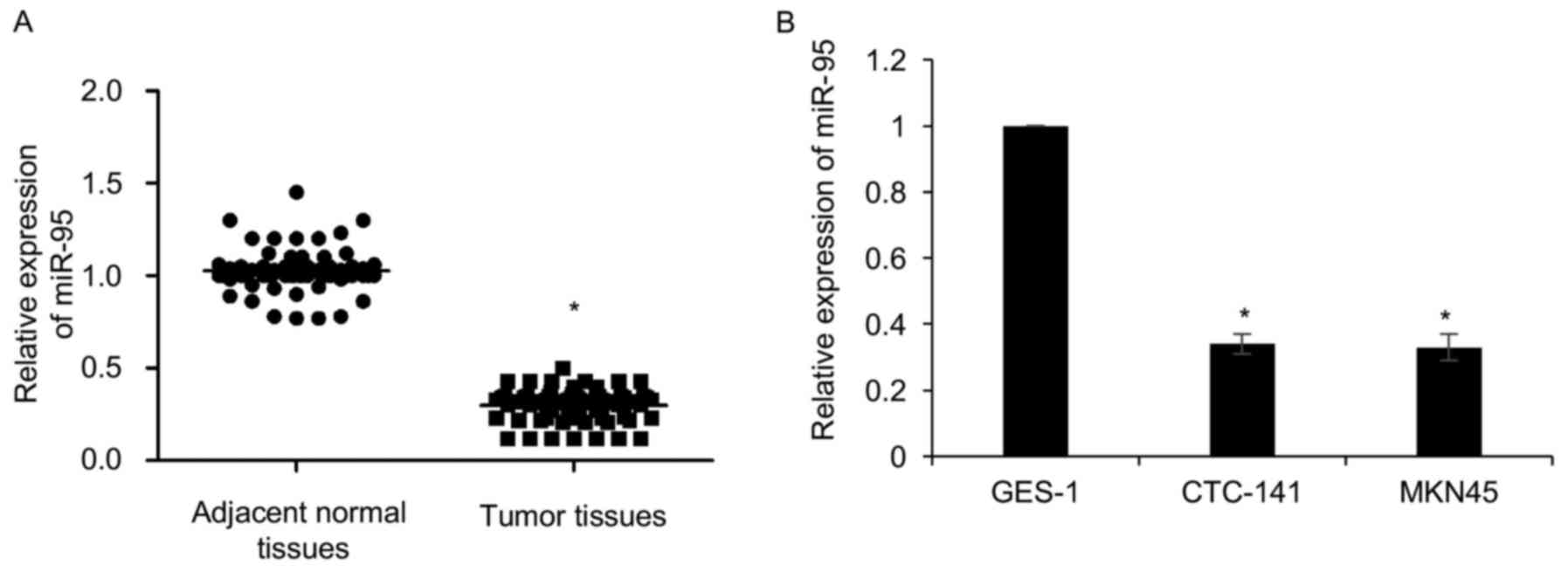
normal tissues samples. As shown in Fig. 1A, our findings revealed that miR-95
expression was significantly downregulated in gastric cancer
tissues compared with adjacent normal tissues (Fig. 1A). In addition, we detected the
expression of miR-95 in gastric cancer cell lines, including
CTC-141 and MKN45. GES-1 was used as a control. The expression of
miR-95 was lower in CTC-141 and MKN45 cells compared to GES-1
(Fig. 1B). Subsequently,
clinicopathological analyses of 63 gastric cancer patients
demonstrated that the expression of miR-95 was closely associated
with tumor size, lymph node metastasis as well as TNM stage
(Table I).
 | Table I.Clinicopathological variables in 63
gastric cancer patients. |
Table I.
Clinicopathological variables in 63
gastric cancer patients.
|
|
| miR-95
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | No. (n=63) | Low (n=42) | High (n=21) | P-value |
|---|
| Age, years |
|
|
| 0.285 |
|
<60 | 33 | 20 | 13 |
|
| ≥60 | 30 | 22 | 8 |
|
| Sex |
|
|
| 0.135 |
| Male | 41 | 30 | 11 |
|
|
Female | 22 | 12 | 10 |
|
| Tumor size (diameter)
(cm) |
|
|
| 0.031 |
| Small
(≤3) | 27 | 14 | 13 |
|
| Large
(≥3) | 36 | 28 | 8 |
|
| TNM stage |
|
|
| 0.020 |
| I–II | 29 | 15 | 14 |
|
|
III–IV | 34 | 27 | 7 |
|
| Lymph node
metastasis |
|
|
| 0.021 |
| No | 32 | 17 | 15 |
|
|
Yes | 31 | 25 | 6 |
|
miR-95 suppresses gastric cancer cell
proliferation
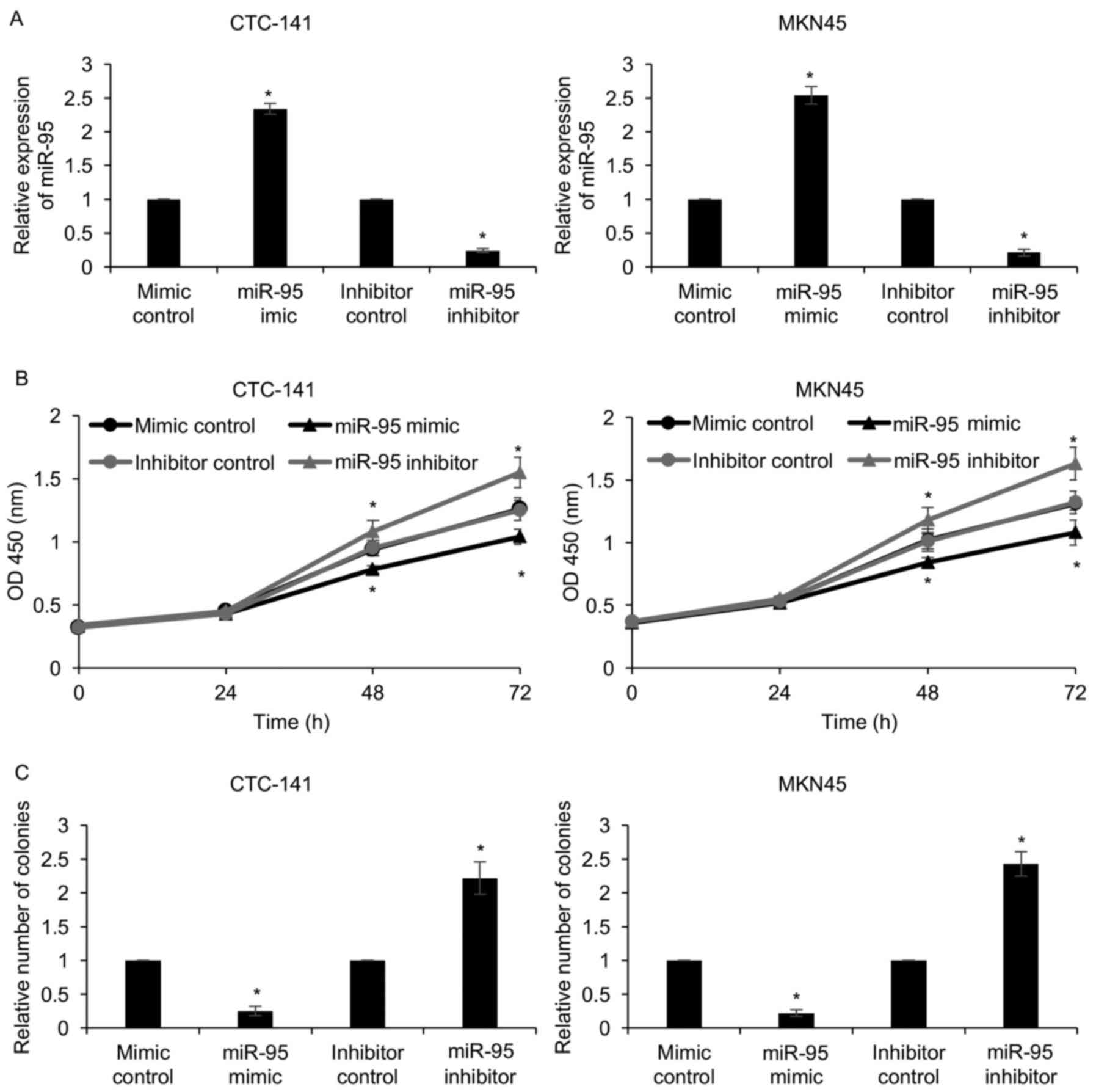
To explore the function of miR-95 in gastric cancer,
we first overexpressed or knocked down miR-95 in CTC-141 and MKN45
cells with miR-95 mimics or miR-95 inhibitor. The expression of
miR-95 was determined by qRT-PCR (Fig.
2A). As shown in Table I, since
the expression of miR-95 was closely associated with tumor size, we
assumed that miR-95 may regulate cell proliferation. To verify our
hypothesis, CCK-8 and colony formation assays were performed. The
results of the CCK-8 assay revealed that compared with that of the
mimic control or inhibitor control groups, the proliferation of
CTC-141 and MKN45 cells in the miR-95-mimic group were
significantly decreased and were clearly increased in the
miR-95-inhibitor group (Fig. 2B).
The colony formation assay also confirmed that ectopic expression
of miR-95 led to a decrease in the number of colonies, and an
inhibition of miR-95 led to an increase in the number of colonies
(Fig. 2C). These results revealed
that miR-95 suppressed gastric cancer cell proliferation.
Downregulation of miR-95 promotes
gastric cancer cell migration and invasion
Our experiments revealed that miR-95 expression was
negatively associated with lymph node metastasis. To determine
whether miR-95 regulated migration and invasion in gastric cancer
cells, we performed wound healing and Transwell assays. The results
of the wound healing assays revealed that ectopic expression of
miR-95 significantly decreased the distance of cell migration
(Fig. 3A). In contrast,
downregulation of miR-95 significantly increased the distance of
cell migration (Fig. 3A). Similar
results were observed in the Transwell assay, revealing that the
ectopic expression of miR-95 resulted in less invading cells than
that in the mimic control group (Fig.
3B). In contrast, the downregulation of miR-95 resulted in more
invading cells than those in the inhibitor control group (Fig. 3B). Since MMP9 is an invasion-related
factor, we next determined whether miR-95 regulated MPP9 secretion
using an ELISA assay, which revealed that the secretion of MMP9 was
decreased when cells were transfected with miR-95 mimics, and
increased when cells were transfected with the miR-95 inhibitor
(Fig. 3C). Our experiments revealed
that downregulation of miR-95 promoted gastric cancer cell
migration and invasion.
miR-95 inhibits the TGF-β1-induced EMT
process of gastric cancer cells
To further decipher the detailed mechanisms of
miR-95 in gastric cancer metastasis, we aimed to explore the
effects of miR-95 on EMT, which was recognized as a main cause for
cell migration and invasion. CTC-141 and MKN45 cells were
transfected with miR-95 mimics and mimic control. After inducing 10
ng/ml of TGF-β1, the expression of EMT-associated proteins was
determined by RT-qPCR and western blotting, respectively. The
results revealed that ectopic expression of miR-95 led to an
increased expression of E-cadherin both at the mRNA level and
protein level, and a decreased expression of N-cadherin and
vimentin both at the mRNA level and protein level (Fig. 4A and B). A suppression of miR-95
reversed these results (Fig. 4C and
D). The mRNA and protein levels of E-cadherin were decreased,
and the levels of N-cadherin as well as vimentin were increased
(Fig. 4C and D). Overall, the
results implied that miR-95 inhibits the TGF-β1-induced EMT process
of gastric cancer cells.
Slug is a target of miR-95 in gastric
cancer cells
miRNAs have been found to participate in multiple
physiological and pathological processes by regulating gene
expression. To further decipher the detailed mechanism of miR-95 on
EMT, we searched for potential targets of miR-95 by bioinformatics
search using TargetScan (www.targetscan.org). We found that Slug, a key
transcription factor of EMT, was a direct target of miR-95. To
verify whether slug was regulated by miR-95, we overexpressed or
knocked down miR-95 in CTC-141 and MKN45 and detected the
expression of Slug. As shown in Fig.
5A, we found that Slug expression was decreased in CTC-141 and
MKN45 cells treated with miR-95 mimics, while Slug expression was
increased in CTC-141 and MKN45 cells treated with miR-95 inhibitors
(Fig. 5A and B). To examine whether
miR-95 directly interacted with the 3′-UTR of Slug, we performed a
luciferase reporter assay. The luciferase activity was
significantly decreased when cells were co-transfected with Slug
3′-UTR and either miR-95 mimics or mimic control (Fig. 5C). These findings indicated that
Slug is a target of miR-95 in gastric cancer.
Discussion
miR-95 has been reported to facilitate cell
proliferation in non-small cell lung cancer (NSCLC) and colorectal
carcinoma, suggesting that miR-95 acts as an oncogenic miRNA
(17,22). Paradoxically, Chen et al
revealed that miR-95 was downregulated in the GSRCC type of gastric
cancer (19). Moreover, miR-95 was
also revealed to regulate chemoresistance and radioresistance in
NSCLC (22). A previous study also
revealed that miR-95 plays a key function in the anticancer
activity of Brucein D which is in contrast to its proliferative
effects (23). Several studies have
indicated that miRNAs are new therapeutic targets for multiple
diseases, such as cancers. In colorectal cancer (CRC), miR-95 was
revealed to be overexpressed in the serum sample of CRC patients
(24), suggesting that miR-95 may
be a potential molecular target for cancer diagnosis.
In this study, we found miR-95 was downregulated in
gastric cancer tissues and cell lines. Additionally, the expression
of miR-95 was significantly associated with tumor size, TNM stage
and lymph node metastasis. Furthermore, we found that miR-95
suppressed proliferation, migration and invasion of gastric cancer
cells.
Metastasis is the main reason for treatment failure
in cancer and results in >90% of cancer-related deaths (25–27).
EMT is a complex process which is associated with the progression
of metastasis, and improves the migration as well as invasive
abilities of cancer cells. Various studies have indicated that
miRNAs are involved in this process (25,28).
In the present study, we found that miR-95 suppressed EMT in
gastric cancer cells. Notably, we found that Slug, a key
transcription factor of EMT, was a target of miR-95. Therefore, we
demonstrated that miR-95 suppressed cell migration and invasion
through regulation of EMT in gastric cancer cells.
However, there are still some limitations in the
present study. The upstream of miR-95 and detailed mechanism of
miR-95 on cellular proliferation in gastric cancer is still
unknown. Moreover, the function of miR-95 in vivo warrants
further investigation.
Collectively, our findings revealed the important
role of miR-95 in regulating EMT of gastric cancer. miR-95
suppressed Slug expression to regulate the progression of EMT,
thereby inhibiting migration and invasion in gastric cancer. In
addition, aberrant expression of miR-95 was closely associated with
tumor size, lymph node metastasis as well as TNM stage, suggesting
that miR-95 may play a key role in gastric cancer development. Our
findings revealed miR-95 as a novel molecular therapeutic target of
gastric cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
WZ and LZ conceived and designed the study. WZ, JS,
CX and JC performed the experiments. WZ and CX wrote the paper. WZ
and LZ reviewed and edited the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The Research Ethics Committee of the Affiliated
Hospital of Jining Medical University, Jining, China approved the
present study. All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shen L, Shan YS, Hu HM, Price TJ, Sirohi
B, Yeh KH, Yang YH, Sano T, Yang HK, Zhang X, et al: Management of
gastric cancer in Asia: Resource-stratified guidelines. Lancet
Oncol. 14:e535–e547. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thrumurthy SG, Chaudry MA, Chau I and
Allum W: Does surgery have a role in managing incurable gastric
cancer? Nat Rev Clin Oncol. 12:676–682. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang G, Fu Y, Liu G, Ye Y and Zhang X:
miR-218 Inhibits Proliferation, Migration, and EMT of Gastric
Cancer Cells by Targeting WASF3. Oncol Res. 25:355–364. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lv H, Zhang Z, Wang Y, Li C, Gong W and
Wang X: MicroRNA-92a promotes colorectal cancer cell growth and
migration by inhibiting KLF4. Oncol Res. 23:283–290. 2016.
View Article : Google Scholar
|
|
9
|
Ji S, Zhang B, Kong Y, Ma F and Hua Y:
miR-326 inhibits gastric cancer cell growth through downregulating
NOB1. Oncol Res. 25:853–861. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang J, Zhang Y, Jiang G, Liu Z, Xiang W,
Chen X, Chen Z and Zhao J: MiR-138 induces renal carcinoma cell
senescence by targeting EZH2 and is downregulated in human clear
cell renal cell carcinoma. Oncol Res. 21:83–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He C, Yu T, Shi Y, Ma C, Yang W, Fang L,
Sun M, Wu W, Xiao F, Guo F, et al: MicroRNA 301A promotes
intestinal inflammation and colitis-associated cancer development
by inhibiting BTG1. Gastroenterology. 152:1434–1448.e15. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun HL, Cui R, Zhou J, Teng KY, Hsiao YH,
Nakanishi K, Fassan M, Luo Z, Shi G, Tili E, et al: ERK Activation
globally downregulates miRNAs through phosphorylating exportin-5.
cancer cell. 30:723–736. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jin Y, Tao LP, Yao SC, Huang QK, Chen ZF,
Sun YJ and Jin SQ: MicroRNA-582-5p suppressed gastric cancer cell
proliferation via targeting AKT3. Eur Rev Med Pharmacol Sci.
21:5112–5120. 2017.PubMed/NCBI
|
|
14
|
Qu Y, Zhang H, Sun W, Han Y, Li S, Qu Y,
Ying G and Ba Y: MicroRNA-155 promotes gastric cancer growth and
invasion by negatively regulating transforming growth factor-β
receptor 2. Cancer Sci. 109:618–628. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guan H, Li W, Li Y, Wang J, Li Y, Tang Y
and Lu S: MicroRNA-93 promotes proliferation and metastasis of
gastric cancer via targeting TIMP2. PLoS One. 12:e01894902017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hwang SJ, Lee HW, Kim HR, Song HJ, Lee DH,
Lee H, Shin CH, Joung JG, Kim DH, Joo KM, et al: Overexpression of
microRNA-95-3p suppresses brain metastasis of lung adenocarcinoma
through downregulation of cyclin D1. Oncotarget. 6:20434–20448.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang Z, Huang S, Wang Q, Liang L, Ni S,
Wang L, Sheng W, He X and Du X: MicroRNA-95 promotes cell
proliferation and targets sorting Nexin 1 in human colorectal
carcinoma. Cancer Res. 71:2582–2589. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fan B, Jiao BH, Fan FS, Lu SK, Song J, Guo
CY, Yang JK and Yang L: Downregulation of miR-95-3p inhibits
proliferation, and invasion promoting apoptosis of glioma cells by
targeting CELF2. Int J Oncol. 47:1025–1033. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen J, Sun D, Chu H, Gong Z, Zhang C,
Gong B, Li Y, Li N and Jiang L: Screening of differential microRNA
expression in gastric signet ring cell carcinoma and gastric
adenocarcinoma and target gene prediction. Oncol Rep. 33:2963–2971.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jun KH, Lee JS, Kim JH, Kim JJ, Chin HM
and Park SM: The rationality of N3 classification in the 7th
edition of the International Union Against Cancer TNM staging
system for gastric adenocarcinomas: A case-control study. Int J
Surg. 12:893–896. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Chen S, Hang W, Huang H and Ma H:
MiR-95 induces proliferation and chemo- or radioresistance through
directly targeting sorting nexin1 (SNX1) in non-small cell lung
cancer. Biomed Pharmacother. 68:589–595. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiao Z, Ching Chow S, Han Li C, Chun Tang
S, Tsui SK, Lin Z and Chen Y: Role of microRNA-95 in the anticancer
activity of Brucein D in hepatocellular carcinoma. Eur J Pharmacol.
728:141–150. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ng EK, Chong WW, Jin H, Lam EK, Shin VY,
Yu J, Poon TC, Ng SS and Sung JJ: Differential expression of
microRNAs in plasma of patients with colorectal cancer: A potential
marker for colorectal cancer screening. Gut. 58:1375–1381. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu W, Li M, Chen X, Zhang D, Wei L, Zhang
Z, Wang S, Meng L, Zhu S and Li B: MicroRNA-373 promotes migration
and invasion in human esophageal squamous cell carcinoma by
inhibiting TIMP3 expression. Am J Cancer Res. 6:1–14.
2015.PubMed/NCBI
|
|
26
|
Yang F, Liu X, Liu Y, Liu Y, Zhang C, Wang
Z, Jiang T and Wang Y: miR-181d/MALT1 regulatory axis attenuates
mesenchymal phenotype through NF-κB pathways in glioblastoma.
Cancer Lett. 396:1–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shishodia G, Shukla S, Srivastava Y,
Masaldan S, Mehta S, Bhambhani S, Sharma S, Mehrotra R, Das BC and
Bharti AC: Alterations in microRNAs miR-21 and let-7a correlate
with aberrant STAT3 signaling and downstream effects during
cervical carcinogenesis. Mol Cancer. 14:1162015. View Article : Google Scholar : PubMed/NCBI
|