Introduction
Lung cancer is the leading cause of cancer-related
death worldwide (1). It includes
two major types: Small cell lung cancer (SCLC) and non-small cell
lung cancer (NSCLC). Although some lung cancer tumors are
resectable or initially responsive to traditional therapies, drug
resistance and poor prognosis remain high, leading to a poor 5-year
survival rate of less than 15% (2–5).
Approximately, non-small cell lung cancer (NSCLC) accounts for 80%
of all lung cancer cases (6).
Traditional and progressive treatments have been applied
clinically, including chemotherapy, radiotherapy and biotherapy,
but the resistance to radiotherapy and chemotherapy remains a
critical issue for lung cancer therapy (7,8).
Recently, it has been suggested that a subpopulation termed cancer
stem cells (CSCs) cause initiation, drug resistance, metastasis and
recurrence of cancer cells. CSCs are divided asymmetrically to stem
cells that have the capacity of self-renewal, and the other cells
that will differentiate and produce phenotypically diverse
tumor-constitutive cancer cells (9). Researchers first reported CSCs in
leukemia (10), and subsequently in
solid tumors, such as colon, brain, breast and lung cancer
(11–14). A small subpopulation of lung cancer
cells called cancer stem-like cells (CSCs) have been certified in
many studies using different isolation assays, including
accumulation in specific medium and cell sorting by certain markers
such as CD133+ (15–18).
Human 95-D cells are highly invasive and metastatic lung cancer
cells (19). In previous research,
cultured in specific medium, 95-D cells can be used to accumulated
lung cancer stem cells in spheres, which were called LCSCs
(20). Conventional and traditional
therapies that fail to eradicate CSCs may reduce tumor cells
temporarily; however, resistance, metastasis and relapse are more
likely to occur when treatment is suspended. After the discovery of
cancer stem cells, efficient approaches targeting CSCs is
considered to be indispensable for eradicating cancer cells
(21).
Neural EGFL like 1 (NELL1) was originally cloned
from a human fetal-brain cDNA library (22). Previous research has found that
NELL1 plays an important role in osteogenic differentiation
(23). NELL1 is highly expressed in
patients with craniosynostosis, and NELL1 can induce bone
regeneration in calvarial defects (24). Overexpression of NELL1 was found to
induce apoptosis in osteoblasts during craniofacial development
(25). On a cellular level, NELL1
is suggested to promote osteoblast differentiation. Thus, we aimed
to ascertain whether NELL1 could induce CSC differentiation. In the
present study, we investigated the effects of NELL1 on 95-D
LCSCs.
Moreover, expression of the NELL1 gene has been
studied in several types of cancer. In several cancers, NELL1 has
been found to be related with poor prognosis. In human renal cell
carcinoma, it was shown that NELL1 was significantly downregulated
in renal cell carcinoma, NELL1 gene was hypermethylation in renal
cell carcinoma cell lines (26).
NELL1 was also found to be downregulated in esophageal squamous
cell carcinoma (27). In
glioblastoma cell lines, NELL1 is lowly expressed (28). However, little is known concerning
the function of NELL1 in NSCLC. In the present study, we
investigated the effects of NELL1 on lung cancer stem-like
cells.
Materials and methods
Cell culture and reagents
The human NSCLC 95-D cell line, which is a
commercial cell line, was obtained from the Chinese Academy of
Science (Shanghai, China). 95-D cells were cultured with Hyclone™
RPMI-1640 medium (GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with Gibco™ 10% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Cells were incubated at 37°C
with 5% CO2. The 95-D LCSCs, after sorting using a BD
fluorescence-activated cell sorting FACSAria flow cytometer (BD
Biosciences, San Jose, CA, USA) were culture in neuroblast medium
with 20 ng/ml basic fibroblast growth factor (bFGF) (Thermo Fisher
Scientific, Inc.), 20 ng/ml epidermal growth factor (EGF) (Thermo
Fisher Scientific, Inc.) and B27 supplement (Thermo Fisher
Scientific, Inc.).
Oncomine database analysis
Oncomine (https://www.oncomine.org/resource/login.html) is a
bioinformatics online cancer microarray database aimed at
facilitating and promoting the discovery of the functions from
genome-wide expression. The differential expression of NELL1
between lung adenocarcinoma and normal counterparts was analyzed
using the Oncomine database by searching ‘Gene:NELL1’; ‘Analysis
Type: Cancer vs. normal analysis’; ‘Cancer Type: Lung cancer’.
Sphere formation
The 95-D LCSCs were accumulated by sphere formation.
Basically, the 95-D cells were digested into single cells with
0.25% trypsin and washed with phosphate-buffered saline (PBS). The
cells were seeded in ultra-low attachment dishes and cultured in
neuroblast medium with 20 ng/ml bFGF, 20 ng/ml EGF and B27 for 7
days to form spheres. The spheres were centrifuged and digested
into single cells for further studies.
Flow cytometry
To analyze the CD133 relative expression in the
accumulated LCSCs in spheres, cells were harvested and washed with
FACS buffer [PBS containing FBS (1%; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany)] and sodium azide (0.05%; Sigma-Aldrich; Merck
KGaA). The CD133-PE/Cy7 (anti-human; dilution 1:100; cat. no.
372810; BioLegend, Shanghai, China) antibody diluted in FACS buffer
was added directly to this mixture and incubated for 30 min at 4°C
in the dark. Meanwhile the IgG1-PE/Cy7 (anti-human; dilution 1:100;
cat. no. 401908; BioLegend) antibody was used as a negative
control. Cells were washed and resuspended in FACS buffer. The
cells were maintained on ice until the analysis using the
FACSCalibur flow cytometer (BD Biosciences) and sorting using
FACSAria flow cytometer (BD Biosciences). Results were obtained by
analyzing data with FlowJo version 7.6.1 software (FlowJo LLC,
Ashland, OR, USA). The results represent the mean value of three
independent experiments. The experiments were performed
independently three times and a representative is shown.
Plasmid and transfection
To prepare the NELL1 expression construct, the NELL1
cDNA fragment (2932 bp, gene accession #BC069674.1) was amplified
by PCR in vitro. The PCR products were digested with
NheI and SgsI restriction enzymes and inserted into
corresponding sites of the PDS023_PL_IRES vector. The
PDS023_PL_IRES lentivirus empty vector or vectors for NELL1 were
co-transfected with lentivirus packing vector pMDLg/pRRE, RSV-rev
and pMD2.G into 293T cells to obtain lentiviral supernatant. The
viral supernatant was collected after 48 and 72 h. Wild-type early
passage 95-D LCSCs and 95-D cells were incubated with
virus-containing medium in the presence of 4 mg/ml polybrene
(Sigma-Aldrich; Merck KGaA). Stable cell lines were established
after 4 days of blasticidine 5 µg/ml selection.
Cell invasion assay
To investigate cell invasion, 20 µl Matrigel (BD
Biosciences) and 80 µl RPMI-1640 medium were added to the upper
layer of a Transwell chamber (Corning Inc., Corning, NY, USA). A
total of 1×104 95-D vector (95-D EV) cells, 95-D LCSC
vector (95-D LCSCs EV) and 95-D LCSC NELL1-overexpressing cells
(95-D LCSCs NELL1) were gently added to the Matrigel medium, while
600 µl PRMI-1640 medium supplemented with 20% FBS was added to the
lower layer of the chamber. The chambers were placed in an
incubator at 37°C. After 48 h, the chambers were fixed with liquid
methanol and stained with 0.5% crystal violet at 37°C for 30 min,
and the chambers were washed with PBS several times. Cotton swabs
were gently used to remove the cells on the upper layers of the
chamber. The cells on the lower layers were counted within 5
randomly observed images using a Nikon microscope (Nikon, Tokyo,
Japan) and statistically analyzed.
Soft agar colony formation assay
The colony formation assay was performed as
previously described (29).
Briefly, a 5% (w/v) base agar (Sigma-Aldrich; Merck KGaA) solution
was prepared and autoclaved. For the bottom agar layer, 2 ml of the
0.4% agar/RPMI-1640 bottom agar layer was added to each well of the
6-well plates and was cooled to semisolid status. Single cells
(2,000) were seeded in the 0.3% top agar layer into each well.
Cells were cultured at 37°C for two or three weeks. Colonies were
stained with 0.05% crystal violet (Sigma-Aldrich; Merck KGaA). All
assays were performed in triplicate.
Western blot analysis
For western blot analysis, samples of the 95-D EV,
95-D LCSCs EV and 95-D LCSCs NELL1 cells were harvested. An amount
of 30 µg of the total protein samples was prepared, denatured at
100°C for 5 min, and inserted into the 10% gel wells for
SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto
polyvinylidine fluoride (PVDF) membranes (Millipore, Darmstadt,
Germany). For immunolabeling, the membranes were blocked with 5%
milk in Tris-buffered saline with Tween 20 (TBS-T). The membranes
were incubated with primary antibodies overnight at 4°C. These
antibodies included anti-CD133 (rabbit monoclonal antibody,
anti-human; dilution 1:1,000; cat. no. 64326; Cell Signaling
Technology, Danvers, MA, USA), anti-Oct4 (rabbit monoclonal
antibody, anti-human; dilution 1:1,000; cat. no. 2890; Cell
Signaling Technology), anti-Sox2 (rabbit monoclonal antibody,
anti-human; dilution 1:1,000; cat. no. 3579; Cell Signaling
Technology), anti-NELL1 (rabbit polyclonal antibody, anti-human;
dilution 1:500; cat. no. ab197315; Abcam, Cambridge, UK),
anti-ABCG2 (rabbit monoclonal antibody, anti-human; dilution
1:1,000; cat. no. 42078; Cell Signaling Technology), anti-ABCB1
(rabbit monoclonal antibody, anti-human; dilution 1:1,000; cat. no.
13342; Cell Signaling Technology), anti-ABCC1 (rabbit monoclonal
antibody, anti-human; dilution 1:1,000; cat. no. 14685; Cell
Signaling Technology), anti-β-catenin (rabbit polyclonal antibody,
anti-human; dilution 1:5,000; cat. no. ab32572; Abcam), anti-shh
(mouse monoclonal antibody, anti-human; dilution 1:500; cat. no.
sc-365112; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
anti-p-MET (Tyr1349), (rabbit monoclonal antibody, anti-human;
dilution 1:1,000; cat. no. 3133; Cell Signaling Technology), c-MET
(rabbit monoclonal antibody, anti-human; dilution 1:1,000; cat. no.
8041; Cell Signaling Technology), anti-Notch3 (rabbit monoclonal
antibody, anti-human; dilution 1:1,000; cat. no. 5276; Cell
Signaling Technology), anti-HES1 (rabbit monoclonal antibody,
anti-human; dilution 1:1,000; cat. no. 11988; Cell Signaling
Technology) and anti-α-tubulin (mouse monoclonal antibody,
anti-human; dilution 1:2,000; cat. no. 3873; Cell Signaling
Technology). Then anti-mouse (rabbit polyclonal antibody; dilution
1:5,000; cat. no. ab6728; Abcam) or anti-rabbit (goat polyclonal;
dilution 1:5,000; cat. no. ab6721; Abcam) horseradish peroxidase
(HRP) conjugated antibodies were used as secondary antibodies and
incubated with membranes for 1 h and blots were developed using
Electrochemiluminescence (ECL)-Plus Western detection system
(Thermo Fisher Scientific, Inc.) for visualization.
Quantitative real-time polymerase
chain reaction (qPCR)
Total RNA of 95-D EV, 95-D LCSCs EV and 95-D LCSCs
EV cells were extracted using Invitrogen™ TRIzol®
reagent (Thermo Fisher Scientific, Inc.). Samples were treated with
TRIzol followed by chloroform and then centrifuged for 10 min at
12,000 × g at 4°C. The supernatant was discarded and the pellet was
washed in cold 75% ethanol. Finally, the RNA samples were diluted
with 40 µl RNase-free water. A total of 1 µg RNA was reverse
transcribed using a Bio-Rad script cDNA synthesis kit (Bio-Rad
Laboratories, Hercules, CA, USA). Real-time PCR analysis of CD133,
Oct4, Sox2, NELL1, ABCG2, ABCB1 and ABCC1 were performed for
quantification using a Mx3000P qPCR system (Stratagene, San Diego,
CA, USA) with qPCR cycling conditions (an initial denaturation at
95°C for 1 min and then 45 cycles of 15 sec at 95°C, 31 sec at
60°C). The sequences of primers used for the qRT-PCR are listed in
Table I. SYBR® Premix Ex
Taq™ II (Takara Bio, Inc., Tokyo, Japan) was used and 20 µl per
gene was analyzed. The relative fold change was quantified by
2−ΔΔCq (30), and
β-actin was used as a housekeeping control.
 | Table I.Primers used for qPCR. |
Table I.
Primers used for qPCR.
| Primer name | Primer sequence
(5′-3′) | Length (bp) |
|---|
| CD133-F |
ACACTACCAAGGACAAGGCGTTCA | 154 |
| CD133-R |
CTCAGTTCAGGGTTGCTATTCA |
|
| Oct4-F |
AGCAGCGACTATGCACAACGAG | 196 |
| Oct4-R |
TGACGGAGACAGGGGGAAAGGCTTC |
|
| Sox2-F |
AGACGCTCATGAAGAAGGAT | 183 |
| Sox2-R |
TGGTCCTGCATCATGCTGTAGC |
|
| NELL1-F |
TATGGTGTTTCACGGCTTAGTG | 234 |
| NELL1-R |
ATCACGTTGGATGGTCATTTCG |
|
| ABCG2-F |
ATAAAGTGGCAGACTCCAAGGT | 264 |
| ABCG2-R |
ATAAGGTGAGGCTATCAAACAA |
|
| ABCB1-F |
ACTCGTAGGAGTGTCCGTGGAT | 286 |
| ABCB1-R |
CAAGGGCTAGAAACAATAGTGAAA |
|
| ABCC1-F |
CAGGCGAGTGTCTCCCTCAAAC | 124 |
| ABCC1-R |
CATTCCTCACGGTGATGCTGTT |
|
| β-actin-F |
TTTTCCAGCCTTCCTT | 150 |
| β-actin-R |
TTGGCATACAGGTCTTT |
|
Cell proliferation assay
Cell Counting Kit-8 (CCK-8) was used to conduct the
proliferation assays. 95-D EV, 95-D LCSCs EV and 95-D LCSCs NELL1
cells were seeded at 1×104 cells per well in 96-well
microtiter plate and were maintained at 37°C for 24 h. A mixture of
200 µl RPMI-1640 medium with different concentrations of
carboplatin and cisplatin was added into each well. After 48 h of
incubation, a mixture of 190 µl RPMI-1640 with 10% FBS and 10 µl of
CCK-8 was added into each well and measured at 450 nm. Each
experiment was performed in replicates of six and background
reading of the media was subtracted.
Statistical analysis
All of the experiments were performed three times
independently. Statistical analysis (simple or plural) was
performed using Statistical Package for Social Science (SPSS)
software, version 19 (IBM Corp., Armonk, NY, USA). Groups were
compared with Student's t-test or one-way analysis of variance
(ANOVA) with Tukey's honestly significant difference (HSD) post hoc
test where groups were more than two. Results were considered to be
statistically significant at P<0.05.
Results
Putative 95-D stem cells express
stemness genes
To evaluate how transcription of NELL1 correlates
with NSCLC, we analyzed clinical cohorts of NSCLC patients using
the Oncomine database. It was found that NELL1 expression was lower
in lung adenocarcinoma (Fig. 1A).
Previous research demonstrated that NELL1 plays an important role
in osteogenic differentiation (31). Thus, we aimed to ascertain whether
overexpression of NELL1 could induce the differentiation of LCSCs.
To measure the effect of NELL1 on LCSCs, specific medium was
utilized to accumulate the LCSCs in spheres. The 95-D cancer
stem-like cells were accumulated by sphere formation. To test
whether the formed spheres were indeed cancer stem cells, flow
cytometry was used to examine the expression of stemness gene
CD133. It was found that CD133 was highly expressed in the sphere
cells (Fig. 1B). These highly
expressed CD133 cells were next obtained through flow cytometric
cell sorting of the CD133-stained cells and these cells were termed
95-D lung cancer stem-like cells (95-D LCSCs). Then, the expression
levels of stemness genes (CD133, Oct4 and Sox2) was assessed using
western blot analysis. It was found that these genes were highly
expressed in LCSCs (Fig. 1C). The
results were confirmed using qPCR (Fig.
1D). These results demonstrated that 95-D LCSCs express tumor
stem-like cell-related genes and may display tumor stem cell
characteristics.
NELL1 overexpression in 95-D LCSCs
results in decreased colony formation and invasion
To test whether NELL1 inhibits 95-D LCSC growth,
95-D cells were transfected with a lentivirus empty vector and 95-D
LCSCs cells with a lentivirus empty vector or a lentivirus carrying
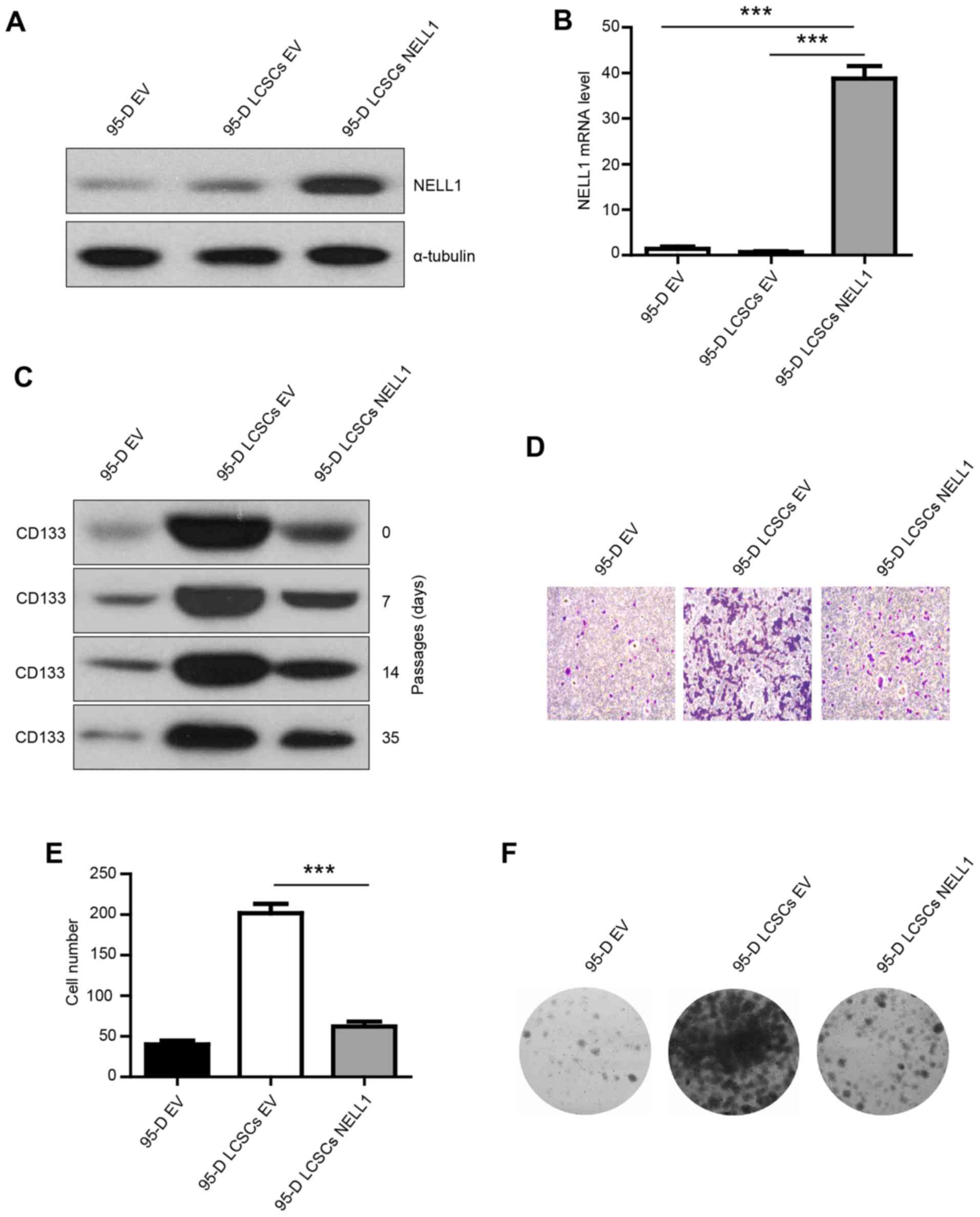
NELL1. The transfection efficiency was confirmed (Fig. 2A and B). NELL1 was highly expressed
in the 95-D LCSCs NELL1 group. To test whether the isolated 95-D
LCSCs could maintain high expression of CD133 in different
passages, the CD133 expression was evaluated in early and multiple
passages. We found that CD133 was highly expressed in the different
passages (Fig. 2C). Then, we
examined the effect of NELL1 on 95-D LCSCs. Transwell invasion
assay was used to examine the change in the invasive ability.
Overexpression of NELL1 reduced the invasive capability of the 95-D
LCSCs compared with the control cells, and the number of invasive
cells was significantly decreased (Fig.
2D and E). Overexpression of NELL1 also significantly inhibited
the colony formation and growth of 95-D LCSCs (Fig. 2F). These results indicate that NELL1
expression decreased the colony formation and invasion of 95-D
LCSCs.
Overexpression of NELL1 inhibits the
proliferation of 95-D LCSCs cells
One of the major points of cancer stem cells is
their chemoresistance. Carboplatin and cisplatin are common
chemotherapeutic drugs. To investigate the role of NELL1 expression
in chemotherapeutic drug resistance, a CCK-8 assay was performed
(Fig. 3A and B). Our results showed
that the chemosensitivity of 95-D LCSCs NELL1 cells was
significantly increased compared with the 95-D LCSCs. Subsequently,
we analyzed the protein levels of common multi-drug resistance
markers ABCG2, ABCB1 and ABCC1. As shown in Fig. 3C and D, the expression of ABCG2,
ABCB1 and ABCC1 was significantly decreased in the 95-D LCSCs NELL1
cells. Upon mRNA analysis, the same trends for ABCG2, ABCB1 and
ABCC1 protein expression were also identified (Fig. 3E). These data show that
overexpression of NELL1 in 95-D LCSCs cells increased the
chemosensitivity of these cells.
 | Figure 3.Cell viability of 95-D EV, 95-D LCSCs
EV and 95-D LCSCs NELL1 cells. (A and B) Cell viability analysis of
95-D EV, 95-D LCSCs EV and 95-D LCSCs NELL1 cells following
treatment with chemotherapeutic agent carboplatin or cisplatin for
48 h (25, 50, 100 and 150 µM). (C and D) Western blot analysis
results of ABCG2, ABCB1 and MDR1 proteins in 95-D EV, 95-D LCSCs EV
and 95-D LCSCs NELL1 cells. (E) Quantitative RT-PCR results of
ABCG2, ABCB1 and MDR1 in 95-D EV, 95-D LCSCs EV and 95-D LCSCs
NELL1 cells. *P<0.5, **P<0.01. NELL1, neural EGFL like 1;
LCSCs, lung cancer stem-like cells. |
NELL1 induces differentiation of 95-D
LSCSs
From the aforementioned results, we confirmed that
95-D LCSCs expressed stem cell genes, CD133, Oct4 and Sox2, and
overexpression of NELL1 could inhibit 95-D cell growth. However,
the mechanism remained unclear. Studies have shown that Sonic
Hedgehog and Wnt pathways are important in cancer stem cells
(32,33). We assessed Sonic Hedgehog (Shh) and
β-catenin which are important proteins in these two pathways
(34,35). Yet, we did not find any change of
expression in these two proteins (Fig.
4A). c-MET was found to be activated in cancer stem cells
(36,37). We examined the expression of these
proteins. As shown in Fig. 4B and
C, we found that NELL1 overexpression downregulated p-MET.
Previous research has demonstrated that Notch signaling plays an
important role in cancer stem cells (38). In the present study, it was also
demonstrated that Notch3 and HES1 were downregulated in the 95-D
LCSCs NELL1 cells. These results demonstrated that NELL1 may affect
95-D LCSC growth through c-MET/Notch signaling. In addition,
overexpression of NELL1 decreased CD133, Oct4 and Sox2 at the
protein and mRNA levels (Fig.
4D-F). Based on these data, we conclude that NELL1 may induce
differentiation of 95-D LCSCs by inhibiting c-MET/Notch signaling
(Fig. 5). Our results showed that
NELL1 could be a potential target for lung cancer stem-like
cells.
 | Figure 4.Relative expression of β-catenin,
Shh, p-MET, Notch3 and HES proteins and stemness genes in 95-D EV,
95-D LCSCs EV and 95-D LCSCs NELL1 cells. (A) Western blot results
of β-catenin and Shh in 95-D EV, 95-D LCSCs EV and 95-D LCSCs NELL1
cells. (B and C) Western blot results of p-MET, c-MET, NOTCH3 and
HES1 proteins in 95-D EV, 95-D LCSCs EV and 95-D LCSCs NELL1 cells.
(D and E) Western blot results of CD133, Oct4 and Sox2 proteins in
95-D EV, 95-D LCSCs EV and 95-D LCSCs NELL1 cells. (F) Quantitative
RT-PCR results of CD133, Oct4 and Sox2 in 95-D EV, 95-D LCSCs EV
and 95-D LCSCs NELL1 cells. *P<0.5, **P<0.01. NELL1, neural
EGFL like 1; LCSCs, lung cancer stem-like cells; Shh, Sonic
Hedgehog; HES1, hairy and enhancer of split-1. |
Discussion
Despite significant advances in the field of
oncology over the previous decade, lung cancer mortality rates
remain high (39). Currently,
traditional therapies for lung cancer are confronted with issues
such as cancer resistance and poor prognosis. Searching for an
effective and safe method is urgent for lung cancer therapy. The
cancer stem cell theory hypothesizes that small populations of
cancer cells may play critical roles in cancer (40). Researchers are looking for an
effective way to target these cancer stem cells.
NELL1 is a novel growth factor that promotes
osteoblast differentiation (41).
NELL1 may exert significant activity in enhancing mesenchymal stem
cells into osteoblast cells. A previous study reported that NELL1
plays an important role in human renal cell carcinoma (26), liver cancer (42) and esophageal adenocarcinoma
(27). Notably, overexpression of
NELL1 was found to induce cancer stem cell differentiation and
decrease their proliferation in glioblastoma (43). However, how NELL1 affects lung
cancer stem cells has never been reported. The 95-D human cell line
is a highly invasive and metastatic lung carcinoma cell line
(19). It has been found that 95-D
cells cultured in specific medium can be used to accumulate lung
cancer stem-like cells in spheres, which are called lung cancer
stem cells (LCSCs) (20). Thus, we
investigated the role of NELL1 in 95-D cancer stem-like cell
differentiation.
In the present study, we investigated the
significance of NELL1 overexpression in the differentiation of 95-D
stem-like cell line, and the molecular mechanisms underlying the
effects of NELL1 overexpression.
Firstly, NELL1 overexpression was suggested to
result in decreased colony formation and invasion (Fig. 2C-E). NELL1 has been previously
reported in other cancers using cell lines and clinical samples.
For example, NELL1 gene loss was noted in more than 40% Hodgkin's
lymphoma patients (44). In gastric
cancer, NELL1 expression was lower in cancer tissues relative to
normal tissue (45). In colon
cancer, researchers found that NELL1 is a promising
tumor-suppressor gene candidate (46). NELL1 protein also participates in
the growth, differentiation, and oncogenesis of cancer cell lines
(47). NELL1 was found to inhibit
renal cell carcinoma cell migration and adhesion (26). These data suggest that NELL1 may be
involved in cancer development.
In order to investigate the effect of NELL1 on
chemotherapeutic resistance, we detected the resistance of these
cells to cisplatin and carboplatin. It was demonstrated that 95-D
LCSCs overexpressing NELL1 had increased sensitive to cisplatin and
carboplatin (Fig. 3A and B) and
exhibited lower expression of ABCG2 and ABCC1 (Fig. 3C and D) compared with the 95-D LCSCs
EV cells. 95-D LCSCs NELL1 cells differed from the control 95-D
LCSCs EV cells following treatment with 50 µM carboplatin and
cisplatin. These results demonstrated that 95-D LCSCs
overexpressing NELL1 were sensitive to chemotherapeutic drugs.
Another confounding issue was to determine the
signaling pathway associated with NELL1. In osteogenesis
differentiation, studies have shown that NELL1 activates MAPK
signaling cascades (48,49), while in the present study we did not
detect change in the expression of these proteins (data not shown).
However, it was found that NELL1 inhibited the expression of p-MET,
Notch3 and HES1. We also found that after overexpression of NELL1,
CD133, Oct4 and Sox2 expression was downregulated. Based on these
data, we conclude that NELL1 may induce 95-D LCSC differentiation
by inhibiting c-MET-Notch signaling.
In summary, our findings revealed that NELL1 may
induce 95-D LCSC differentiation, resulting in decreased invasion,
migration and proliferation abilities. Therefore, NELL1 is a
promising potential target for lung cancer stem-like cells.
Acknowledgements
The authors thank Professor Tianshu Yang for the
technical support.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81272433, 81472732
and 8177315).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GL, XJ and HW conceived and designed the study; YZ
wrote the manuscript and performed most of the experiments; RW, SS
and CL assisted with the quantitative RT-PCR and the western blot
analysis. All authors read and approved the manuscript and agree to
be accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriate
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of
interest.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alamgeer M, Peacock CD, Matsui W, Ganju V
and Watkins DN: Cancer stem cells in lung cancer: Evidence and
controversies. Respirology. 18:757–764. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kelsey CR, Marks LB, Hollis D, Hubbs JL,
Ready NE, D'amico TA and Boyd JA: Local recurrence after surgery
for early stage lung cancer. Cancer. 115:5218–5227. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Murray N, Coy P, Pater JL, Hodson I,
Arnold A, Zee B, Payne D, Kostashuk EC, Evans WK and Dixon P:
Importance of timing for thoracic irradiation in the combined
modality treatment of limited-stage small-cell lung cancer. The
national cancer institute of Canada clinical trials group. J Clin
Oncol. 11:336–344. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wisnivesky JP, Yankelevitz D and Henschke
CI: Stage of lung cancer in relation to its size: Part 2. Evidence.
Chest. 127:1136–1139. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Devesa SS, Bray F, Vizcaino AP and Parkin
DM: International lung cancer trends by histologic type:
Male:female differences diminishing and adenocarcinoma rates
rising. Int J Cancer. 117:294–299. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Donnenberg VS and Donnenberg AD: Multiple
drug resistance in cancer revisited: The cancer stem cell
hypothesis. J Clin Pharmacol. 45:872–877. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lapidot T, Sirard C, Vormoor J, Murdoch B,
Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA and
Dick JE: A cell initiating human acute myeloid leukaemia after
transplantation into SCID mice. Nature. 367:645–648. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
13
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
Biffoni M, Todaro M, Peschle C and De Maria R: Identification and
expansion of human colon-cancer-initiating cells. Nature.
445:111–115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eramo A, Lotti F, Sette G, Pilozzi E,
Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C and De
Maria R: Identification and expansion of the tumorigenic lung
cancer stem cell population. Cell Death Differ. 15:504–514. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bertolini G, Roz L, Perego P, Tortoreto M,
Fontanella E, Gatti L, Pratesi G, Fabbri A, Andriani F, Tinelli S,
et al: Highly tumorigenic lung cancer CD133+ cells
display stem-like features and are spared by cisplatin treatment.
Proc Natl Acad Sci USA. 106:16281–16286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Leung EL, Fiscus RR, Tung JW, Tin VP,
Cheng LC, Sihoe AD, Fink LM, Ma Y and Wong MP: Non-small cell lung
cancer cells expressing CD44 are enriched for stem cell-like
properties. PLoS One. 5:e140622010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qiu X, Wang Z, Li Y, Miao Y, Ren Y and
Luan Y: Characterization of sphere-forming cells with stem-like
properties from the small cell lung cancer cell line H446. Cancer
Lett. 323:161–170. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang F, Qiu Q, Khanna A, Todd NW, Deepak
J, Xing L, Wang H, Liu Z, Su Y, Stass SA and Katz RL: Aldehyde
dehydrogenase 1 is a tumor stem cell-associated marker in lung
cancer. Mol Cancer Res. 7:330–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang X, Yang Y, Tang S, Tang H, Yang G, Xu
Q and Wu J: Anti-tumor effect of polysaccharides from
Scutellaria barbata D. Don on the 95-D xenograft model via
inhibition of the C-met pathway. J Pharmacol Sci. 125:255–263.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yue H, Huang D, Qin L, Zheng Z, Hua L,
Wang G, Huang J and Huang H: Targeting lung cancer stem cells with
antipsychological drug thioridazine. Biomed Res Int 2016.
67098282016.
|
|
21
|
Dalerba P, Cho RW and Clarke MF: Cancer
stem cells: Models and concepts. Annu Rev Med. 58:267–284. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Watanabe TK, Katagiri T, Suzuki M, Shimizu
F, Fujiwara T, Kanemoto N, Nakamura Y, Hirai Y, Maekawa H and
Takahashi Ei: Cloning and characterization of two novel human cDNAs
(NELL1 and NELL2) encoding proteins with six EGF-like repeats.
Genomics. 38:273–276. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Kuroda S, Carpenter D, Nishimura
I, Soo C, Moats R, Iida K, Wisner E, Hu FY, Miao S, et al:
Craniosynostosis in transgenic mice overexpressing Nell-1. J Clin
Invest. 110:861–870. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aghaloo T, Cowan CM, Chou YF, Zhang X, Lee
H, Miao S, Hong N, Kuroda S, Wu B, Ting K and Soo C: Nell-1-induced
bone regeneration in calvarial defects. Am J Pathol. 169:903–915.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang X, Carpenter D, Bokui N, Soo C, Miao
S, Truong T, WU B, Chen I, Vastardis H, Tanizawa K, et al:
Overexpression of Nell-1, a craniosynostosis-associated gene,
induces apoptosis in osteoblasts during craniofacial development. J
Bone Miner Res. 18:2126–2134. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakamura R, Oyama T, Tajiri R, Mizokami A,
Namiki M, Nakamoto M and Ooi A: Expression and regulatory effects
on cancer cell behavior of NELL1 and NELL2 in human renal cell
carcinoma. Cancer Sci. 106:656–664. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jin Z, Mori Y, Yang J, Sato F, Ito T,
Cheng Y, Paun B, Hamilton JP, Kan T, Olaru A, et al:
Hypermethylation of the nel-like 1 gene is a common and early event
and is associated with poor prognosis in early-stage esophageal
adenocarcinoma. Oncogene. 26:6332–6340. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Maeda K, Matsuhashi S, Tabuchi K, Watanabe
T, Katagiri T, Oyasu M, Saito N and Kuroda S: Brain specific human
genes, NELL1 and NELL2, are predominantly expressed in
neuroblastoma and other embryonal neuroepithelial tumors. Neurol
Med Chir (Tokyo). 41:582–589. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shohet JM, Ghosh R, Coarfa C, Ludwig A,
Benham AL, Chen Z, Patterson DM, Barbieri E, Mestdagh P, Sikorski
DN, et al: A genome-wide search for promoters that respond to
increased MYCN reveals both new oncogenic and tumor suppressor
microRNAs associated with aggressive neuroblastoma. Cancer Res.
71:3841–3851. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ting K, Vastardis H, Mulliken JB, Soo C,
Tieu A, Do H, Kwong E, Bertolami CN, Kawamoto H, Kuroda S and
Longaker MT: Human NELL-1 expressed in unilateral coronal
synostosis. J Bone Miner Res. 14:80–89. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cochrane CR, Szczepny A, Watkins DN and
Cain JE: Hedgehog signaling in the maintenance of cancer stem
cells. Cancers (Basel). 7:1554–1585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
de Sousa e Melo F and Vermeulen L: Wnt
signaling in cancer stem cell biology. Cancers (Basel). 8:E602016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu X, Lu Y, Li Y and Prinz RA: Sonic
hedgehog signaling in thyroid cancer. Front Endocrinol (Lausanne).
8:2842017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Behrens J, von Kries JP, Kühl M, Bruhn L,
Wedlich D, Grosschedl R and Birchmeier W: Functional interaction of
β-catenin with the transcription factor LEF-1. Nature. 382:638–642.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun J, Zhang C, Liu G, Liu H, Zhou C, Lu
Y, Zhou C, Yuan L and Li X: A novel mouse CD133 binding-peptide
screened by phage display inhibits cancer cell motility in vitro.
Clin Exp Metastasis. 29:185–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Firtina Karagonlar Z, Koç D, Şahin E, Avci
ST, Yilmaz M, Atabey N and Erdal E: Effect of adipocyte-secreted
factors on EpCAM+/CD133+ hepatic stem cell population. Biochem
Biophys Res Commun. 474:482–490. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Suman S, Das TP and Damodaran C: Silencing
NOTCH signaling causes growth arrest in both breast cancer stem
cells and breast cancer cells. Br J Cancer. 109:2587–2596. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Eyler CE and Rich JN: Survival of the
fittest: Cancer stem cells in therapeutic resistance and
angiogenesis. J Clin Oncol. 26:2839–2845. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang J, Liao J, Zhang F, Song D, Lu M, Liu
J, Wei Q, Tang S, Liu H, Fan J, et al: NEL-like molecule-1 (Nell1)
is regulated by bone morphogenetic protein 9 (BMP9) and potentiates
BMP9-induced osteogenic differentiation at the expense of
adipogenesis in mesenchymal stem cells. Cell Physiol Biochem.
41:484–500. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ding D, Lou X, Hua D, Yu W, Li L, Wang J,
Gao F, Zhao N, Ren G, Li L and Lin B: Recurrent targeted genes of
hepatitis B virus in the liver cancer genomes identified by a
next-generation sequencing-based approach. PLoS Genet.
8:e10030652012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jiang X, Xu M, Yin D, Zhang Z, Yu J, Black
K and Liu G: Expression and functional analysis of Nell-1 on cancer
stem cells and glioma patients' survival. Am Association Cancer
Res. 68:12–16. 2008.
|
|
44
|
Slovak ML, Bedell V, Hsu YH, Estrine DB,
Nowak NJ, Delioukina ML, Weiss LM, Smith DD and Forman SJ:
Molecular karyotypes of Hodgkin and Reed-Sternberg cells at disease
onset reveal distinct copy number alterations in chemosensitive
versus refractory Hodgkin lymphoma. Clin Cancer Res. 17:3443–3454.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gao C and Zhang Q, Kong D, Wu D, Su C,
Tong J, Chen F and Zhang Q: MALDI-TOF mass array analysis of Nell-1
promoter methylation patterns in human gastric cancer. Biomed Res
Int 2015. 1369412015.
|
|
46
|
Mori Y, Cai K, Cheng Y, Wang S, Paun B,
Hamilton JP, Jin Z, Sato F, Berki AT, Kan T, et al: A genome-wide
search identifies epigenetic silencing of somatostatin,
tachykinin-1, and 5 other genes in colon cancer. Gastroenterology.
131:797–808. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kuroda S, Oyasu M, Kawakami M, Kanayama N,
Tanizawa K, Saito N, Abe T, Matsuhashi S and Ting K: Biochemical
characterization and expression analysis of neural
thrombospondin-1-like proteins NELL1 and NELL2. Biochem Biophys Res
Commun. 265:79–86. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bokui N, Otani T, Igarashi K, Kaku J, Oda
M, Nagaoka T, Seno M, Tatematsu K, Okajima T, Matsuzaki T, et al:
Involvement of MAPK signaling molecules and Runx2 in the
NELL1-induced osteoblastic differentiation. FEBS Lett. 582:365–371.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cowan CM, Jiang X, Hsu T, Soo C, Zhang B,
Wang JZ, Kuroda S, Wu B, Zhang Z, Zhang X and Ting K: Synergistic
effects of Nell-1 and BMP-2 on the osteogenic differentiation of
myoblasts. J Bone Miner Res. 22:918–930. 2007. View Article : Google Scholar : PubMed/NCBI
|